Abstract
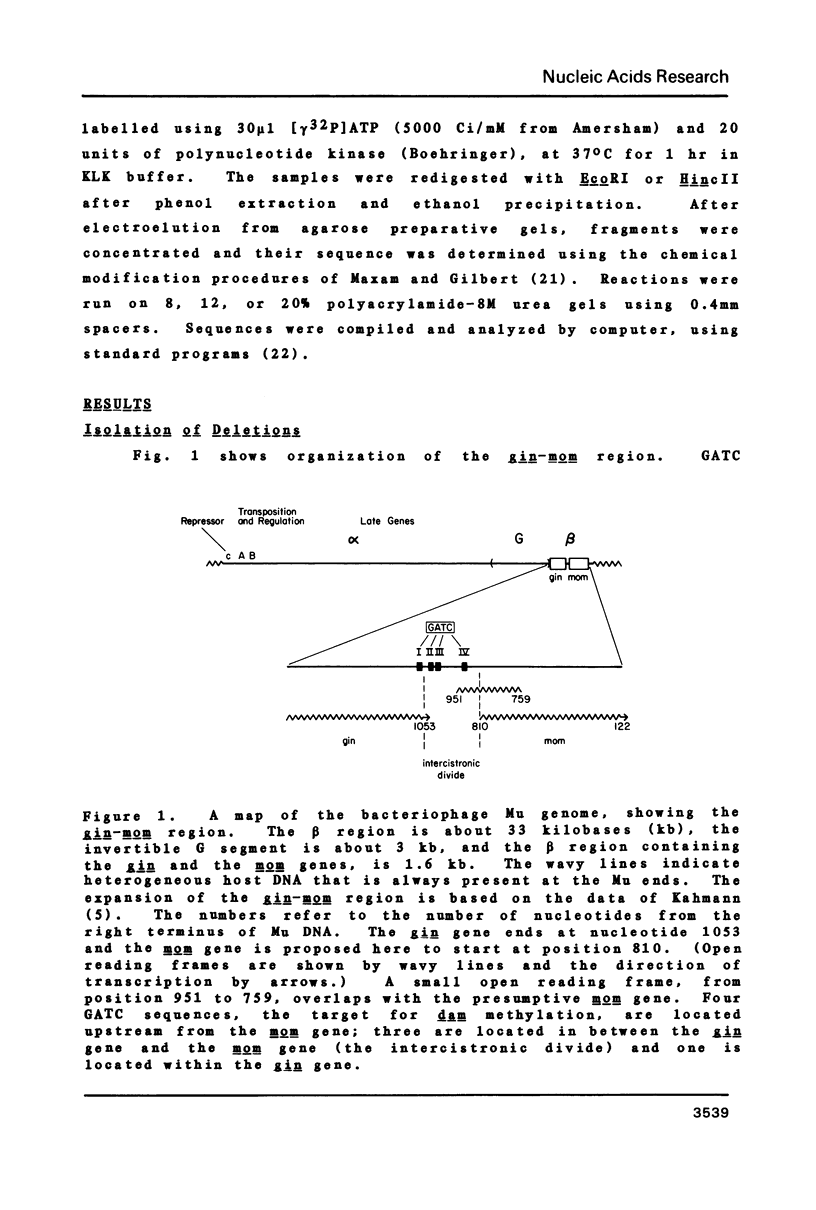
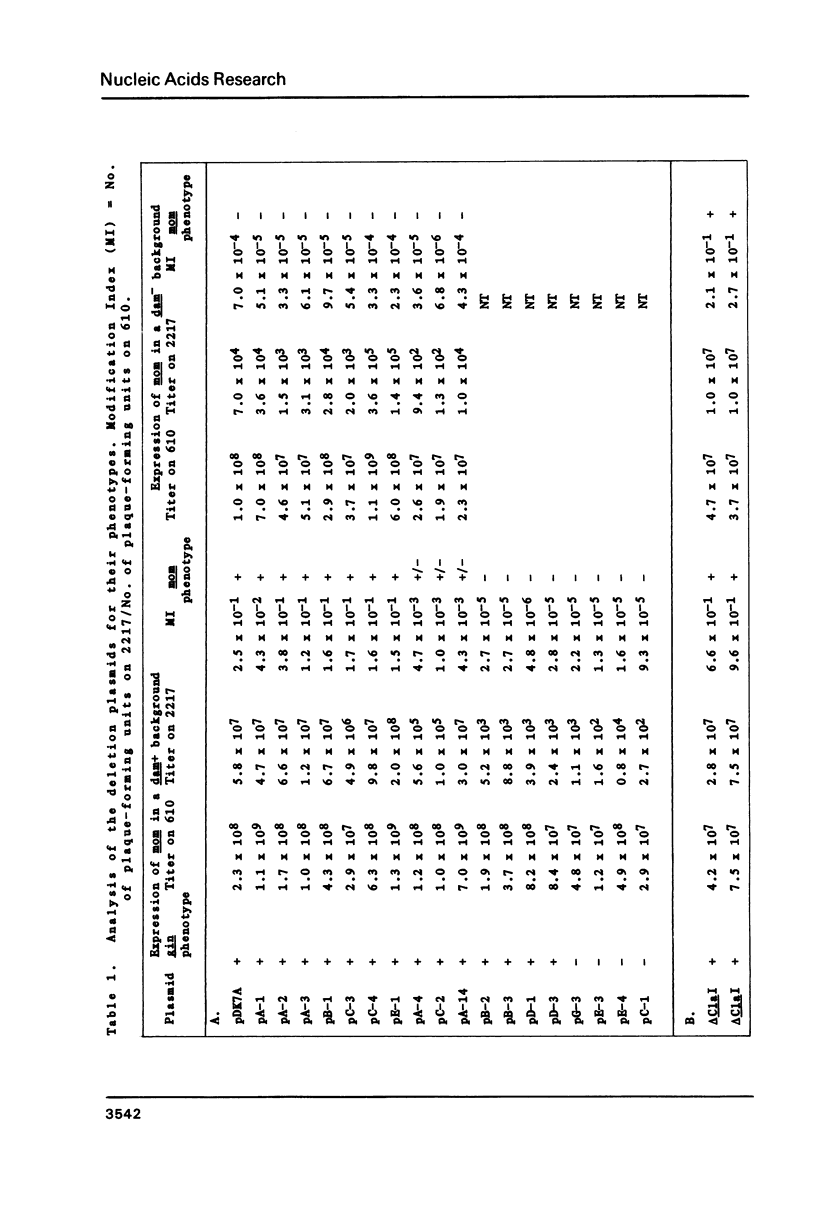
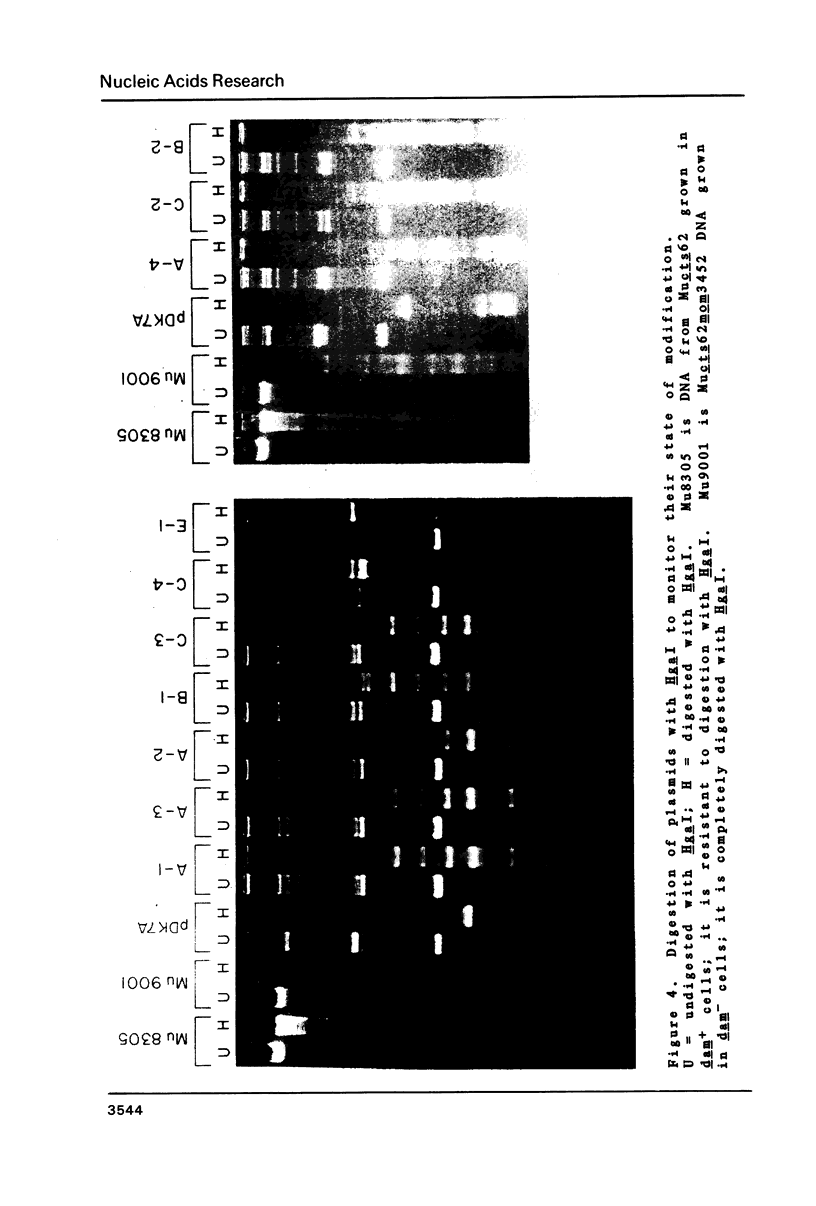
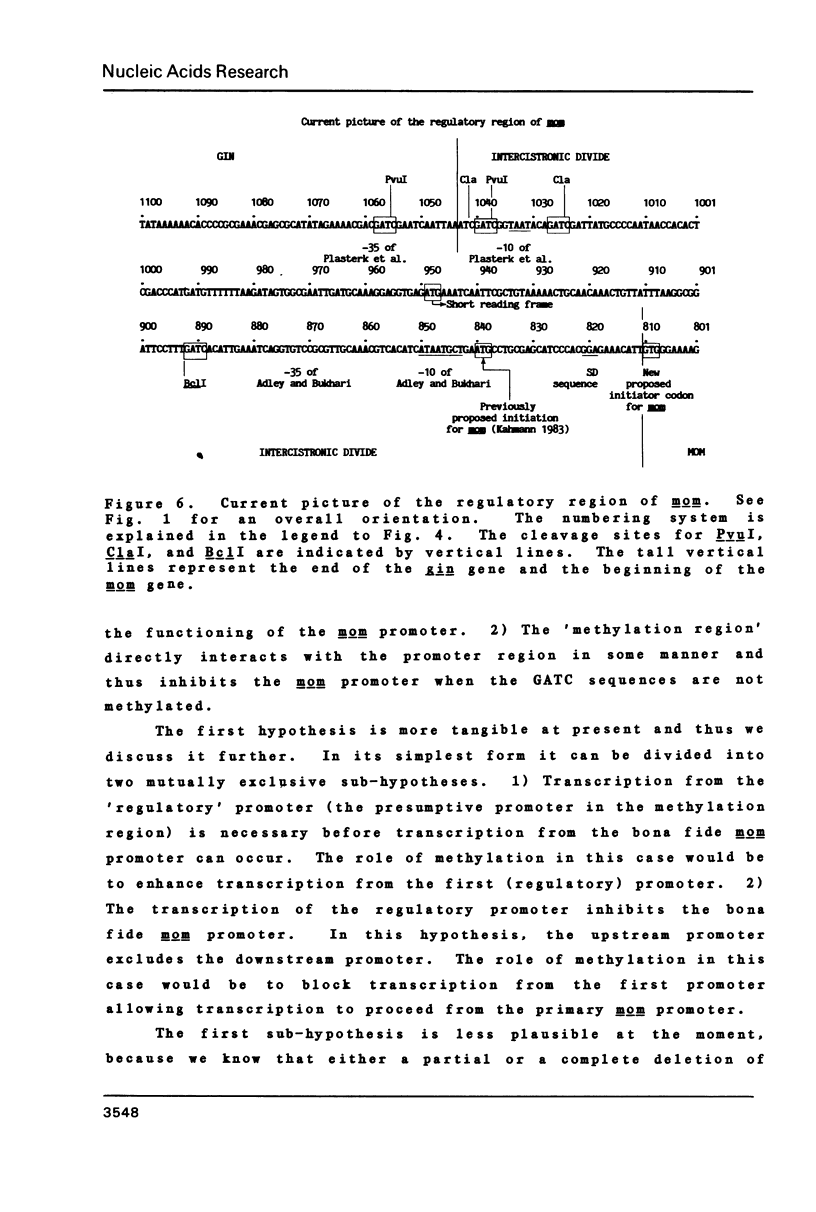
The expression of the DNA modification gene (mom) of bacteriophage Mu requires the cellular deoxyadenosine methylase (dam) and a transactivation factor from the phage. By hypothesis, the transcription of mom is activated by methylation of three GATC sequences upstream from the mom gene. We have introduced small deletions at a fourth GATC site located about 140 base pairs downstream from the primary methylation region. Some of the deletions severely affect the mom gene expression. We propose from this analysis that (1) some important elements, possibly the promoter, concerned with the expression of mom are located between nucleotides 840 and 880 from the right end of Mu and (2) the mom protein starts with the codon GTG located at position 810. We favor the hypothesis that methylation turns off transcription upstream, thereby allowing the main mom promoter to function.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adhya S., Gottesman M. Promoter occlusion: transcription through a promoter may inhibit its activity. Cell. 1982 Jul;29(3):939–944. doi: 10.1016/0092-8674(82)90456-1. [DOI] [PubMed] [Google Scholar]
- Allet B., Bukhari A. I. Analysis of bacteriophage mu and lambda-mu hybrid DNAs by specific endonucleases. J Mol Biol. 1975 Mar 15;92(4):529–540. doi: 10.1016/0022-2836(75)90307-1. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukhari A. I., Metlay M. Genetic mapping of prophage Mu. Virology. 1973 Jul;54(1):109–116. doi: 10.1016/0042-6822(73)90120-7. [DOI] [PubMed] [Google Scholar]
- Chaconas G., de Bruijn F. J., Casadaban M. J., Lupski J. R., Kwoh T. J., Harshey R. M., DuBow M. S., Bukhari A. I. In vitro and in vivo manipulations of bacteriophage Mu DNA: cloning of Mu ends and construction of mini-Mu's carrying selectable markers. Gene. 1981 Jan-Feb;13(1):37–46. doi: 10.1016/0378-1119(81)90041-x. [DOI] [PubMed] [Google Scholar]
- Chow L. T., Kahmann R., Kamp D. Electron microscopic characterization of DNAs of non-defective deletion mutants of bacteriophage Mu. J Mol Biol. 1977 Jul 15;113(4):591–609. doi: 10.1016/0022-2836(77)90224-8. [DOI] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an opern circular DNA form. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper D. N. Eukaryotic DNA methylation. Hum Genet. 1983;64(4):315–333. doi: 10.1007/BF00292363. [DOI] [PubMed] [Google Scholar]
- Gingeras T. R., Milazzo J. P., Sciaky D., Roberts R. J. Computer programs for the assembly of DNA sequences. Nucleic Acids Res. 1979 Sep 25;7(2):529–545. doi: 10.1093/nar/7.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattman S. DNA methyltransferase-dependent transcription of the phage Mu mom gene. Proc Natl Acad Sci U S A. 1982 Sep;79(18):5518–5521. doi: 10.1073/pnas.79.18.5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattman S., Goradia M., Monaghan C., Bukhari A. I. Regulation of the DNA-modification function of bacteriophage Mu. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):647–653. doi: 10.1101/sqb.1983.047.01.076. [DOI] [PubMed] [Google Scholar]
- Kamp D., Kahmann R., Zipser D., Broker T. R., Chow L. T. Inversion of the G DNA segment of phage Mu controls phage infectivity. Nature. 1978 Feb 9;271(5645):577–580. doi: 10.1038/271577a0. [DOI] [PubMed] [Google Scholar]
- Khatoon H., Bukhari A. I. Bacteriophage Mu-induced modification of DNA is dependent upon a host function. J Bacteriol. 1978 Oct;136(1):423–428. doi: 10.1128/jb.136.1.423-428.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwoh D. Y., Zipser D. Identification of the gin protein of bacteriophage mu. Virology. 1981 Oct 15;114(1):291–296. doi: 10.1016/0042-6822(81)90280-4. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasterk R. H., Vrieling H., Van de Putte P. Transcription initiation of Mu mom depends on methylation of the promoter region and a phage-coded transactivator. Nature. 1983 Jan 27;301(5898):344–347. doi: 10.1038/301344a0. [DOI] [PubMed] [Google Scholar]
- Razin A., Riggs A. D. DNA methylation and gene function. Science. 1980 Nov 7;210(4470):604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toussaint A. DNA modification of bacteriophage Mu-1 requires both host and bacteriophage functions. J Virol. 1977 Sep;23(3):825–826. doi: 10.1128/jvi.23.3.825-826.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toussaint A., Desmet L., Faelen M. Mapping of the modification function of temperate phage Mu-1. Mol Gen Genet. 1980 Jan;177(2):351–353. doi: 10.1007/BF00267450. [DOI] [PubMed] [Google Scholar]
- Toussaint A. The DNA modification function of temperate phage Mu-1. Virology. 1976 Mar;70(1):17–27. doi: 10.1016/0042-6822(76)90232-4. [DOI] [PubMed] [Google Scholar]