TPX2 promotes mitotic spindle formation by enhancing Eg5 accumulation on microtubules and limiting motor activity.
Abstract
Mitotic spindle assembly requires the regulated activity of numerous spindle-associated proteins. In mammalian cells, the Kinesin-5 motor Eg5 interacts with the spindle assembly factor TPX2, but how this interaction contributes to spindle formation and function is not established. Using bacterial artificial chromosome technology, we generated cells expressing TPX2 lacking the Eg5 interaction domain. Spindles in these cells were highly disorganized with multiple spindle poles. The TPX2–Eg5 interaction was required for kinetochore fiber formation and contributed to Eg5 localization to spindle microtubules but not spindle poles. Microinjection of the Eg5-binding domain of TPX2 resulted in spindle elongation, indicating that the interaction of Eg5 with TPX2 reduces motor activity. Consistent with this possibility, we found that TPX2 reduced the velocity of Eg5-dependent microtubule gliding, inhibited microtubule sliding, and resulted in the accumulation of motor on microtubules. These results establish a novel function of TPX2 in regulating the location and activity of the mitotic motor Eg5.
Introduction
Recently, genome-wide RNAi screens have shown that hundreds of genes are involved in cell division; to date, however, a detailed mechanistic understanding of how these genes function, including their localization, dynamics, and interactions, is available for only a small subset (Neumann et al., 2010). A further complication is that many proteins are multifunctional, contributing to more than one aspect of mitosis. An example of such a multifunctional mitotic protein is the Ran-regulated spindle assembly factor TPX2, identified as a factor required for the dynein-dependent localization of the kinesin Xklp2 to spindle poles (Wittmann et al., 1998). TPX2 activates and targets the kinase Aurora A to spindle microtubules and is required for chromosome-associated microtubule formation during spindle assembly (Garrett et al., 2002; Gruss et al., 2002; Kufer et al., 2002; Bayliss et al., 2003; Tulu et al., 2006; Bird and Hyman, 2008). TPX2 is transported toward spindle poles in a manner that is sensitive to inhibition of Eg5, dynein, or microtubule flux (Ma et al., 2010), and, in Xenopus laevis, TPX2 is part of a larger complex, the Hepatoma up-regulated protein (HURP) complex, that contributes to Ran-dependent bipolar spindle formation (Koffa et al., 2006; Silljé et al., 2006; Wong and Fang, 2006).
Elucidating the functions of TPX2 has been challenging because either depletion or overexpression of the protein results in spindles with multiple defects, including collapsed spindles and multiple poles (Garrett et al., 2002; Gruss et al., 2002; Eckerdt et al., 2008). To address this problem and to specifically study TPX2-dependent activation of Aurora A during spindle assembly in HeLa cells, Bird and Hyman (2008) used bacterial artificial chromosome (BAC)–based recombineering. With this approach, the gene of interest is expressed from a BAC, which includes all of the endogenous gene regulatory elements. In addition, deletions and point mutations can be made in the gene using homologous recombination in bacteria. Cells expressing the protein from a BAC are then treated with siRNA to deplete the endogenous transcript, thus effecting gene replacement in mammalian cells (Kittler et al., 2005). Using this method to express TPX2 that could not interact with Aurora A, Bird and Hyman (2008) showed that this interaction was critical for microtubule nucleation near chromosomes and to establish spindle length and demonstrated proof of principle for BAC recombineering.
We and others recently showed that TPX2 interacts with the mitotic motor Eg5 in vitro and that this interaction requires the C-terminal 35 amino acids (CT-35) of TPX2 (Eckerdt et al., 2008; Ma et al., 2010), but the physiological significance of this interaction is not yet established. Eg5 is a slow plus-end–directed homotetrameric kinesin that cross-links and slides microtubules in vitro and generates outward forces to separate spindle poles in early mitosis (Kapitein et al., 2005; Ferenz et al., 2010). Eg5-mediated outward forces are antagonized by dynein-dependent inward forces (Ferenz et al., 2009). In cells, Eg5 is enriched at spindle poles, indicating that the motor can cross-link both antiparallel and parallel spindle microtubules in vivo, an activity that has been demonstrated in vitro (Sawin et al., 1992; Blangy et al., 1995; van den Wildenberg et al., 2008) To determine whether and how the interaction of TPX2 and Eg5 contributes to mitosis in vivo, we used BAC recombineering to replace full-length TPX2 with TPX2 lacking the domain that contributes to the interaction with Eg5. Our results show that the TPX2–Eg5 interaction is critical for proper organization and stability of spindle microtubules and for targeting Eg5 to spindle microtubules. Using in vitro assays with purified proteins, we further demonstrate that activity and microtubule targeting of Eg5 are regulated by TPX2.
Results
TPX2–Eg5 interaction is required for spindle microtubule organization
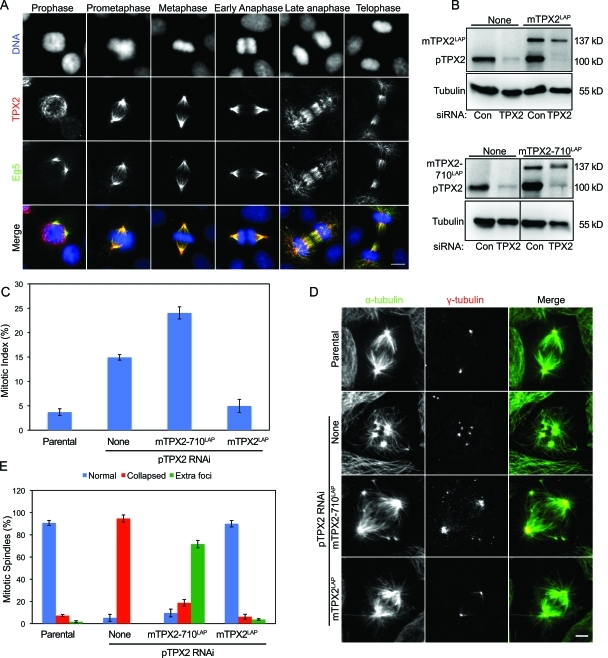
We examined the distribution of endogenous Eg5 and TPX2 in LLC-Pk1 cells and found that the proteins colocalized along spindle microtubules in prometaphase and metaphase and were enriched at spindle poles (Fig. 1 A). In prophase, Eg5 localized to centrosomes, whereas TPX2 also interacted with microtubules surrounding the nucleus. In late anaphase and telophase, both proteins were distributed along interzonal microtubules with a region of reduced accumulation in the center of the interzone and were present along microtubules extending toward each daughter cell. These results show that endogenous Eg5 and TPX2 colocalize in mitotic cells.
Figure 1.
Spindle microtubule organization requires the CT-35 of TPX2. (A) Endogenous TPX2 and Eg5 colocalize on spindle microtubules in LLC-Pk1 cells. Immunofluorescence images with anti-TPX2 and -Eg5 antibodies are shown. DNA was stained with DAPI. (B) Western blots of extracts of the cell lines used in these experiments. The blot was stained with anti-TPX2, which recognizes both pig and mouse TPX2. None, no transgene. Control (Con) or siRNA-targeting pig TPX2 is shown. (C) mTPX2LAP, but not mTPX2-710LAP, rescues mitotic index after siRNA to deplete pig TPX2. (D) Spindle microtubule organization in parental cells (top row) and in cells expressing the indicated transgene and treated with siRNA-targeting pig TPX2. Cells were stained for γ- and α-tubulin. (E) Quantification of the spindle phenotype in fixed parental cells and cells expressing the indicated transgene and treated with pTPX2 siRNA. (C and E) For each condition, n = 400; error bars represent SD. Bars, 5 µm.
Previous work using purified proteins showed that TPX2 and Eg5 interact and that this interaction requires the CT-35 of TPX2 (Eckerdt, et al., 2008; Ma et al., 2010). To determine how the interaction of TPX2 with Eg5 contributes to mitosis, we used BAC-based recombineering to replace full-length TPX2 with a construct lacking the CT-35 so that other functions of the protein (e.g., activation of Aurora A and microtubule formation near chromosomes) would not be compromised (Kittler et al., 2005; Bird and Hyman, 2008). We added a C-terminal localization and affinity purification (LAP) tag to full-length mouse TPX2 (mTPX2LAP) and mouse TPX2 lacking the final 35 amino acids (mTPX2-710LAP), transfected the constructs into LLC-Pk1 cells, and generated clonal cell lines (Wadsworth et al., 2005). Using the same method, a stable cell line expressing full-length mouse Eg5 (mEg5LAP) was made. Cells containing these transgenes, without additional siRNA treatment, progressed through mitosis without any obvious abnormalities (Fig. S1 A).
In multiple cell lines that we generated, we found that the level of expression of mTPX2LAP or mTPX2-710LAP was less than the endogenous pig TPX2 (pTPX2; Fig. 1 B). This is in contrast to cells expressing Eg5LAP from a BAC, suggesting that LLC-Pk1 cells are sensitive to the total level of TPX2 but not Eg5. Next, we designed an siRNA that targets pTPX2 but not the mouse transcript. Western blots verified that the siRNA specifically reduced pig, but not mouse, TPX2 (Fig. 1 B). Treatment of parental cells with this siRNA resulted in an increased mitotic index and collapsed spindles (Figs. 1 [C–E] and S1 D). Cells expressing mTPX2LAP and treated with pTPX2-specific siRNA had normal spindle length and morphology and a mitotic index comparable with parental cells, indicating that the full-length mouse protein rescued loss of the endogenous pig TPX2 (Figs. 1 [C–E] and S1 [B and C]). This result also demonstrates that the level of mTPX2LAP is sufficient for normal spindle morphology and function. In contrast, the mitotic index was increased in cells expressing mTPX2-710LAP and depleted of endogenous pTPX2, and spindles were abnormal (Figs. 1 [C–E] and S1 B). The most striking defect was the presence of extra foci of microtubules, located near the spindle poles or in the equatorial region adjacent to the metaphase plate, a phenotype that is distinct from the collapsed spindles that we observe when pTPX2 is depleted from parental cells using siRNA (Fig. 1, D and E). The truncated protein bound to spindle microtubules, and spindle length, measured as the distance between the two dominant poles, was not significantly different from parental cells (Fig. S1 C).
We quantified the mitotic phenotype and found that after depletion of endogenous TPX2, 71.7 ± 3.4% of spindles in mTPX2-710LAP cells had extra foci compared with 3.8 ± 0.7% of spindles in mTPX2LAP cells. Immunofluorescence staining showed that γ-tubulin was present at each spindle pole and at the center of extra foci in cells expressing mTPX2-710LAP, indicating that the foci contain microtubule minus-ends (Fig. 1 D). We noticed that the total γ-tubulin–stained area per cell in mTPX2-710LAP cells depleted of endogenous TPX2 was significantly larger than in mTPX2LAP cells depleted of endogenous TPX2 (6.4 ± 2.3 vs. 2.1 ± 0.4 µm2, respectively). Together, these results indicate that the interaction between TPX2 and Eg5 regulates spindle microtubule organization. Further, we demonstrate that gene replacement using BAC recombineering (Kittler et al., 2005) can be performed using LLC-Pk1 pig cells, which have favorable morphology for observation of spindle assembly and mitosis.
Disorganized spindles and spindle pole fragmentation in cells lacking the TPX2–Eg5 interaction
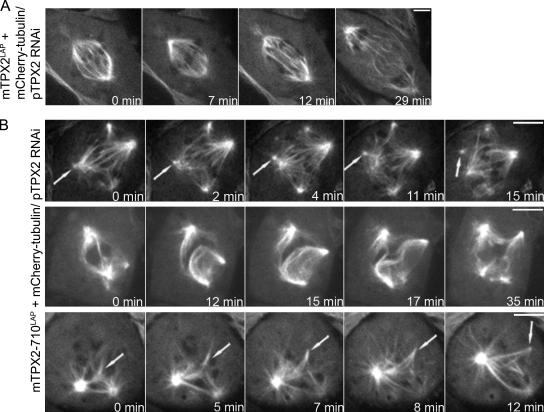
To examine mitosis in live cells in which the TPX2–Eg5 interaction was prevented, we transfected cells expressing mTPX2LAP and mTPX2-710LAP with mCherry-tubulin and generated stable cell lines (see Materials and methods). Unless otherwise indicated, all of the following experiments were performed in cells expressing the indicated BAC and treated with pig-specific TPX2 siRNA. Cells expressing mTPX2LAP formed bipolar spindles and progressed through mitosis without any observable defects (Fig. 2 A and Video 1). In cells expressing mTPX2-710LAP, clusters of microtubules moved away from spindle poles, forming extra foci (Fig. 2 B [top row] and Video 2), indicating that the loss of the TPX2–Eg5 interaction results in spindle pole fragmentation. In some cells, bundles of spindle microtubules at the spindle equator appeared to slide apart, resulting in bent or buckled microtubules (Fig. 2 B [middle row] and Video 3). We also observed microtubule nucleation in the chromosome region (see Chromosome-mediated microtubule nucleation… section; Fig. 2 B [bottom row] and Video 4). These chromosome-derived microtubules coalesced, forming foci, but they were not successfully incorporated into a bipolar spindle (Fig. 2 B). The majority of spindles in live cells expressing mTPX2-710 contained extra foci of microtubules (Fig. S1 F), which is consistent with the quantification of fixed cells. However, the organization of spindle microtubules was very dynamic, and extra foci could appear and then coalesce with other foci during the course of observation. Collectively, our observations show that the organization and stability of the mitotic spindle are compromised in cells in which the TPX2–Eg5 interaction is prevented.
Figure 2.
Disorganized spindles and spindle pole fragmentation in cells lacking TPX2–Eg5 interaction. (A and B) Selected frames from time-lapse imaging of cells expressing mCherry α-tubulin and either mTPX2LAP (A) or mTPX2-710LAP (B) and treated with siRNA-targeting pig TPX2. (B, top row) Clusters of microtubules moving away from spindle poles, forming extra foci (arrows). (middle row) Elongation of microtubule bundles in the spindle midzone. (bottom row) Microtubule nucleation near chromosomes (arrows). Bars, 5 µm.
TPX2–Eg5 interaction is required for kinetochore fiber formation and chromosome congression
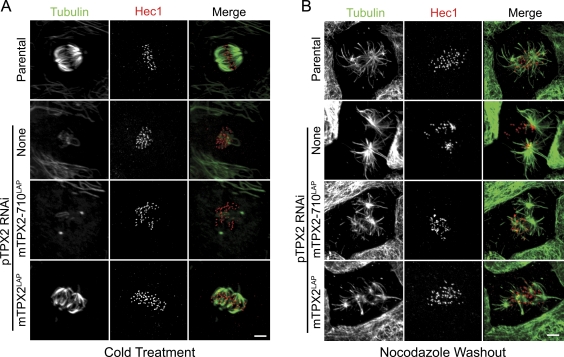
In cells expressing mTPX2-710LAP and mCherry-tubulin, spindle microtubules appeared highly dynamic and disorganized, suggesting that kinetochore fiber formation might be defective when the interaction between TPX2 and Eg5 is prevented. To test this possibility, we treated cells at 4°C for 10 min, a treatment that disassembles labile nonkinetochore microtubules but not stable kinetochore microtubules (Rieder, 1981). In parental cells or mTPX2LAP cells treated with pig-specific siRNA, cold-stable kinetochore fibers were clearly detected, which is consistent with prior work (Fig. 3 A; DeLuca et al., 2002). In contrast, in mTPX2-710LAP cells, few or no stable microtubules were present after cold treatment (Fig. 3 A). Similarly, in parental cells treated with siRNA-targeting pTPX2, cold-stable kinetochore microtubules were not detected. These data demonstrate that an interaction between TPX2 and Eg5 is required for kinetochore fiber formation. Because mTPX2-710LAP retains the ability to bind to microtubules even after depletion of endogenous TPX2 (Fig. S1 B), these data demonstrate that TPX2 binding to microtubules is not sufficient to generate cold-stable microtubules in the absence of an interaction with Eg5.
Figure 3.
TPX2–Eg5 interaction is required for kinetochore fiber formation but not microtubule nucleation near chromosomes. (A and B) Parental cells and cells expressing either no transgene (none) or the indicated transgene and treated with pig TPX2 siRNA. Cells were stained for Hec1 and α-tubulin. (A) Cold-stable microtubules are observed in parental cells and cells expressing mTPX2LAP and depleted of pTPX2 (bottom two rows) but not after depletion of pTPX2 from parental cells or cells expressing mTPX2-710LAP. (B) After release from nocodazole, microtubules form at centrosomes and near kinetochores in parental cells and cells expressing mTPX2LAP or mTPX2-710LAP and depleted of endogenous TPX2 (bottom two rows) but not in parental cells depleted of TPX2. Bars, 5 µm.
Because defects in kinetochore fiber formation could alter chromosome behavior in mitotic cells, we examined chromosome alignment in parental cells and cells expressing mTPX2LAP or mTPX2-710LAP. Quantification showed that 73.8 ± 6.8% of spindles in mTPX2-710LAP cells contained unaligned chromosomes, compared with 10.2 ± 3.1% of spindles in mTPX2LAP cells (Fig. S2 A). Consistent with the defects in chromosome alignment, the checkpoint protein Mad2 was retained on kinetochores in cells expressing mTPX2-710LAP (Fig. S2 B); however, the extent of Mad2 enrichment in mTPX2-710LAP was less than in parental cells depleted of TPX2. These results show that the TPX2–Eg5 interaction is important to establish kinetochore fibers and for chromosome alignment during mitosis.
Chromosome-mediated microtubule nucleation does not require the TPX2–Eg5 interaction
TPX2 is required for microtubule nucleation near kinetochores in mammalian cells (Tulu et al., 2006); this function of TPX2 requires an interaction between the N terminus of TPX2 and Aurora A (Bird and Hyman, 2008). To determine whether mTPX2-710LAP could support microtubule formation near kinetochores, we disassembled microtubules with nocodazole, removed the drug to allow microtubule regrowth, fixed the cells, and stained for microtubules and kinetochores (Tulu et al., 2006). In control cells, microtubules were observed at centrosomes and near kinetochores (Fig. 3 B). In cells depleted of TPX2, microtubule formation was observed at centrosomes, but not at kinetochores/chromatin, consistent with previous work (Tulu et al., 2006). In cells expressing either mTPX2LAP or mTPX2-710LAP and depleted of endogenous TPX2, microtubules were observed at centrosomes and associated with kinetochores (Fig. 3 B). This result demonstrates that the microtubule-nucleating function of TPX2 is not compromised by truncation of the CT-35.
Interaction between TPX2 and Eg5 contributes to the localization of Eg5 to spindle microtubules
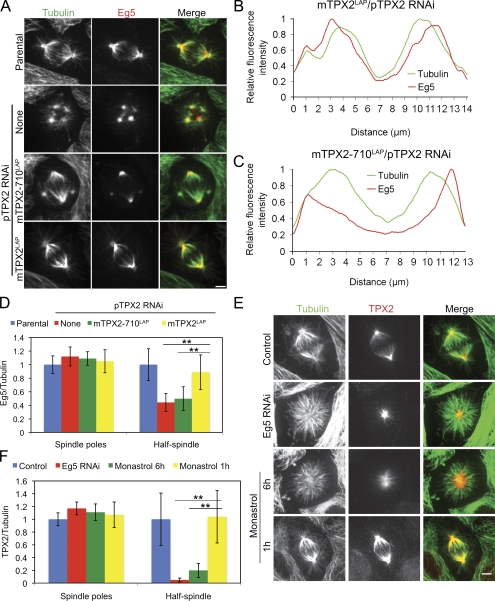
Next, we asked whether the localization of either TPX2 or Eg5 was perturbed when the interaction between TPX2 and Eg5 was prevented. In cells expressing mTPX2LAP, Eg5 localized to centrosomes, spindle poles, and along spindle microtubules, a distribution indistinguishable from the endogenous protein in parental cells (Fig. 4, A and B). However, in mTPX2-710LAP cells, Eg5 was present at the spindle poles and at the center of microtubule foci but was greatly reduced on microtubules in the half-spindle, which is defined as the region between the pole and equator (Fig. 4, A and C). We quantified the distribution of Eg5 by measuring the Eg5/tubulin ratio in different spindle regions (Fig. 4 D). In the half-spindle, but not at spindle poles, the ratio of Eg5/tubulin was reduced in cells expressing mTPX2-710LAP as compared with cells expressing mTPX2LAP. We also examined the localization of p150, a subunit of dynactin, in cells expressing mTPX2LAP and mTPX2-710LAP. The localization of p150 was not altered when the TPX2–Eg5 interaction was prevented (Fig. S3). These data show that the CT-35 of TPX2 are specifically required to target Eg5 to spindle microtubules but not to spindle poles.
Figure 4.
Spindle localization of Eg5 is dependent on an interaction with TPX2. (A) Distribution of Eg5 and α-tubulin in parental cells and parental cells expressing no transgene (none) or the indicated transgene and treated with siRNA-targeting endogenous pig TPX2. (B and C) Corresponding fluorescence intensity profiles of cells in A (bottom two rows). (D) Ratio of Eg5/tubulin fluorescence intensity (n > 20). Error bars represent SD. **, P < 0.01. (E) Distribution of TPX2 and α-tubulin in parental (control) or cells depleted of Eg5 or treated with monastrol for 1 or 6 h. (F) Ratio of TPX2/tubulin fluorescence intensity (n > 20). Error bars represent SD. **, P < 0.01. Bars, 5 µm.
To determine whether the distribution of endogenous TPX2 in parental cells required either Eg5 or Eg5 activity, we depleted Eg5 or inhibited Eg5 activity with monastrol. On monopolar spindles in cells depleted of Eg5 or treated with monastrol for 6 h, TPX2 was localized to spindle poles but did not extend along bundles of microtubules associated with chromosomes (Figs. 4 E and S1 E). This indicates that Eg5 is required to target TPX2 to microtubules in monopoles. However, in mammalian cells, Eg5 activity is dispensable for maintenance of spindle bipolarity (Tanenbaum et al., 2009; Vanneste et al., 2009), so we also examined the distribution of TPX2 on bipolar spindles in cells treated with monastrol for 1 h. Under these conditions, TPX2 was present at spindle poles and along spindle microtubules in a distribution indistinguishable from the protein in control untreated cells (Fig. 4 E). Quantification of the ratio of TPX2/tubulin confirmed that targeting of TPX2 to microtubules was greatly reduced on monopolar arrays but not bipolar spindles (Fig. 4 F). This demonstrates that in a bipolar spindle, TPX2 is retained on spindle microtubules even in the absence of Eg5 activity.
Maintenance of extra foci requires Eg5 activity
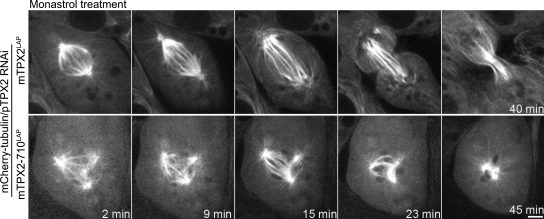
To determine whether the activity of Eg5 contributed to the disorganized spindle phenotype observed in cells expressing mTPX2-710LAP, we treated cells with monastrol to inhibit Eg5 (Mayer et al., 1999). Metaphase cells expressing mTPX2LAP completed mitosis after the addition of low concentrations of monastrol, which is consistent with prior work showing that other motors (i.e., Kif15) function redundantly with Eg5 to maintain spindle bipolarity (Tanenbaum et al., 2009; Vanneste, et al., 2009). In contrast, treatment of cells expressing mTPX2-710LAP with monastrol resulted in spindle collapse (Fig. 5 and Video 5). Extra foci of microtubules at the spindle periphery were drawn inward, eventually forming a monopolar array of microtubules. This result demonstrates that maintenance of extra foci requires Eg5 activity. The observed increased sensitivity could result from less Eg5 targeted to spindle microtubules. The formation of extra foci could result from increased activity of Eg5 in the absence of an interaction with TPX2 (see below).
Figure 5.
Maintenance of extra foci requires Eg5 activity. Selected frames from time-lapse imaging after monastrol treatment of cells expressing mCherry–α-tubulin and mTPX2LAP or mTPX2-710LAP and treated with siRNA-targeting pig TPX2. The times shown on the bottom are applicable to the top unless indicated otherwise. Bar, 5 µm.
TPX2–Eg5 interaction regulates motor activity
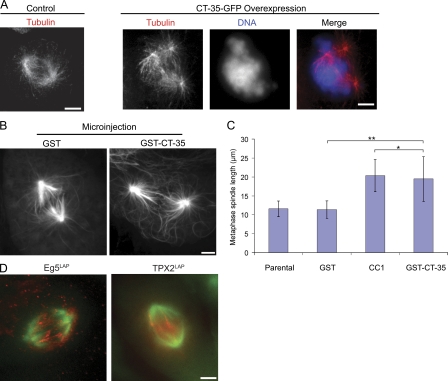
In Xenopus S3 cells, overexpression of a large C-terminal fragment of TPX2, but not full-length TPX2, blocks centrosome separation, which can be overcome by an addition of excess Eg5. These observations suggest that TPX2 C terminus inhibits Eg5 activity (Eckerdt et al., 2008). To test this possibility, we performed two experiments. First, we expressed the CT-35 in parental cells and observed spindle morphology. In these cells, spindle formation was defective, with disorganized microtubules and fragmentation of spindle poles, a phenotype similar to that observed when spindles assemble in cells expressing mTPX2-710LAP (Fig. 6 A). This result demonstrates that the excess C-terminal fragment interferes with spindle formation.
Figure 6.
TPX2–Eg5 interaction regulates motor function. (A) Defective spindles in cells overexpressing CT-35–GFP and stained for α-tubulin and DNA. (B) Spindle lengthening in LLC-Pk1 cells microinjected with GST or GST–CT-35. Cells are expressing EGFP–α-tubulin. (C) Spindle length for the indicated conditions (GST alone, n = 6; CC1, n = 13; GST–CT-35, n = 21; uninjected parental, n = 21; error bars represent SD). *, P > 0.05; **, P < 0.05. (D) Eg5 and TPX2 localize to midzone microtubules by TIRF microscopy. (left) Overlay of Eg5LAP visualized by widefield (green) and by TIRF (red). (right) Overlay of TPX2LAP imaged in TIRF and mCherry microtubules in widefield (green). Bars, 5 µm.
Next, we purified the CT-35 and microinjected the protein into mitotic cells so that we could interfere with the Eg5–TPX2 interaction with precise temporal control after spindle formation had occurred. After the microinjection, spindles elongated, and microtubules near the middle of the spindle were buckled or wavy (Fig. 6 B), a phenotype that is strikingly similar to that observed in cells microinjected with CC1 to inhibit dynein (Gaetz and Kapoor, 2004; Ferenz et al., 2009). We measured spindle length in parental cells and cells microinjected with GST–CT-35, GST alone, or CC1 and found that spindle length was statistically significantly increased in both GST–CT-35 and CC1-injected cells, as compared with spindles in parental cells or cells injected with GST alone (Fig. 6 C). Spindles in microinjected cells remained bipolar, and no fragmentation of spindle poles was detected, indicating that redundant mechanisms maintain pole focusing in the steady-state metaphase spindle (Compton, 1998).
In mammalian cells, the minus-end–directed motor dynein and Eg5 contribute to a balance of forces that regulates spindle length; this balance depends on overlapping antiparallel microtubules (Sharp et al., 2000; Ferenz et al., 2009). The observed spindle elongation after the microinjection of CT-35 is consistent with the possibility that an excess of the C-terminal fragment interferes with the TPX2–Eg5 interaction, relieving the inhibition of TPX2 on Eg5 and generating the observed spindle lengthening. According to this model, TPX2 and Eg5 should localize to microtubules in the spindle midzone. However, in both live and fixed cells, the fluorescence signal of Eg5 and TPX2 in the spindle midzone is very weak. To determine whether TPX2 and Eg5 localize to midzone microtubules, we imaged cells expressing mEg5LAP or mTPX2LAP using total internal reflection fluorescence (TIRF) microscopy. The results reveal discrete punctae of each protein along microtubules in the spindle midzone (Fig. 6 D), which is consistent with the idea that the proteins interact on antiparallel midzone microtubules.
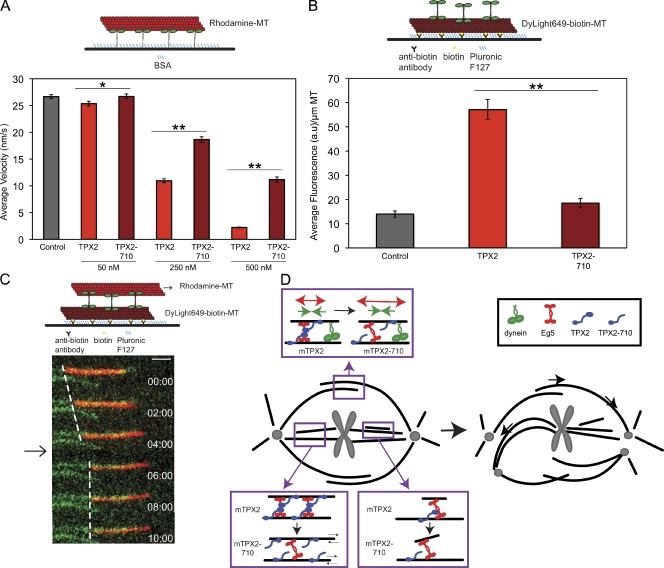
To directly determine whether TPX2 regulates Eg5, we measured Eg5-dependent microtubule gliding in vitro using purified Eg5 and TPX2 (Fig. S4 A). Full-length TPX2 at a concentration of 500 nM reduced the mean rate of microtubule gliding by dimeric Eg5 from 26.7 ± 0.4 to 2.2 ± 0.1 nm/s. When tested at the same concentration, TPX2-710 was less effective at reducing the rate of microtubule gliding (to 11.2 ± 0.5 nm/s), supporting the view that the interaction between TPX2 and Eg5 contributes to the inhibition of gliding (mean ± SEM; Fig. 7 A and Video 6). Microtubule gliding driven by Kinesin-1 was unaffected by TPX2, and the microtubule-associated protein Tau induced microtubule release from both Kinesin-1 and Eg5 in gliding assays (Fig. S4, B and C; Dixit et al., 2008). Because Eg5 can cross-link and slide adjacent microtubules, we also examined the effect of TPX2 in Eg5-dependent microtubule–microtubule sliding assays. We found that sliding was abolished upon addition of TPX2 (Fig. 7 C and Video 7). Finally, we examined the accumulation of Eg5 on microtubules in the presence and absence of TPX2 and TPX2-710. Consistent with in vivo results, TPX2 targeted Eg5 to microtubules in vitro, whereas TPX2-710 did not (Fig. 7 B). These results directly demonstrate that TPX2 regulates Eg5 activity and localization in vitro.
Figure 7.
TPX2 regulates Eg5 activity and accumulation on microtubules in vitro. (A, top) A diagram of gliding assay. (bottom) Microtubule gliding velocity of Eg5-513 alone (control) and with the addition of 50, 250, and 500 nM TPX2 or TPX2-710. *, P < 0.05; **, P < 0.01. n = 174–618 microtubules from at least three independent experiments. Error bars represent SEM. (B, top) A diagram of TIRF assay. (bottom) Mean fluorescence intensity of Eg5-EGFP in arbitrary units (a.u.) along the length of the microtubule. **, P < 0.01. n = 290–374 microtubules from three independent experiments. Error bars represent SEM. (C, top) A diagram of microtubule–microtubule sliding assay. (bottom) Overlay from time-lapse imaging of immobilized microtubules (green) and mobile microtubules (red). The arrow indicates when TPX2 was added. Dashed lines show the location of the trailing microtubule end over time (time = minutes/seconds). n = 3 microtubules from two independent experiments. Bar, 1 µm. (D) A schematic model of the contribution of the TPX2–Eg5 interaction to spindle formation. Boxed regions depict how the TPX2–Eg5 interaction could regulate Eg5 behavior on antiparallel and parallel microtubules.
Discussion
Here, we used BAC transgenomics to determine whether and how the interaction between TPX2 and Eg5 contributes to mitosis in mammalian cells. In this approach, TPX2 lacking only the CT-35 was expressed under the control of the native promoter so that the replacement gene was expressed with appropriate regulation, and RNAi was used to deplete the endogenous protein. The results of these experiments demonstrate that proper spindle formation is dependent on an interaction between TPX2 and Eg5. In addition, we demonstrate that this interaction is required to target Eg5 to spindle microtubules and for the formation of cold-stable kinetochore fibers. By acute inhibition of the TPX2–Eg5 interaction after spindle formation had occurred and by in vitro measurement of motor activity, we further show that TPX2 regulates Eg5.
TPX2-dependent localization of spindle proteins
We found that the localization of Eg5 to spindle microtubules was regulated by an interaction with TPX2. Interestingly, previous work has shown that TPX2 is also required to localize several other spindle-associated proteins to spindle microtubules, including the kinase Aurora A, the plus-end–directed kinesin Kif15/Hklp2, and the nuclear scaffold protein scaffold attachment factor A (Kufer et al., 2002; Tanenbaum, et al., 2009; Vanneste et al., 2009; Ma et al., 2011). Because Aurora A, Eg5, and Kif15 each interacts directly with TPX2 at unique sites, the data suggest that TPX2 can simultaneously target multiple proteins to the spindle, perhaps by acting as a scaffold. These observations are consistent with a model in which TPX2 binds to microtubules during spindle assembly and subsequently recruits additional microtubule regulators to the spindle (Tulu et al., 2006).
Although TPX2 contributes to the targeting of Eg5 to spindle microtubules, both Eg5 and TPX2 accumulate at spindle poles. Poleward motion of Eg5 requires dynein activity (Uteng et al., 2008); poleward motion of TPX2 requires Eg5, dynein, and microtubule flux (Ma et al., 2010), and dynein-dependent poleward accumulation of Xklp2 requires TPX2 (Wittmann, et al., 1998). This raises the possibility that dynein carries both TPX2 and Eg5 poleward along fluxing microtubules and that TPX2 may function to coordinate the activity of plus- and minus-end–directed spindle motors.
Kinetochore fiber formation requires TPX2–Eg5 interaction
During spindle formation, microtubules associate with kinetochores to form a bundle of differentially stable microtubules that dynamically link each kinetochore to the spindle pole. Although truncated TPX2 retains the ability to bind to spindle microtubules, it cannot generate cold-stable microtubules in the absence of an interaction with Eg5. This indicates that the interaction of both proteins with microtubules is required for generating cold-stable kinetochore fibers. TPX2 that cannot interact with Aurora A shows defects in cold-stable microtubule formation in prometaphase, but not in metaphase (Bird and Hyman, 2008), showing that TPX2 and its binding partners contribute to the formation of stable microtubules during early and late spindle assembly.
In addition to TPX2, the Ran-regulated microtubule-associated protein HURP is also required for the formation of cold-stable kinetochore fiber microtubules, indicating that these proteins function nonredundantly in this process (Silljé et al., 2006). In Xenopus extracts, TPX2 and Eg5 coimmunoprecipitate with HURP, and Eg5 activity regulates the association of HURP with midzone microtubules in meiotic spindles (Koffa et al., 2006; Breuer et al., 2010), raising the possibility that Eg5 is additionally regulated by HURP. However, in mitotic spindles, HURP is enriched near chromosomes, whereas TPX2 and Eg5 are enriched near spindle poles, suggesting region-specific regulation of Eg5 (Koffa et al., 2006; Silljé et al., 2006; Wong and Fang, 2006).
Regulation of motor localization and spindle organization
Our results demonstrate that TPX2 regulates Eg5 activity. Is the inhibition of Eg5-dependent microtubule–microtubule sliding in vitro by TPX2 consistent with the established role of Eg5 to generate outward forces in mitosis? Our in vivo data support the view that TPX2 functions to reduce Eg5 activity; however, in vitro sliding assays may not reflect the more complex situation in live cells, where TPX2 could dampen, but not eliminate, Eg5 activity. Interestingly, recent work shows that the tail domain of Eg5 increases the association time of the motor with the microtubule and reduces the velocity of individual motors on microtubules (Weinger et al., 2011), and, in budding yeast, the chromosome passenger complex regulates the localization of the Kinesin-5 motors Cin8 and Kip1 during spindle elongation to promote sliding and prevent braking (Rozelle et al., 2011). Collectively, these results demonstrate that Eg5 activity is regulated to achieve appropriate microtubule sliding behavior.
Based on our observations, we propose the following model to explain how the TPX2–Eg5 interaction contributes to spindle assembly. First, TPX2 can function to restrict Eg5 activity on overlapping antiparallel microtubules in the midzone (Fig. 7 D, top box). In cells lacking an interaction between TPX2 and Eg5 (mTPX2-710LAP), an increase in Eg5 activity on antiparallel microtubules would lead to an imbalance of spindle microtubule–generated forces, resulting in the observed disruption of spindle poles and the formation of bent or buckled microtubules in the spindle (Fig. 7 D, top box; Manning and Compton, 2007). The observed spindle elongation in cells injected with CT-35 after spindle formation is also consistent with this model. In this situation, the C-terminal fragment would interfere with the TPX2–Eg5 interaction, relieving inhibition of the motor. Second, our results demonstrate that an interaction between TPX2 and Eg5 is necessary to form cold-stable kinetochore fibers, incorporate newly assembled microtubules into the spindle, and stabilize bipolar spindles. The observed defects could result from reduced targeting of Eg5 to parallel microtubules in the half-spindle (Fig. 7 D, bottom left box) or from an alteration in Eg5 activity (Fig. 7 D, bottom right box) and are consistent with TPX2–Eg5 interactions mediating microtubule cross-linking. In summary, our results reveal a novel role of TPX2 in the targeting of Eg5 to spindle microtubules and regulating motor activity.
Materials and methods
Materials
All materials for cell culture were obtained from Sigma-Aldrich with the exception of Opti-MEM (Invitrogen) and FBS (Atlanta Biologicals). BACs and BAC cloning reagents were obtained from BACPAC Resources and Gene Bridges, respectively; electroporation cuvettes were received from Molecular BioProducts, Inc. The C-terminal LAP tag (C-terminal R6K-Amp-LAP) was a gift from the Max Planck Institute of Molecular Cell Biology and Genetics Hyman Laboratory. All other chemical reagents, unless specified, were obtained from Sigma-Aldrich.
Cell culture, immunofluorescence, microinjection, and inhibitors
Parental LLC-Pk1 pig kidney epithelial cells and LLC-Pk1 cells expressing EGFP–α-tubulin were grown as previously described (Tulu et al., 2003). For immunofluorescence, cells were rinsed twice in warm calcium- and magnesium-free PBS (PBS−/−) followed by fixation in ice-cold methanol or 3.2% PFA, 0.1% glutaraldehyde, and 0.5% Triton X-100 in PBS−/−. Fixed cells were rinsed in PBS−/− containing 0.02% sodium azide and 0.1% Tween 20. The following primary antibodies were used in these experiments: Mad2 (a gift from A. Khodjakov (Wadsworth Center, Albany, NY); YL1/2 (Accurate Chemical); Hec1, TPX2, and Eg5 (Novus Biologicals); and p150 (BD). Incubations with primary antibodies were performed for 1 h at 37°C using the manufacturer’s recommended dilution. Cy3- (Jackson ImmunoResearch Laboratories, Inc.) or fluorescein isothiocyanate–labeled (Sigma-Aldrich) secondary antibodies were used at the recommended dilution for 1 h at 37°C. Coverslips were mounted in Vectashield containing DAPI to visualize DNA (Vector Laboratories) and were sealed with nail polish.
For nocodazole treatment, cells were incubated with 3.3 µM nocodazole for 3–5 h, washed with non–CO2 culture medium, and imaged. To inhibit dynein, cells were microinjected with p150 CC1, prepared as previously described (Wadsworth, 1999; Ferenz et al., 2009). Before microinjection, all solutions were centrifuged at maximum speed in a microcentrifuge at 4°C through a syringe tip filter held in a microcentrifuge tube. Images shown in Fig. 6 were acquired 30 min after injection. The number of cells was as follows: GST alone, n = 6; CC1, n = 13; GST–CT-35, n = 21; and uninjected parental cells, n = 21.
For cold treatment, growth medium was removed from cells and replaced with ice-cold medium; cells were incubated on ice for 10 min, rinsed with ice-cold PBS−/−, and fixed and stained for microtubules and DNA. Monastrol was used at 200 µM.
Fixed and live-cell imaging and analysis
Images were acquired using a microscope (Eclipse TE300; Nikon) equipped with a 100× phase 1.4 NA objective lens, a spinning disc confocal scan head (PerkinElmer), and a cooled charge-coupled device camera (Orca ER; Hamamatsu Photonics). All images were taken using a dual wavelength filter cube. Image acquisition was controlled by MetaMorph software (Molecular Devices).
TIRF microscopy of live cells was performed using a microscope (Ti-E; Nikon) equipped with a 60× 1.4 NA objective. The laser for TIRF was a blue diode laser (488 nm and 50 mW). The system was run by Elements software (Nikon). Images were acquired using a 512 × 512–pixel camera (Cascade II; Photometrics). A 4× image expansion telescope in front of the camera was used. For imaging live cells, time-lapse videos were taken using a 100-ms exposure with no shutter delay between images. All imaging was performed at 23.5–30°C. During imaging, cells were maintained in non–CO2 MEM conditioned with 0.3 U/ml EC-Oxyrase oxygen scavenging system (Oxyrase). TIRF images are SD maximum intensity projections of a time series of images.
To quantify spindle phenotypes, cells were stained with α- and γ-tubulin. Collapsed spindles lacked bipolar spindle structure and had closely associated spindle poles with positive γ-tubulin staining and associated microtubule bundles (parental cells treated with pTPX2 siRNA; Fig. 1 D); spindles with extra foci had two major γ-tubulin signals with several minor signals of γ-tubulin, and the bipolar structure of the spindle was partially maintained (TPX2-710LAP treated with pTPX2 siRNA; Fig. 1 D); disorganized spindles had two spindle poles with positive γ-tubulin staining and disorganized microtubule organization; and monopolar spindles had one spindle pole with two close γ-tubulin--positive signals. Monopolar and disorganized spindle phenotypes accounted for <5 and <2.5% of the total phenotypes observed, respectively, and were considered collapsed in the final quantification. To quantify the ratio of Eg5/tubulin and TPX2/tubulin, fluorescence in a 2 × 2–pixel square was measured and background subtracted; at least four fibers in five cells were measured.
For mitotic index, values are the means of three independent experiments (n = 400). For chromosome alignment, values are the means of three independent experiments (n = 400).
In vitro assays
10-µl flow chambers were assembled from a glass slide attached to a coverslip with double-stick tape. For gliding assays, the coverslips were not treated. For all other assays, the coverslips were biologically cleaned and then treated with 2% (weight/volume) dimethyldichlorosilane solution (GE Healthcare). For Eg5-513 gliding assays with the addition of TPX2 or TPX2-710, 5 µM Eg5-513 was incubated for 2 min, and then the surface was blocked using a BSA wash (PEM-100 + 5 mg/ml BSA, 20 µM paclitaxel, and 20 mM DTT). Next, the chamber was incubated with 0.05 mg/ml rhodamine microtubules in PEM-100 + 20 µM paclitaxel for 2 min. Finally, the chamber was perfused with activation mix 1 (AM1 [PEM-100 + 5 mM MgATP, 50 µM paclitaxel, 5 mM DTT, 15 mg/ml glucose, 1.25 mg/ml glucose oxidase, 0.375 mg/ml catalase, and various concentrations of TPX2 or TPX2-710 where indicated]) and imaged immediately. For Eg5-513 and Kinesin-1 gliding assays with the addition of Tau and for Kinesin-1 gliding assays with the addition of TPX2, the same chamber setup was followed but with the following changes: MAPs were omitted from AM1, a control time lapse was acquired, and then a second round of AM1 with the indicated MAP was added to the same chamber. This change accounts for the increase in Kinesin-1 gliding velocity with the addition of TPX2, as more ATP was added to the chamber. For TIRF and sliding assays, 10% antibiotin antibody (Sigma-Aldrich) was added to the chamber and incubated for 2 min followed by 0.1 mg/ml DyLight649 biotin microtubules for 2 min. Next, the surface was blocked with 5% Pluronic F-127 (Sigma-Aldrich) for 2 min followed by incubation with 50 nM Eg5-EGFP for 2 min. For TIRF assays, activation mix 2 (AM2 [PEM-70 + 10 mM MgATP, 0.5% Pluronic F-127, 50 µM paclitaxel, 5 mM DTT, 15 mg/ml glucose, 1.23 mg/ml glucose oxidase, 0.375 mg/ml catalase, and 50 nM TPX2 or TPX2-710 where indicated]) was added to the chamber and imaged immediately. For sliding assays, the chamber was incubated for 2 min with 0.2 mg/ml rhodamine microtubules before adding AM2 lacking TPX2. While acquiring the time lapse of a sliding microtubule, AM2 with 50 nM TPX2 was added.
In vitro imaging and analysis
Assays were performed on a microscope (Ti-E) equipped with a 60× 1.4 NA objective. The laser for TIRF was a blue diode laser (488 nm and 50 mW). The system was run by Elements software. Images were acquired using a 512 × 512–pixel camera (Cascade II) with a 4× image expansion telescope in front of the camera. All imaging was performed at room temperature. For gliding assays, epifluorescence images using exposure times of 500–800 ms were acquired every 10 or 30 s for 2 or 5 min, respectively. For TIRF assays, epifluorescence images of the microtubules were taken with a 2-s exposure time before imaging Eg5-EGFP molecules in the TIRF field. TIRF images were collected at 50 frames per second for 5 s. For sliding assays, rhodamine microtubule and DyLight649 biotin microtubule images were taken with 500–800-ms and 2-s exposure times, respectively, every 30 s for 15 min. All microscopy was performed at room temperature. Image sequences were saved as nd2 files and exported as 16-bit TIF files. Imaged sequences were imported to ImageJ (National Institutes of Health). Gliding velocities were determined using MTrackJ. For TIRF analysis, Eg5-EGFP intensities were based on a z project of the mean fluorescence intensity. A segmented line was drawn along the length of the microtubule in the DyLight649-acquired image, and that line was transposed to the same position on the z project image. The mean fluorescence and length of the line were measured, and a second measurement of the same line next to the molecules was taken to subtract background. The background-subtracted fluorescence was divided by the length of the microtubule in micrometers.
BAC cloning
Mouse TPX2 BAC clone RP24370E11 and Eg5 BAC clone RP23-117H14 (BACPAC Resources) were obtained as stab cultures and were streaked onto Luria broth plates conditioned with chloramphenicol. Subsequent cloning steps were performed according to the Counter-Selection BAC Modification kit (version 3.1; Gene Bridges). Preparation of cells for electroporation was performed at 4°C, and electroporation voltage was 1.8 kV. Primers for addition of the LAP tag were designed following the Mitocheck BACPAC resources website. The LAP-tagged BAC was purified using NucleoBond BAC 100 Maxi prep protocol (Takara Bio Inc.).
Mammalian transfection, siRNA, and Western analysis
LLC-Pk1 cells were plated at a density of 1.0 × 105 and transfected 48 h after plating with each LAP-tagged BAC using an Amaxa nucleofector (Lonza) following the protocol recommended by the manufacturer. Cells were selected in 2.0 g/L G418 for 2 wk. To isolate clonal cell lines expressing LAP-tagged mTPX2 or mTPX2-710, cells were plated at low density so that single cells would give rise to individual colonies (Wadsworth et al., 2005). Colonies were isolated using cloning rings. The level of transgene expression was determined by Western blotting. Clonal cell lines expressing mTPX2 or mTPX2-710 were nucleofected with mCherry-tubulin using an Amaxa nucleofector, and cell lines were prepared as previously described.
Whole-cell extracts were prepared by lysis in 0.5% SDS, 1 mM EDTA, and protease inhibitors and sonicated twice for 10 s, with cooling in between sonications. A sample was saved for protein determination, and extracts were boiled for 5 min after the addition of 6× electrophoresis sample buffer. Extracts were run on an 8% polyacrylamide gel and then transferred to Hybond-P membrane (GE Healthcare). Blots were probed with a rabbit anti-TPX2 antibody (1:1,000) for 1 h at room temperature and a goat anti–rabbit IgG HRP (Jackson ImmunoResearch Laboratories, Inc.) as a secondary antibody (1:5,000) for 1 h at room temperature. Blots were detected using chemiluminescence.
To deplete endogenous pig TPX2, we used the sequence 5′-GGACAAAACUCCUCUGAGA-3′. For transfections, cells in 24-well plates were added to prewarmed media, and transfection complexes containing 3 µl Oligofectamine and 60 pmol RNA were added immediately afterward. Reagents used for mammalian transfection were obtained from Invitrogen. To overexpress CT-35, mammalian cells were nucleofected with pCMV-CT-35–GFP and fixed after 48 h.
Protein purification
A human Eg5 construct truncated at residue 513 with a C-terminal 5× His tag cloned into bacterial expression vector pRSET-a was a gift from S.P. Gilbert (Rensselaer Polytechnic Institute, Troy, NY). The plasmid was transformed into E. coli BL21(DE3)pLysS for protein expression and purification. A Kinesin-1 construct truncated at residue 560 with a C-terminal GFP and N-terminal His tag and Tau4RL were expressed and purified from E. coli BL21(DE3)pLysS using affinity chromatography and boiling, respectively (Dixit et al., 2008; Ross et al., 2008). p150-CC1 was expressed and purified from E. coli BL21(DE3)pLysS by boiling and ammonium sulfate precipitation (King et al., 2003; Ferenz et al., 2009). To prepare C-terminal TPX2 (GST–CT-35), truncated TPX2 in pGEX4T-1 and N-terminally tagged with GST was expressed in E. coli BL21(DE3)pLysS and purified using affinity chromatography (Ma et al., 2010).
Full-length Eg5 C-terminally tagged with EGFP, TPX2, and TPX2 truncated at residue 710, each containing an N-terminal 6× His tag, was expressed and purified from Sf9 insect cells using the Bac-to-Bac Expression system (Invitrogen). pFastBac plasmids and TPX2 baculovirus were a gift from A. Wilde (University of Toronto, Toronto, Canada). Protein concentrations were calculated for monomers.
Unlabeled tubulin was purified from porcine brain (Peloquin et al., 2005). DyLight649 (Thermo Fisher Scientific)-labeled tubulin was prepared using porcine brain tubulin as previously described (Desai and Mitchison, 1998); rhodamine tubulin and biotin tubulin were obtained from Cytoskeleton. Cytoskeleton tubulin was resuspended to 5 mg/ml in PEM-100 (100 mM K-Pipes, pH 6.8, 2 mM MgSO4, and 2 mM EGTA).
Rhodamine microtubules were prepared using a 50-µl mixture of unlabeled tubulin with 8% rhodamine tubulin in PEM-100. DyLight649 biotin microtubules were prepared using a 50-µl mixture of unlabeled tubulin with 20% DyLight649 tubulin and 4% biotin tubulin in PEM-100. The mixtures were centrifuged for 10 min at 300,000 g at 4°C, 1 mM GTP was added to the supernatant, and microtubules were polymerized at 37°C for 20 min. 50 µM paclitaxel was added and incubated for 20 min at 37°C. Microtubules were pelleted at 14,100 g for 15 min at room temperature. The microtubule pellet was resuspended in 50 µl PEM-100 + 40 µM pactlitaxel.
Online supplemental material
Fig. S1 shows the distribution of mTPX2LAP and mTPX2-710LAP in LLC-Pk1 cells, along with metaphase spindle length and quantification of the spindle phenotypes in live cells. Fig. S2 shows the chromosome alignment defects in cells lacking the TPX2–Eg5 interaction. Fig. S3 shows the distribution of dynactin subunit p150 in cells expressing mTPX2LAP or mTPX2-710LAP and treated with siRNA-targeting pTPX2. Fig. S4 shows a Coomassie-stained gel of the proteins used in the in vitro experiments as well as graphs showing differential regulation of Eg5-513 and Kinesin-1 motor behavior by TPX2 and Tau. Video 1 shows live-cell imaging of a cell expressing mTPX2LAP, treated with siRNA-targeting pTPX2, and progressing through mitosis. Videos 2–4 show live-cell imaging of abnormal spindle morphologies in cells expressing mTPX2-710LAP with pTPX2 knocked down. Video 5 shows the effect of monastrol on a spindle with extra foci in a cell expressing mTPX2-710LAP with pTPX2 knocked down. Videos 6 and 7 show time lapses from gliding and sliding in vitro assays. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201106149/DC1.
Supplementary Material
Acknowledgments
The authors thank Dr. W.-L. Lee and members of his lab for sharing their expertise and for the use of lab equipment. We thank all the members of our laboratory for discussions and Drs. T. Maresca and W.-L. Lee for comments on the manuscript. We especially thank Drs. A. Bird, I. Poser, and A. Hyman for their generous assistance with BAC cloning methods.
This work was supported by funds from the National Science Foundation Materials Research Science and Engineering Center, Institute for Cellular Engineering Integrative Graduate Education and Research Traineeship (DGE-065412), and National Institutes of Health (GM-59057).
Footnotes
Abbreviations used in this paper:
- BAC
- bacterial artificial chromosome
- HURP
- Hepatoma up-regulated protein
- LAP
- localization and affinity purification
- TIRF
- total internal reflection fluorescence
References
- Bayliss R., Sardon T., Vernos I., Conti E. 2003. Structural basis of Aurora-A activation by TPX2 at the mitotic spindle. Mol. Cell. 12:851–862 10.1016/S1097-2765(03)00392-7 [DOI] [PubMed] [Google Scholar]
- Bird A.W., Hyman A.A. 2008. Building a spindle of the correct length in human cells requires the interaction between TPX2 and Aurora A. J. Cell Biol. 182:289–300 10.1083/jcb.200802005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blangy A., Lane H.A., d’Hérin P., Harper M., Kress M., Nigg E.A. 1995. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell. 83:1159–1169 10.1016/0092-8674(95)90142-6 [DOI] [PubMed] [Google Scholar]
- Breuer M., Kolano A., Kwon M., Li C.C., Tsai T.F., Pellman D., Brunet S., Verlhac M.H. 2010. HURP permits MTOC sorting for robust meiotic spindle bipolarity, similar to extra centrosome clustering in cancer cells. J. Cell Biol. 191:1251–1260 10.1083/jcb.201005065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton D.A. 1998. Focusing on spindle poles. J. Cell Sci. 111:1477–1481 [DOI] [PubMed] [Google Scholar]
- DeLuca J.G., Moree B., Hickey J.M., Kilmartin J.V., Salmon E.D. 2002. hNuf2 inhibition blocks stable kinetochore–microtubule attachment and induces mitotic cell death in HeLa cells. J. Cell Biol. 159:549–555 10.1083/jcb.200208159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A., Mitchison T.J. 1998. Preparation and characterization of caged fluorescein tubulin. Methods Enzymol. 298:125–132 10.1016/S0076-6879(98)98014-4 [DOI] [PubMed] [Google Scholar]
- Dixit R., Ross J.L., Goldman Y.E., Holzbaur E.L.F. 2008. Differential regulation of dynein and kinesin motor proteins by tau. Science. 319:1086–1089 10.1126/science.1152993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckerdt F., Eyers P.A., Lewellyn A.L., Prigent C., Maller J.L. 2008. Spindle pole regulation by a discrete Eg5-interacting domain in TPX2. Curr. Biol. 18:519–525 10.1016/j.cub.2008.02.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenz N.P., Paul R., Fagerstrom C., Mogilner A., Wadsworth P. 2009. Dynein antagonizes eg5 by crosslinking and sliding antiparallel microtubules. Curr. Biol. 19:1833–1838 10.1016/j.cub.2009.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenz N.P., Gable A., Wadsworth P. 2010. Mitotic functions of kinesin-5. Semin. Cell Dev. Biol. 21:255–259 10.1016/j.semcdb.2010.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaetz J., Kapoor T.M. 2004. Dynein/dynactin regulate metaphase spindle length by targeting depolymerizing activities to spindle poles. J. Cell Biol. 166:465–471 10.1083/jcb.200404015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett S., Auer K., Compton D.A., Kapoor T.M. 2002. hTPX2 is required for normal spindle morphology and centrosome integrity during vertebrate cell division. Curr. Biol. 12:2055–2059 10.1016/S0960-9822(02)01277-0 [DOI] [PubMed] [Google Scholar]
- Gruss O.J., Wittmann M., Yokoyama H., Pepperkok R., Kufer T., Silljé H., Karsenti E., Mattaj I.W., Vernos I. 2002. Chromosome-induced microtubule assembly mediated by TPX2 is required for spindle formation in HeLa cells. Nat. Cell Biol. 4:871–879 10.1038/ncb870 [DOI] [PubMed] [Google Scholar]
- Kapitein L.C., Peterman E.J.G., Kwok B.H., Kim J.H., Kapoor T.M., Schmidt C.F. 2005. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 435:114–118 10.1038/nature03503 [DOI] [PubMed] [Google Scholar]
- King S.J., Brown C.L., Maier K.C., Quintyne N.J., Schroer T.A. 2003. Analysis of the dynein-dynactin interaction in vitro and in vivo. Mol. Biol. Cell. 14:5089–5097 10.1091/mbc.E03-01-0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittler R., Pelletier L., Ma C., Poser I., Fischer S., Hyman A.A., Buchholz F. 2005. RNA interference rescue by bacterial artificial chromosome transgenesis in mammalian tissue culture cells. Proc. Natl. Acad. Sci. USA. 102:2396–2401 10.1073/pnas.0409861102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koffa M.D., Casanova C.M., Santarella R., Köcher T., Wilm M., Mattaj I.W. 2006. HURP is part of a Ran-dependent complex involved in spindle formation. Curr. Biol. 16:743–754 10.1016/j.cub.2006.03.056 [DOI] [PubMed] [Google Scholar]
- Kufer T.A., Silljé H.H.W., Körner R., Gruss O.J., Meraldi P., Nigg E.A. 2002. Human TPX2 is required for targeting Aurora-A kinase to the spindle. J. Cell Biol. 158:617–623 10.1083/jcb.200204155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N., Tulu S., Ferenz N., Fagerstrom C., Wilde A., Wadsworth P. 2010. Poleward transport of TPX2 in the mammalian spindle requires dynein, Eg5, and microtubule flux. Mol. Biol. Cell. 21:979–988 10.1091/mbc.E09-07-0601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N., Matsunaga S., Morimoto A., Sakashita G., Urano T., Uchiyama S., Fukui K. 2011. The nuclear scaffold protein SAF-A is required for kinetochore-microtubule attachment and contributes to the targeting of Aurora-A to mitotic spindles. J. Cell Sci. 124:394–404 10.1242/jcs.063347 [DOI] [PubMed] [Google Scholar]
- Manning A.L., Compton D.A. 2007. Mechanisms of spindle-pole organization are influenced by kinetochore activity in mammalian cells. Curr. Biol. 17:260–265 10.1016/j.cub.2006.11.071 [DOI] [PubMed] [Google Scholar]
- Mayer T.U., Kapoor T.M., Haggarty S.J., King R.W., Schreiber S.L., Mitchison T.J. 1999. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 286:971–974 10.1126/science.286.5441.971 [DOI] [PubMed] [Google Scholar]
- Neumann B., Walter T., Hériché J.-K., Bulkescher J., Erfle H., Conrad C., Rogers P., Poser I., Held M., Liebel U., et al. 2010. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 464:721–727 10.1038/nature08869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peloquin J., Komarova Y., Borisy G. 2005. Conjugation of fluorophores to tubulin. Nat. Methods. 2:299–303 10.1038/nmeth0405-299 [DOI] [PubMed] [Google Scholar]
- Rieder C.L. 1981. The structure of the cold-stable kinetochore fiber in metaphase PtK1 cells. Chromosoma. 84:145–158 10.1007/BF00293368 [DOI] [PubMed] [Google Scholar]
- Ross J.L., Shuman H., Holzbaur E.L.F., Goldman Y.E. 2008. Kinesin and dynein-dynactin at intersecting microtubules: motor density affects dynein function. Biophys. J. 94:3115–3125 10.1529/biophysj.107.120014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozelle D.K., Hansen S.D., Kaplan K.B. 2011. Chromosome passenger complexes control anaphase duration and spindle elongation via a kinesin-5 brake. J. Cell Biol. 193:285–294 10.1083/jcb.201011002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin K.E., LeGuellec K., Philippe M., Mitchison T.J. 1992. Mitotic spindle organization by a plus-end-directed microtubule motor. Nature. 359:540–543 10.1038/359540a0 [DOI] [PubMed] [Google Scholar]
- Sharp D.J., Brown H.M., Kwon M., Rogers G.C., Holland G., Scholey J.M. 2000. Functional coordination of three mitotic motors in Drosophila embryos. Mol. Biol. Cell. 11:241–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silljé H.H.W., Nagel S., Körner R., Nigg E.A. 2006. HURP is a Ran-importin beta-regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes. Curr. Biol. 16:731–742 10.1016/j.cub.2006.02.070 [DOI] [PubMed] [Google Scholar]
- Tanenbaum M.E., Macůrek L., Jeanssen N., Geers E.F., Alvarez-Fernández M., Medema R.H. 2009. Kif15 cooperates with eg5 to promote bipolar spindle assembly. Curr. Biol. 19:1703–1711 10.1016/j.cub.2009.08.027 [DOI] [PubMed] [Google Scholar]
- Tulu U.S., Rusan N.M., Wadsworth P. 2003. Peripheral, non-centrosome-associated microtubules contribute to spindle formation in centrosome-containing cells. Curr. Biol. 13:1894–1899 10.1016/j.cub.2003.10.002 [DOI] [PubMed] [Google Scholar]
- Tulu U.S., Fagerstrom C., Ferenz N.P., Wadsworth P. 2006. Molecular requirements for kinetochore-associated microtubule formation in mammalian cells. Curr. Biol. 16:536–541 10.1016/j.cub.2006.01.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uteng M., Hentrich C., Miura K., Bieling P., Surrey T. 2008. Poleward transport of Eg5 by dynein–dynactin in Xenopus laevis egg extract spindles. J. Cell Biol. 182:715–726 10.1083/jcb.200801125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Wildenberg S.M., Tao L., Kapitein L.C., Schmidt C.F., Scholey J.M., Peterman E.J. 2008. The homotetrameric kinesin-5 KLP61F preferentially crosslinks microtubules into antiparallel orientations. Curr. Biol. 18:1860–1864 10.1016/j.cub.2008.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste D., Takagi M., Imamoto N., Vernos I. 2009. The role of Hklp2 in the stabilization and maintenance of spindle bipolarity. Curr. Biol. 19:1712–1717 10.1016/j.cub.2009.09.019 [DOI] [PubMed] [Google Scholar]
- Wadsworth P. 1999. Microinjection of mitotic cells. Methods Cell Biol. 61:219–231 10.1016/S0091-679X(08)61983-4 [DOI] [PubMed] [Google Scholar]
- Wadsworth P., Rusan N.M., Tulu U.S., Fagerstrom C. 2005. Stable expression of fluorescently tagged proteins for studies of mitosis in mammalian cells. Nat. Methods. 2:981–987 10.1038/nmeth1205-981 [DOI] [PubMed] [Google Scholar]
- Weinger J.S., Qiu M., Yang G., Kapoor T.M. 2011. A nonmotor microtubule binding site in kinesin-5 is required for filament crosslinking and sliding. Curr. Biol. 21:154–160 10.1016/j.cub.2010.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann T., Boleti H., Antony C., Karsenti E., Vernos I. 1998. Localization of the kinesin-like protein Xklp2 to spindle poles requires a leucine zipper, a microtubule-associated protein, and dynein. J. Cell Biol. 143:673–685 10.1083/jcb.143.3.673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J., Fang G. 2006. HURP controls spindle dynamics to promote proper interkinetochore tension and efficient kinetochore capture. J. Cell Biol. 173:879–891 10.1083/jcb.200511132 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.