The Rab11 GTPase-binding protein FIP5 collaborates with the sorting nexin 18 to transport proteins to the apical surface and to tubulate membranes during epithelial apical lumen formation.
Abstract
During the morphogenesis of the epithelial lumen, apical proteins are thought to be transported via endocytic compartments to the site of the forming lumen, although the machinery mediating this transport remains to be elucidated. Rab11 GTPase and its binding protein, FIP5, are important regulators of polarized endocytic transport. In this study, we identify sorting nexin 18 as a novel FIP5-interacting protein and characterize the role of FIP5 and SNX18 in epithelial lumen morphogenesis. We show that FIP5 mediates the transport of apical proteins from apical endosomes to the apical plasma membrane and, along with SNX18, is required for the early stages of apical lumen formation. Furthermore, both proteins bind lipids, and FIP5 promotes the capacity of SNX18 to tubulate membranes, which implies a role for FIP5 and SNX18 in endocytic carrier formation and/or scission. In summary, the present findings support the hypothesis that this FIP5-SNX18 complex plays a pivotal role in the polarized transport of apical proteins during apical lumen initiation in epithelial cells.
Introduction
The misregulation of polarized transport within epithelial cells can result in a loss of apico-basolateral polarity. The importance of this cellular asymmetry is clear, as its loss is the first step of the epithelial-to-mesenchymal transition (EMT; Townsend et al., 2008), which in addition to being a critical developmental mechanism, is also a well-established prerequisite for tumorigenesis and metastasis (Wang et al., 2008a; López-Novoa and Nieto, 2009; Strizzi et al., 2009). The appropriate regulation of epithelial polarity depends on the regulation of endocytic and biosynthetic transport routes. Consequently, polarized transport is a very complex process that is highly regulated throughout development and tissue morphogenesis (Zurzolo et al., 1992). The plasma membrane (PM) of epithelial cells is separated into the apical and basolateral domains, distinct in both lipid and protein composition. This asymmetry is essential for the proper function of these specialized cells, which line the internal face of many organs and act as a barrier between the internal and external environments of the organism. The “holy grail” of epithelial organogenesis is to determine how individual cells are capable of coordinating and acting as a multicellular unit with a defined architecture that enables this organ to carry out complex processes that are beyond the capabilities of any single cell.
The formation of the apical lumen is a key step in organogenesis, required for the establishment of the organ’s architecture, and thereby its function. Although there is a high degree of morphogenetic diversity between organisms, the end result of lumen formation is always a structure in which the apical surface of the cell is facing the epithelial lumen. Thereby, the establishment and expansion of the apical lumen is a key step during tissue morphogenesis. Much effort has been dedicated to deciphering the machinery responsible for the establishment and expansion of the epithelial lumen, with one of the more recent areas of interest being apical targeting, which has been shown to play a key role in the establishment of the epithelial lumen during development (Lubarsky and Krasnow, 2003; Martin-Belmonte and Mostov, 2008).
As the result of work from many laboratories, we are beginning to decipher the machinery that mediates lumen formation during tissue morphogenesis. One model, proposed by Dr. Mostov and colleagues, suggests that at the single-cell stage, the apical domain markers, such as glycoprotein 135 (gp135) and Crumbs3, are localized at the PM facing the ECM (Martin-Belmonte and Mostov, 2008; Schlüter et al., 2009; Bryant et al., 2010). After initial cell division, apical proteins are endocytosed and are transported to the site of the forming apical lumen. Here, apical cargo–containing transport vesicles fuse with the PM to initiate the formation and expansion of the apical lumen. The identity of these apical cargo–containing transport vesicles remains to be fully understood. Originally they were referred to as vacuolar apical compartments (VACs), as they contain apical PM protein gp135. However, recent work has shown that these organelles also contain Rab11 and Rab8 GTPases, and thus likely represent a subpopulation of apical recycling endosomes (Bryant et al., 2010). We will henceforth refer to these organelles as gp135-containing endosomes.
The machinery that targets gp135-containing endosomes to the site of the forming apical lumen involves interplay between multiple proteins (Bryant et al., 2010). Among such proteins, Rab11 plays a key role in regulating the polarized transport (Desclozeaux et al., 2008; Bryant et al., 2010). However, the precise molecular mechanisms mediating Rab11 function remain to be determined. It is thought that Rab11 binding to specific effector proteins is vital for determining Rab11’s functions, as well as its specificity for individual endocytic recycling pathways. Rab11 family interacting proteins (Rab11-FIPs, referred to henceforth as FIPs) were identified as Rab11 effector proteins (Prekeris et al., 2000; Hales et al., 2001). FIPs serve as scaffolds that recruit specific sets of endocytic proteins. One member of the FIP family, FIP5 (also known as Rip11), is required for protein targeting to the apical PM (Prekeris et al., 2000). A potential role of FIP5 in mediating Rab11-dependent apical lumen formation has never been investigated and is the focus of these studies. Here, we investigate the mechanism of lumen formation and demonstrate that gp135 is transported via apical recycling endosomes, a process that is dependent on the FIP5. Furthermore, we identify sorting nexin 18 (SNX18) as a FIP5-binding protein, and demonstrate that interaction between these proteins regulates membrane tubulation in vitro, as well as epithelial lumen formation in vivo, while having no effect on basolateral protein transport. Thereby, we propose that the FIP5, via interaction with SNX18, mediates the formation and transport of gp135-containing endosomes, and consequently plays a role in the establishment of a single lumen during epithelial cyst morphogenesis.
Results
FIP5 is a Rab11-effector protein that is enriched in apical recycling endosomes in polarized epithelial cysts
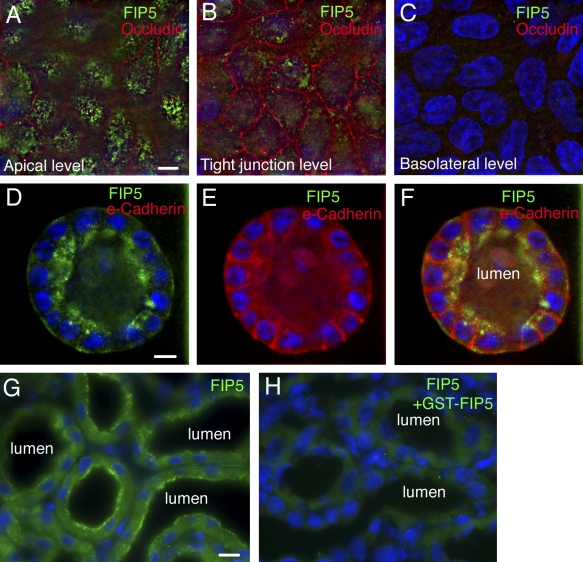
The establishment of the epithelial lumen is a complex process known to depend on Rab11 GTPase (Desclozeaux et al., 2008), as well as on the polarized transport of apical proteins (Vega-Salas et al., 1987; Davis and Bayless, 2003; Kamei et al., 2006; Martin-Belmonte and Mostov, 2008). Because Rab11, like all Rab GTPases, works via the recruitment and activation of its effector proteins, we set out to elucidate the role of FIPs in mediating epithelial lumen formation. Previous studies have shown that FIP5, a member of the FIP family of proteins, is required for transcytosis of the polymeric IgA receptor (pIgA-R) in filter-grown MDCK cells (Prekeris et al., 2000). Because transport of apical PM proteins during lumen formation resembles transcytosis, we decided to test whether FIP5 may also be involved in the transport and targeting of apical membrane proteins during lumen formation. First, we tested the localization of FIP5 in 3D cultures of polarized MDCK cysts. As shown in Fig. 1 (D–F), FIP5 localizes to the apical domain of the cysts, although some FIP5 staining can also be observed close to the basolateral PM. Similarly, in rat kidney proximal tubules, FIP5 is enriched at the apical pole of the cells (Fig. 1, G and H). This localization in both 2D and 3D cultures is fully consistent with the observed staining of FIP5 in filter-grown MDCK cells, where FIP5 is enriched in the endosomal organelles close to the apical PM (Fig. 1, A–C). Furthermore, in filter-grown MDCK cells, FIP5 partially colocalizes with the pIgA-R but not the transferrin receptor (TfR; unpublished data), which is consistent with a role for FIP5 in apically directed protein transport.
Figure 1.
FIP5 is enriched at the apical pole of polarized epithelial cells. (A–F) MDCK cells were grown in 2D (A–C) or 3D (D–F) cultures. Cells were fixed with 4% paraformaldehyde and stained with anti-FIP5 (A–F, green), anti-Occludin (A–C, red) or anti–e-Cadherin (D–F, red) antibodies. In A–C, images were taken at the apical, tight junction and basolateral levels of polarized MDCK cells. (G and H) Rat kidney sections were stained with anti-FIP5 antibody. In H, antibody was preincubated with a 20-fold excess of purified recombinant FIP5. Bars: (A–F) 8 µm; (G and H) 10 µm.
FIP5 knockdown disrupts lumen morphogenesis during the formation of epithelial cysts
To further understand the role of FIP5 in epithelial lumen morphogenesis, we used the pHUSH conditional short hairpin RNA (shRNA) system (Gray et al., 2007) to create a tetracycline (tet)-inducible FIP5 shRNA-expressing MDCK cell line (MDCK-shFIP5). The use of tet-inducible shRNA allows us to knock down FIP5 expression before or after the formation of epithelial cysts, thus separating the roles of FIP5 in the establishment and maintenance of the epithelial lumen. As shown in Fig. S1 (A and B), incubation of MDCK-shFIP5 cells with 1 µg/ml doxycycline (dox) resulted in the specific depletion of FIP5 by >85% while having no effect on the expression of other FIPs, such as FIP1. Interestingly, after FIP5 knockdown, filter-grown cells were still able to form tight junctions, as observed by the unchanged trans-epithelial resistance and localization of the tight junction marker Occludin (Fig. S1 C). We also performed transferrin (Tf) basolateral recycling assays, and have shown that FIP5 knockdown had no significant effect on the rate or fidelity of basolateral endocytic recycling (Fig. S1 D). Thus, filter-grown MDCK cells lacking FIP5 are capable of polarization and the formation of intact epithelial monolayers.
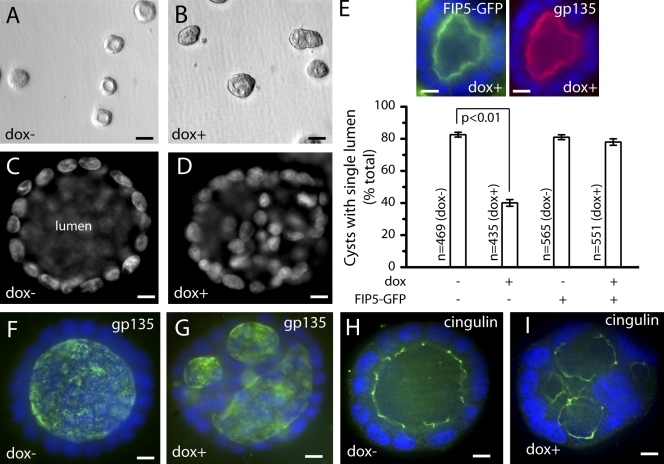
To test whether FIP5 is required for epithelial lumen morphogenesis, we grew MDCK-shFIP5 cells in Matrigel 3D cultures to allow for the development of polarized epithelial cysts. As shown in Fig. 2 (A and C), after 12 d, untreated MDCK-shFIP5 cells formed normal epithelial cysts with a single lumen that is surrounded by a monolayer of polarized cells. In contrast, upon FIP5 knockdown, the resulting cysts have multiple lumens that are surrounded in some areas by several layers of cells (Fig. 2, B, D, and E). The phenotype could be rescued by the expression of shRNA-resistant FIP5-GFP (Fig. 2 E), which demonstrates that multiple lumens are the result of FIP5 knockdown rather than off-target effects. To determine whether cells in FIP5-lacking cysts still polarize, we stained control and dox-treated cysts with antibodies against gp135. Glycoprotein 135 (gp135) is a 135-kD cell surface membrane protein that has been found to localize to the apical pole of MDCK cells, where it is thought to associate with the actin cytoskeleton and is a hallmark of the apical PM in epithelial cells. As shown in Fig. 2 F, control cysts contain a single lumen outlined by anti-gp135 staining. In contrast, in FIP5 knockdown cysts, gp135 marks the presence of multiple lumens (Fig. 2 G). To further confirm that dox-treated cells form multiple lumens, we also tested the localization of ezrin. Ezrin is a well-characterized member of the ezrin-radixin-moesin (ERM) family of proteins shown to concentrate on the apical surface of polarized epithelial cells. Consistent with our observation that FIP5 regulates lumen formation, anti-ezrin staining also confirmed the presence of multiple lumens in knockdown cysts (unpublished data). To test whether FIP5 knockdown cells form tight junctions in 3D cultures, we used anti-cingulin antibody to visualize tight junctions. As shown in Fig. 2 I, after FIP5 depletion, 3D cysts could still form tight junctions, although these tight junctions are now found to be associated with each of the multiple lumens.
Figure 2.
FIP5 is required for the formation of a single lumen in 3D epithelial cysts. (A–E) MDCK-shFIP5 cells were grown for 9 d in the presence (B and D) or absence (A and C) of 1 µg/ml of dox. Cells were then fixed with 4% paraformaldehyde, stained with Hoechst DNA stain, and imaged by bright-field (A and B) or fluorescence (C and D) microscopy. E shows the quantitation of epithelial cysts with single lumen. Data shown are the means and standard deviations derived from three independent experiments (error bars). n is the total number of cysts analyzed. Insets show dox+ cyst expressing FIP5-GFP. (F–I) MDCK-shFIP5 cells were grown for 9 d in the presence (G and I) or absence (F and H) of 1 µg/ml of dox. Cells were then fixed with 4% paraformaldehyde and stained with anti-gp135 (F and G) or anti-cingulin (H and I) antibodies. Bars: (A and B) 40 µm; (C and D) 8 µm; (E) 16 µm (F–I) 8 µm.
FIP5 is required for gp135 targeting during early stages of apical lumen formation
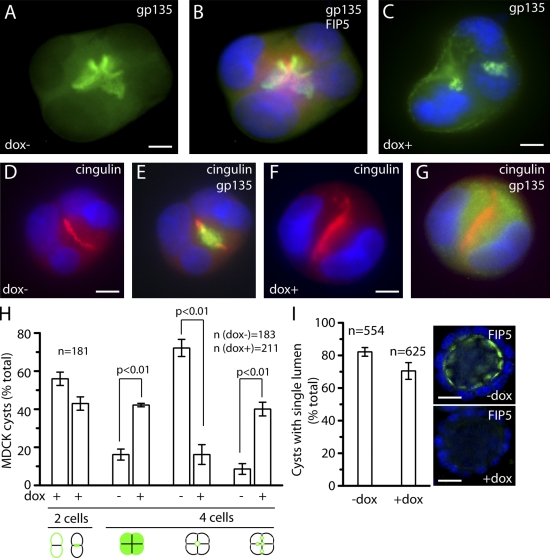
Although our data demonstrate that FIP5-mediated transport is required for epithelial lumen formation, it remains unclear whether FIP5 is required for the establishment of the lumen or for its maintenance after the polarization of the epithelial cyst has occurred. Although the mechanisms of epithelial lumen establishment remain to be fully understood, one existing model proposes that in the early stages of MDCK cell lumen morphogenesis, apical proteins are localized in the PM facing the ECM (Martin-Belmonte and Mostov, 2008; Schlüter et al., 2009). At the two/four cell stage, the apical proteins are transported to the site of the forming apical lumen (known as apical membrane initiation site [AMIS]). Fusion of these transport organelles initiates lumen formation and establishes the apical-basolateral polarity of the cyst (Martin-Belmonte and Mostov, 2008). Consistent with this model, at the two-cell stage, almost 40% of cysts displayed gp135 at the PM domain facing the ECM (Fig. S2, A–C and J). At this stage, FIP5 is localized to punctate organelles that are diffusely scattered throughout the cell (Fig. S2, B and C). In the remaining ∼60% of forming cysts, gp135 was localized to the area of cell–cell contact, with the basolateral proteins facing the ECM (Fig. S2, D–F; and Fig. 3 J). These cells likely represent the initial stages of lumen formation after the inversion of polarity. By this stage, FIP5 also concentrates at the site of the forming lumen between the cells and exhibits a substantial overlap with gp135 staining (Fig. S2, E and F). By the four-cell stage, >80% of cysts have formed a single lumen that is situated at the center of the dividing cells with the basolateral proteins facing the ECM, with FIP5 staining concentrated around the site of the initiated lumen (Fig. S2, G–J).
Figure 3.
FIP5 is required for the establishment of the apical lumen during the early stages of epithelial cyst formation. (A–G) MDCK-shFIP5 cells were grown for 74 h in the presence (C, F, and G) or absence (A, B, D, and E) of 1 µg/ml of dox to pre-knockdown FIP5. Cells were then seeded in 3D cultures and grown for 24 h. Cells were fixed with 4% paraformaldehyde and stained with anti-gp135 (A–C, E, and G, green), anti-cingulin (D–G, red), or anti-FIP5 (B, red) antibodies. (H) The quantitation of 3D cyst polarization at the two and four cell stages from the experiment shown in A–F. Data shown are the means and standard deviations derived from three independent experiments. n is the number of cysts analyzed. (I) The quantitation of fully matured (9 d) epithelial cysts with a single lumen in cells incubated with or without 1 µg/ml of dox supplementation beginning 24 h after seeding in 3D cultures. Data shown are the means and standard deviations derived from three independent experiments (error bars). n is the number of cysts analyzed. Insets show the extent of FIP5 (green) knockdown in the presence of doxycycline. Bars: (A–G) 8 µm; (I) 20 µm.
Protein transport to the apical membrane resembles transcytosis of the pIgA-R that occurs in filter-grown MDCK cells. Because FIP5 is known to be required for transcytosis and is localized to the site of the forming lumen (Fig. S2, E and H), we tested whether FIP5 may be required for transport of gp135 to the apical PM. MDCK-shFIP5 cells were preincubated for 74 h in the presence or absence of dox, seeded, allowed to grow for 24 h in 3D cultures, then imaged to analyze the cysts at the two- and four-cell stages. As shown in Fig. 3 (A, B, C, and H), in the FIP5-depleted cells, only 20% of the cysts at the four-cell stage had formed a single lumen, as compared with 70% of the control cysts. In the remaining 80% of FIP5-depleted cysts, half endocytosed gp135, but these gp135-containing endosomes accumulated at multiple sites within the cells (Fig. 3, C and H). In the other half of the cells, gp135 was still present at PM as well as inside the organelles scattered throughout the cytosol (Fig. 3, G and H; and not depicted). Interestingly, when we added dox after the initial lumen had been formed (24 h after seeding), we observed no defects in single apical lumen maintenance (Fig. 3 I).
Multiple lumen formation can be the result of the failure of the AMIS formation or of defects in delivery/fusion of gp135-containing endosomes. Because tight junction proteins have been shown to be a marker for the AMIS at the two-cell stage, we stained cells grown in the presence or absence of dox with anti-cingulin and anti-gp135 antibodies. As shown in Fig. 3 (D–G), FIP5 depletion did not have any effect on AMIS formation. In summary, these results indicate a role for FIP5 in regulating the transport of gp135-containing endosomes to the AMIS during the initiation of the epithelial lumen, but no role in lumen maintenance.
FIP5 is required for scission of the endocytic carriers from recycling endosomes
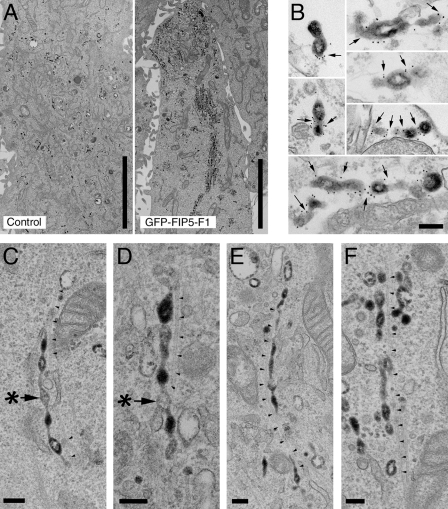
Although our data demonstrate the involvement of FIP5 in the transport of apical proteins and the formation of the apical lumen, it remains unclear what stage of membrane traffic is dependent on FIP5. To gain insight into whether FIP5 regulates endocytic carrier formation, budding, transport, or fusion, we transduced HeLa cells with a FIP5 dominant-negative mutant (GFP-FIP5-F1, also known as GFP-Rip11-F1) and analyzed endosomal morphology by EM. GFP-FIP5-F1 is the GFP-tagged C-terminal end of FIP5, which contains the Rab11-binding domain. This mutant has previously been shown to act as a strong dominant-negative inhibitor by dimerizing with endogenous FIP5 and inhibiting its cellular functions (Junutula et al., 2004). Because FIP5 has been shown to play a role in Tf recycling in nonpolarized HeLa cells (Schonteich et al., 2008), we loaded HeLa cells (for 45 min at 37°C) with Tf-HRP to identify recycling endosomes. As shown in Fig. 4 A (right), overexpression of GFP-FIP5-F1 caused extensive accumulation of tubulated membrane-bound organelles, which are likely recycling endosomes, as they contain Tf-HRP. Interestingly, most of these tubular endosomes exhibited a “beads-on-a-string” phenotype and were often extended along microtubules (Fig. 4, C–F). The association of these endosomes with microtubules is consistent with previous observations that FIP5 also binds Kinesin II and regulates endosomal transport along microtubules. Furthermore, the putative Kinesin II binding domain is located within FIP5’s C terminus (Schonteich et al., 2008), thus allowing GFP-FIP5-F1 to interact with Kinesin II. A similar, although less dramatic, beads-on-a-string phenotype was also observed in HeLa cells transfected with FIP5, but not FIP1, siRNAs (Fig. S3, A and B; and not depicted).
Figure 4.
FIP5 is required for the scission of endocytic carriers at recycling endosomes. (A) Control (left) and GFP-FIP5-F1–expressing (right) HeLa cells were incubated with Tf-HRP for 45 min at 37°C. Cells were then fixed and prepared for ultrastructural EM analysis. Electron-dense dark precipitate indicates DAB reaction product and highlights the localization of Tf-HRP. (B) GFP-FIP5-F1–expressing HeLa cells were incubated with Tf-HRP for 45 min at 37°C. The DAB reaction was performed before permeabilization, antibody incubation, and fixation. Electron-dense dark precipitate indicates the DAB reaction product and highlights the localization of Tf-HRP, whereas 10 nm gold particles indicate the localization of GFP-FIP5-F1 (arrows). (C–F) Higher-magnification images from HeLa cells expressing GFP-FIP5-F1 and loaded with Tf-HRP. Asterisks and arrows in C and D point to the “bead” that lacks Tf-HRP, whereas arrowheads track nearby microtubules. Bars: (A) 5 µm; (B–F) 200 nm.
Perhaps the most interesting observation is that Tf-HRP was not evenly distributed within the GFP-FIP5-F1–induced beads-on-a-string endosomes, as some “beads” contain high levels of Tf-HRP, whereas others appear to completely lack Tf-HRP (Fig. 4, C and D, asterisks). This uneven Tf-HRP distribution within a single endosome suggests that overexpression of the FIP5 dominant-negative mutant allows for cargo sorting, but may block the scission of endocytic carriers, thus resulting in this beads-on-a-string phenotype. Consistent with this hypothesis, the majority of GFP-FIP5-F1 appears to be localized to the necks between the beads, as well as to the necks of the forming buds at the ends of the endosomal tubules (Fig. 4 B).
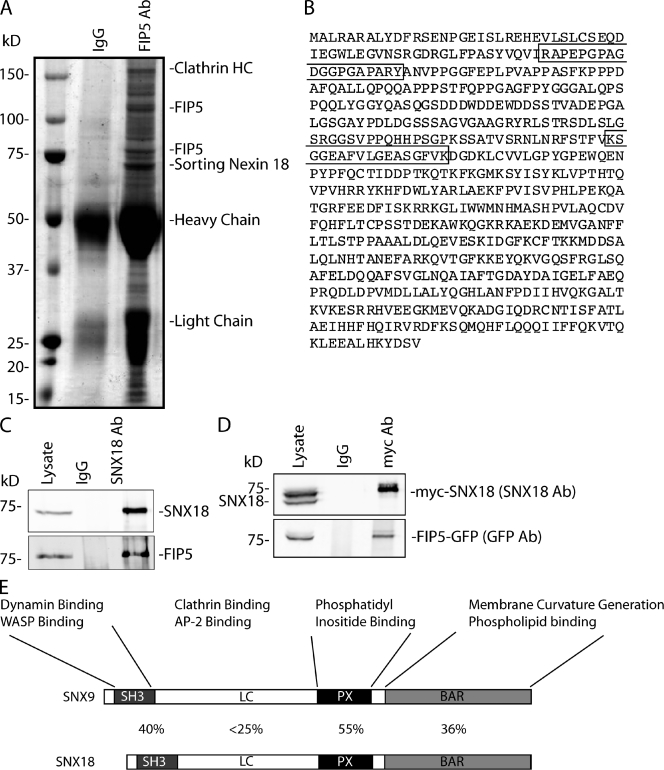
SNX18 is a FIP5-binding protein
Recent studies demonstrate that FIP family members function as scaffolding proteins that recruit various mediators of membrane transport to the recycling endosomes. To identify potential FIP5-binding proteins, we immunoprecipitated FIP5 from HeLa cells using anti-FIP5 antibody (Fig. 5 A). The protein bands present only in the anti-FIP5 antibody lane, and not in the IgG control lane, were cut out and analyzed by mass spectrometry (Fig. 5 A). The corresponding areas from the IgG control were also cut out and analyzed as negative controls. Only proteins that were identified by at least two different peptides solely in the anti-FIP5 immunoprecipitate were considered as potential FIP5-binding proteins. Interestingly, proteomic analysis identified the clathrin heavy chain and SNX18 as FIP5-associating proteins (Fig. 5, A and B). To confirm that SNX18 and FIP5 are part of the same protein complex in vivo, anti-SNX18 and anti-SNX9 antibodies were generated (Fig. S3, C and D). These antibodies demonstrated that FIP5 coimmunoprecipitates from MDCK cell lysates with anti-SNX18 but not anti-SNX9 antibodies (Fig. 5 C and not depicted). Furthermore, FIP5-GFP coimmunoprecipitates with myc-SNX18 from cells coexpressing FIP5-GFP and myc-SNX18 (Fig. 5 D).
Figure 5.
SNX18 is FIP5-binding protein. (A) FIP5 was immunopreciptated from HeLa cell lysates with anti-FIP5 antibody. The immunoprecipitate was then separated by SDS-PAGE and stained with Coomassie dye. Proteins listed in the figure were identified by at least two peptides from the anti-FIP5 immunoprecipitate, and were not present in the IgG control. (B) Human SNX18 sequence. Boxed regions indicate the peptides identified in proteomic analysis of the immunoprecipitate from A. (C) SNX18 or FIP5 were immunoprecipitated from MDCK cell lysates and immunoblotted with anti-SNX18, anti-FIP5, and anti-SNX9 antibodies. (D) HeLa cells were cotransfected with myc-SNX18 and FIP5-GFP. Cells were lysed, and myc-SNX18 was immunoprecipitated with anti-myc antibodies and blotted with anti-SNX18 and anti-GFP antibodies. (E) Schematic representation of the domains present in SNX9 and SNX18 proteins. Numbers between the SNX9 and SNX18 schematics indicate the percentage of homology between the corresponding domains of these proteins.
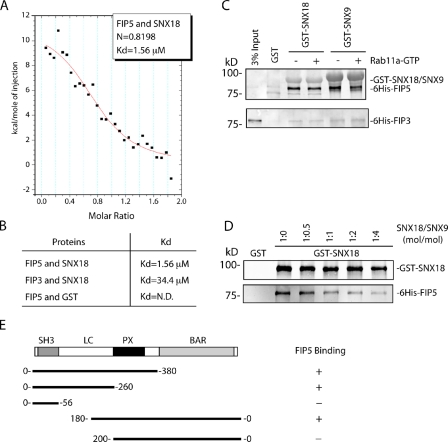
SNX18 is one of >30 members of the SNX family of proteins, many of which have been identified as regulators of endocytic cargo sorting and vesicle formation. SNX family proteins are peripheral membrane proteins that are characterized by the presence of a Phox-homology (PX) domain, which binds phosphatidylinositides (PIs) on membrane surfaces. SNX18, along with its highly similar homologues SNX9 (depicted in Fig. 5 E) and SNX33, are part of a subfamily of SNXs that contain a BAR domain that binds to phospholipids and induces curvature in membranes, thus aiding in tubule and vesicle formation. The SNX9/18/33 family members are highly homologous to each other with the exception of the low complexity (LC) region (Fig. 5 E). SNX9, SNX18, and SNX33 were shown to have distinct subcellular localizations, and are thought to regulate different stages of endocytic trafficking, although recent work has shown that there may be some redundancy between SNX9 and SNX18 (Park et al., 2010). To further characterize the interaction between FIP5 and SNX18, we performed isothermal titration calorimetry (ITC). ITC analysis shows that FIP5 and SNX18 bind with an apparent Kd of 1.56 µM (Fig. 6, A and B). The binding appears to be specific to FIP5, as SNX18 interacts with other FIP family members with considerably lower affinity (Fig. 6, B and C). To test whether Rab11 regulates FIP5 binding to SNX18, purified recombinant FIP5 was incubated with GST-SNX18 beads in the presence or absence of Rab11-GTP. As shown in Fig. 6 C, Rab11-GTP had no effect on the interaction between FIP5 and GST-SNX18, which suggests that FIP5 can interact with Rab11 and SNX18 simultaneously. Surprisingly, FIP5 also bound to GST-SNX9, which suggests that FIP5 can bind in vitro to at least two members of the SNX9/18/33 family. To test if FIP5 binds to SNX9 and SNX18 via the same domain, we tested whether SNX9 and SNX18 compete for binding to FIP5. As shown in Fig. 6 D, soluble SNX9 inhibited binding of FIP5 to GST-SNX18 beads. Furthermore, our competition assay suggests that the affinity of SNX9 binding to FIP5 appears to be similar to that of SNX18 binding to FIP5 (Fig. 6 D).
Figure 6.
FIP5 binds to SNX18-LC domain. (A and B) The affinity of FIP5 and SNX18 binding as determined by ITC. N.D., not detected. (C) Glutathione beads were coated with GST-SNX18, GST-SNX9, or GST alone and incubated with 6His-FIP5 or 6His-FIP3 in the presence or absence of a fivefold excess of Rab11a-GTP. The amount of bound 6His-FIP3 or 6His-FIP5 was determined by immunoblotting with anti-FIP3 or anti-FIP5 antibodies. (D) Glutathione beads were coated with GST-SNX18 or GST alone and incubated with 6His-FIP5 in the presence of increasing concentrations of soluble recombinant SNX9. The amount of bound 6His-FIP5 was determined by immunoblotting with anti-FIP5 antibodies. (E) Schematic representation of the FIP5-binding domain in SNX18 as determined by glutathione bead pull-down assays.
To further characterize the interaction between FIP5 and SNX18, a series of SNX18 truncation mutants were created to determine the site of FIP5 binding to SNX18. The binding site was narrowed to a region of 20 amino acids (aa 180–200), which reside within the LC region of SNX18 (Fig. 6 D). Interestingly, SNX18’s LC domain contains a conserved binding sequence for Arp2/3, called the “A-like domain,” that is also found in SNX9 (Shin et al., 2008), as well as the site for interaction with the AP-1 complex (Håberg et al., 2008), with both of these sites located outside of the FIP5 binding region (aa 180–200). Because Arp2/3 and AP-1 have both been implicated in the regulation of endocytic budding, it is possible that SNX18 binding to FIP5 may be required for the formation and/or scission of endosomal carriers.
FIP5 induces SNX18-dependent tubulation of liposomes
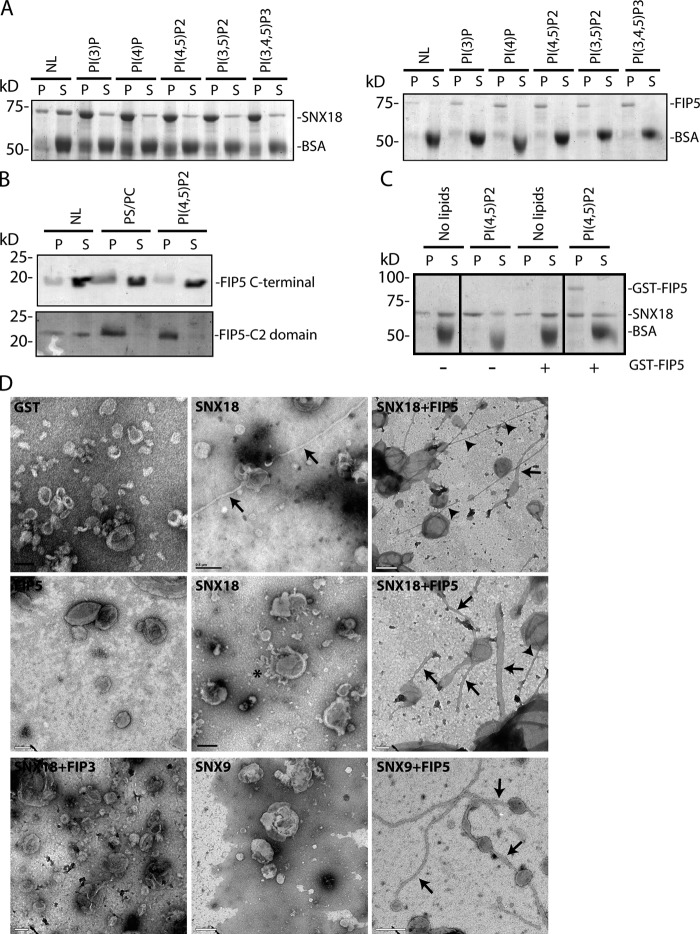
SNX18 and SNX9, being BAR domain–containing proteins known to bind phospholipids and induce curvature in membranes, likely contribute to tubule/vesicle formation and scission. Indeed, it was shown that SNX9 can tubulate liposomes in vivo and is required for AP-2 and clathrin-dependent budding and scission at the PM. Because SNX18 was shown to bind to AP-1, we postulated that SNX18 may play a similar role to SNX9 at the endosomes. The in vitro binding of SNX18 to liposomes was evaluated and showed that SNX18 readily binds liposomes (Fig. 7 A). Because SNX18 contains a PX domain, the ability of SNX18 to bind different PIs was tested. As shown in Fig. 7 A, SNX18 binds to liposomes containing different PIs in a seemingly nondiscriminatory manner. This observation is consistent with previous findings that the PX domains of SNX9 and SNX18 exhibit little selectivity for various PIs (Di Paolo and De Camilli, 2006; Pylypenko et al., 2007; Håberg et al., 2008).
Figure 7.
FIP5 induces SNX18- and SNX9-dependent liposome tubulation. (A) SNX18 or FIP5 were incubated with PS/PC liposomes containing various phosphatidylinositides. Liposomes were then sedimented and levels of bound SNX18 or FIP5 were determined by Coomassie staining. (B) C- (FIP5 C-terminal) or N-terminal (FIP5-C2) domains of FIP5 were tested for their ability to bind PS/PC liposomes containing 5% PI(4,5)P2. The levels of bound proteins were determined by Coomassie staining. (C) SNX18 was incubated with PS/PC/PI(4,5)P2 liposomes in the presence or absence of GST-FIP5. The levels of bound SNX18 and GST-FIP5 were determined by Coomassie staining. Black lines indicate the removal of intervening lanes for presentation purposes. (D) EM analysis of liposomes incubated with GST, GST-SNX9, GST-SNX18, 6His-FIP3, and 6His-FIP5. Arrows point to wide tubules (122.1 ± 32.9 nm, n = 10) and arrowheads point to narrow tubules (54.1 ± 18.1nm, n = 10). The asterisk marks short tubules induced by SNX18 alone.
Because FIP5 contains a C2 domain, which was suggested to bind lipids (Lindsay and McCaffrey, 2004), the ability of FIP5 to bind liposomes containing various PIs was analyzed (Fig. 7, A and B). Consistent with previous findings, FIP5 did bind to liposomes, an interaction that is mediated by its C2 domain (Fig. 7 B). However, in contrast to previous data (Lindsay and McCaffrey, 2004), no selectivity for specific lipid composition was detected (Fig. 7 A). The disparity in the results of these two binding assays may result from the present studies being direct liposome-binding assays, whereas previous work used protein–phospholipid overlay assays.
Next we tested whether FIP5 binding affects the ability of SNX18 to bind lipids. To this end, we incubated SNX18 with 1,2-dipalmitoyl-glycero-3-phosphoserine (PS)/1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (PC)/l-α-phosphatidyinositol-4,5-bisphosphate (PI(4,5)P2) liposomes in the presence or absence of GST-FIP5. As shown in Fig. 7 C, the addition of GST-FIP5 had no effect on SNX18 association with liposomes in vitro. Similarly, FIP5 depletion in MDCK cells had no effect on SNX18’s subcellular localization (unpublished data), which suggests that FIP5 and SNX18 are initially recruited to membranes independently from each other.
Because both SNX18 and FIP5 are capable of independently binding liposomes, the capacity of SNX18 and FIP5 to independently or coordinately tubulate liposomes was evaluated. Interestingly, although SNX18 alone was capable of inducing a small number of membrane tubules (Fig. 7 D), preincubation of SNX18 with FIP5 resulted in a much greater degree of liposome tubulation (Fig. 7 D). Interestingly, SNX18 coincubation with FIP5 resulted in liposome tubules of two distinct sizes. The larger tubules (122.1 ± 32.9 nm, n = 10) were shorter and often connected nontubulated liposomes, thus resembling a beads-on-a-string phenotype (Fig. 7 D, arrows). In contrast, thinner tubules (54.1 ± 18.1 nm, n = 10) were longer and usually not connected to round liposomes (Fig. 7 D, arrowheads). Finally, consistent with in vitro binding experiments, liposomes incubated with SNX9 and FIP5 also showed a higher degree of tubulation as compared with SNX9 alone (Fig. 7 D). In contrast, SNX18 incubated with FIP3 did not induce liposome tubulation, nor did GST or FIP5 alone (Fig. 7 D). These results indicate a role for FIP5 as an activator of SNX9/18’s tubulating activity that may play a role in the formation and/or scission of endocytic transport carriers.
SNX18 localizes to endocytic compartments and is required for lumen morphogenesis
The subcellular localization of proteins often provides clues about their potential functions. Unfortunately, published studies about the localization of SNX18 have been controversial. Håberg et al. (2008) have demonstrated that in HeLa cells, SNX18 is present on endosomes and may regulate endocytic transport. In contrast, recently published work suggested that SNX18 has redundant functions with SNX9, and may actually work with and localize to the PM (Park et al., 2010). To further understand the subcellular distribution of SNX18, we analyzed the localization of SNX18 in filter-grown, polarized MDCK cells. As shown in Fig. S4 (A–D), the majority of endogenous SNX18 was localized in the cytosol and on endosome-like organelles, rather than at the PM. This data support the hypothesis that SNX18 may aid in mediating vesicle budding and/or scission in endosomes. Note that, unlike FIP5, in 2D- or 3D-grown MDCK cells, SNX18 was not only present on apical endosomes, but could also be detected on basolaterally located organelles (Fig. S4). Thus, it is possible that SNX18 regulates basolateral endocytic transport steps in addition to apical transport.
In addition to MDCK cells, we compared the localization of SNX18 and SNX9 in HeLa cells. As shown in Fig. S4 G, GFP-SNX9 was predominately localized to multiple puncta present at the PM. In contrast, GFP-SNX18 was predominately localized to perinuclear organelles (Fig. S4 H). These organelles are likely recycling endosomes, as they could be labeled with Tf–Alexa Fluor 495 (Tf-Alexa495; Fig. S4, I–K). Interestingly, although GFP-SNX18 and Tf-Alexa495 were both present on some of the endocytic organelles, they were often enriched in different recycling endosomal subdomains (Fig. S4, I–K; GFP-SNX18 domains are marked by arrowheads). Thus, SNX18 may be required for the sorting and/or budding of specialized cargo while having no role in the trafficking of the TfR. To test this hypothesis, we analyzed Tf uptake and recycling dynamics in HeLa cells transfected with SNX18 siRNA (Fig. S5 F). Consistent with our localization data in HeLa cells, we see no effect of SNX18 knockdown on either Tf uptake (Fig. S5 F) or its recycling (not depicted).
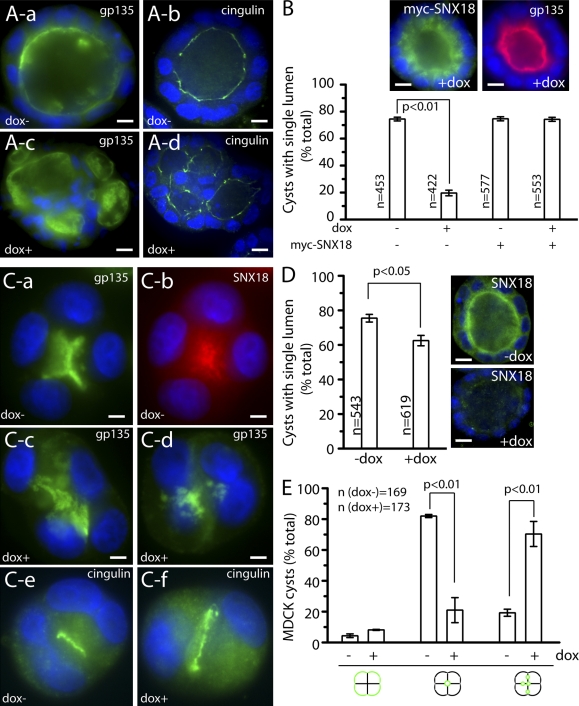
Because SNX18 appears to regulate endocytic transport of specialized cargo, we speculated that in polarized cells it may mediate apical protein targeting during lumen formation. To that end, a tet-inducible SNX18 shRNA-expressing MDCK cell line (MDCK-shSNX18) was created. As shown in Fig. S5 (A–C), after incubation with dox for 96 h, this cell line knocks down SNX18 by >85%, while having no effect on SNX9 levels. Just as with the FIP5 shRNA-expressing cell line, filter-grown SNX18-depleted cells can still form a polarized monolayer with intact tight junctions, as observed by anti-Occludin staining and an unchanged trans-epithelial electrical resistance (Fig. S5 D). Furthermore, consistent with our data using HeLa cells, the knockdown of SNX18 had no significant effect on the rate or fidelity of the Tf receptor’s recycling in MDCK cells (Fig. S5 E). To determine whether SNX18 is required for proper epithelial lumen morphogenesis, MDCK-shSNX18 cells were grown in 3D cultures and allowed to develop into mature cysts. After 9 d, ∼70% of untreated MDCK-shSNX18 cells formed normal cysts with a single lumen (Fig. 8, A and B), whereas ∼75% of SNX18 knockdown cysts were disorganized, mispolarized in areas, and formed multiple lumens (Fig. 8, A and B). This phenotype can be rescued by expressing shRNA-resistant myc-SNX18 (Fig. 8 B). To test whether SNX18 is also required for the early stages of lumen formation, we imaged 3D cultures 12 h after plating to determine whether SNX18 knockdown has any effect on the initiation of lumen formation. Although ∼80% of control cysts formed a single lumen (Fig. 8, C and E), ∼75% of SNX18-depleted cells formed cysts with multiple lumens, with only ∼25% of knockdown cysts forming a single lumen (Fig. 8, C and E). If dox was added after the initial formation of the apical lumen, SNX18 knockdown had a much smaller, although statistically significant, effect on the expansion and maintenance of single lumens (Fig. 8 D). These data collectively imply that SNX18, like FIP5, is involved in the establishment of a single apical lumen during the formation of epithelial cysts.
Figure 8.
SNX18 is required for the establishment of the apical lumen at the early stages of epithelial cyst formation. (A and B) MDCK-shSNX18 cells were grown for 9 d in the presence (A-c and A-d) or absence (A-a and A-b) of 1 µg/ml of dox. Cells were then fixed with 4% paraformaldehyde and stained with anti-gp135 (A-a and A-c) or anti-cingulin (A-b and A-d) antibodies. B shows the quantitation of epithelial cysts with a single lumen. Data shown are the means and standard deviations derived from three independent experiments (error bars). n is the number of cysts analyzed. Insets show dox+ cyst expressing myc-SNX18. (C–E) MDCK-shSNX18 cells were grown for 74 h in the presence (C-c, C-d, and C-f) or absence (C-a, C-b, and C-e) of 1 µg/ml of dox to pre-knockdown SNX18. Cells were then seeded in 3D cultures and grown for 24 h. Cells were fixed with 4% paraformaldehyde and stained with anti-gp135, anti-cingulin, or anti-SNX18 antibodies. E shows the quantitation of 3D cyst polarization at the two and four cell stages of the experiment shown in C. Data shown are the means and standard deviations derived from three independent experiments. n is the number of cysts analyzed. (D) The quantitation of fully matured (9 d) epithelial cysts with a single lumen in cells incubated with or without 1 µg/ml of dox added after 24 h in 3D cultures. Data shown are the means and standard deviations derived from three independent experiments (error bars). n is the number of cysts analyzed. Insets show the extent of SNX18 (green) knockdown in the presence of dox. Bars: (A) 8 µm; (B) 16 µm; (C) 3 µm; (D) 16 µm.
Discussion
Polarized membrane trafficking is critical for the morphogenesis of the apical lumen in epithelial cells (Lubarsky and Krasnow, 2003; Martin-Belmonte and Mostov, 2008). Rab11 GTPase has been shown to regulate endocytic recycling in the polarized epithelia and is thought to be involved in apical-directed transport (for review see Prekeris, 2003). Recent studies have also implicated Rab11 in mediating the proper establishment and expansion of the apical lumen (Desclozeaux et al., 2008). Rab11 functions by interacting with and recruiting several effector proteins (Prekeris, 2003; Tarbutton et al., 2005). Here we demonstrate that FIP5, a known Rab11 effector protein (Prekeris et al., 2000), is required for the initial formation of the apical lumen by regulating the formation and scission of gp135-containing transport vesicles from apical recycling endosomes. We also show that FIP5 functions by binding and activating SNX18, a BAR domain–containing protein involved in membrane remodeling. This work, in combination with a recently published study from Bryant et al. (2010), demonstrates that the Rab11 GTPase mediates epithelial tissue morphogenesis by sequentially regulating the generation, targeting, and fusion of gp135-containing endosomes to the AMIS during the initiation of lumen formation.
Rab11 forms mutually exclusive complexes with various FIPs (Meyers and Prekeris, 2002), a process thought to allow Rab11 to function in many distinct endocytic recycling pathways. Because FIP5 is an apically localized protein that is required for transcytosis to the apical PM, we hypothesized that FIP5 may regulate gp135 transport during the formation of the apical lumen. Indeed, the knockdown of FIP5 results in the formation of multiple lumens, while having no effect on the fidelity or kinetics of the basolateral transport of the TfR. The formation of multilumen cysts is an indication of defects in either the initiation or expansion/maintenance of the apical lumen. The generation of tet-inducible shRNA cell lines allows us to test whether FIP5 is required during or after lumen formation. Interestingly, although the depletion of FIP5 during the formation of the apical lumen resulted in multilumenal epithelial cysts, FIP5 knockdown after the initiation of the lumen had little effect on epithelial cyst organization. These data suggest that FIP5 may only play a minor role in the expansion and maintenance of the apical lumen and are mainly required for the apical lumen initiation.
Our proteomics data identified SNX18 as a putative FIP5-interacting protein, a finding that is further supported by our pull-down and ITC assays. Because SNX9/18/33 family member proteins contain a BAR domain and are implicated in transport carrier formation/scission, this interaction suggests a potential role for SNX18 and FIP5 in the formation of endocytic carriers on apical recycling endosomes. It was previously suggested that in nonpolarized cells, SNX18 is localized to and functions in endocytic organelles (Håberg et al., 2008), although this suggestion remains controversial. Consistent with this hypothesis, we show that SNX18 is capable of liposome tubulation in vitro. Pre-incubation of SNX18 with FIP5 results in a further increase in liposome tubulation, arguing for a role of FIP5 in the activation of SNX18. Indeed, it was previously shown that the overexpression of full-length SNX18 and SNX9 results in less tubulating activity as compared with the overexpression of only the PX-BAR domains (Håberg et al., 2008). Recent studies also suggested that the SH3 domain of SNX9 interacts with the SNX9-BAR domain in vitro (Wang et al., 2008b). Thus, it is likely that SNX18 is autoinhibited by the SNX18-SH3 domain binding to the SNX18-BAR domain. Afterwards, the binding of FIP5 to the LC region of SNX18 likely results in the opening and activation of SNX18. Consistent with the involvement of FIP5 in endocytic carrier scission, overexpression of the FIP5 dominant-negative mutant results in the tubulation of endosomes and the formation of endocytic beads-on-a-string structures. Because inhibition of FIP5 would prevent the activation of endocytic SNX18, this beads-on-a-string phenotype is likely caused by the failure of the scission of endocytic carriers from the recycling endosomes. Indeed, most of the gold labeling for the FIP5 mutant was localized to the necks of the beads and buds, which are the likely scission sites of the forming endocytic carriers.
The physical and functional association between FIP5 and SNX18 brings up the possibility that SNX18 and FIP5 function together during epithelial lumen morphogenesis. Consistent with this, knockdown of SNX18 leads to a multilumen phenotype. Furthermore, SNX18 is also predominantly required during the early stages of lumen formation, and its knockdown leads to a phenotype similar to the one resulting from FIP5 knockdown. Collectively, our data suggest that FIP5 binding to SNX18 induces SNX18-BAR domain-dependent membrane bending, and that SNX18 activation is required for the formation and/or scission of gp135-containing transport vesicles at apical recycling endosomes.
Surprisingly, our data show that in vitro, FIP5 also interacts with SNX9 with an affinity that is similar to SNX18 binding. Furthermore, liposome assays show that FIP5 binding also induces SNX9-dependent membrane tubulation. Although we cannot rule out the possibility that FIP5 interacts with SNX9 in vivo, it is not likely that this interaction plays any role in mediating protein transport from apical recycling endosomes to the apical PM. Multiple studies have shown that SNX9 localizes to the PM, where it regulates protein endocytosis. In contrast, FIP5 is associated with apical recycling endosomes (Fig. 1; Prekeris et al., 2000). Furthermore, although SNX9 is known to regulate Tf endocytosis, FIP5 knockdown has no effect on the rate of Tf uptake at the PM (Schonteich et al., 2008). It is likely that within the cell, differences in the spatial distribution between SNX9 and SNX18 ensure that FIP5 binds only to SNX18. Indeed, FIP5 and SNX18 interact only with micromolar affinity, which is too low for the recruitment of SNX18 to membranes in a FIP5-dependent manner. Thus, lipid binding–mediated recruitment of SNX18 at the recycling endosomes would be required to allow interaction with FIP5. Consistent with this, FIP5 knockdown did not affect SNX18’s localization to endosomes, and binding to FIP5 had no effect on the efficiency of SNX18 binding to liposomes in vitro. What remains unclear is how the specificity of SNX18’s recruitment to endosomes is regulated, as SNX18’s PX domain does not differentiate between different phosphatidylinositides.
In addition to FIPs, Rab11 binds several other effector proteins. Some of them, such as the Exocyst complex and Rabin8, were also implicated in mediating Rab11-dependent targeting and fusion of gp135-containing endosomes with the apical PM (Bryant et al., 2010). What remains unclear is how Rab11 interactions with FIPs, Rabin8, and the Exocyst complexes are regulated. Structural studies of FIPs and Sec15 show that they all bind Rab11 switch motifs (Wu et al., 2005; Eathiraj et al., 2006; Jagoe et al., 2006). Furthermore, because Rabin8 binds Rab11 in a GTP-dependent manner, it also likely interacts via the switch motif of Rab11 GTPase (Knödler et al., 2010). As a result, Rab11 should not be able to bind to Sec15, Rabin8, and FIP5 simultaneously. How then does Rab11 mediate the formation and targeting of gp135-containing endosomes? The likely scenario is that Rab11 sequentially binds to FIP5, Rabin8, and the Exocyst complex in a “hand-me-down” manner. Consistent with this, FIP5 and the Exocyst mediate different steps of apical transport. Although Rab11 binding to Sec15 is required for gp135 targeting to the apical PM (Bryant et al., 2010), our data suggest that FIP5 and SNX18 mediate gp135-containing endosome formation/scission. Future studies will be needed to understand the mechanisms that regulate the sequential interaction of Rab11 GTPase with multiple effector proteins during apical transport.
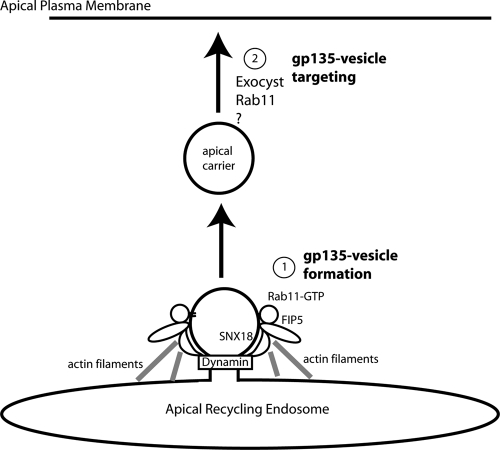
In summary, we propose that during lumen morphogenesis, apical proteins are internalized from the basolateral PM and are transported to apical recycling endosomes. There, apical proteins are sorted into specialized transport organelles that carry cargo to the site of the forming apical lumen. Rab11 plays a key role in regulating multiple steps of this transport pathway. First, the formation/scission of these carriers is mediated by the interaction between the Rab11 effector protein, FIP5, and its binding partner SNX18. This binding initiates the process that leads to the activation of SNX18 and the induction of endocytic membrane bending (Fig. 9). Recent studies have also implicated Arp2/3 (Shin et al., 2008) and N-WASP as SNX9 binding proteins that mediate endocytosis. Interestingly, SNX18 shares SNX9’s conserved Arp2/3 binding “A-like” domain, as well as its N-WASP binding domain, potentially allowing SNX18 to act as a site of interaction with the actin cytoskeleton. Thus, it is possible that SNX18 activation by FIP5 also leads to an increase in Arp2/3-dependent actin polymerization and scission, although this remains to be demonstrated (Fig. 9). Newly formed transport organelles are then targeted to the apical PM, possibly via Rab11 binding to the Exocyst complexes (Fig. 9; Bryant et al., 2010). Recent studies have shown that Rab11-dependent apical transport also results in the activation of Cdc42 and the formation of the Par3–Par6 complex (McCaffrey and Macara, 2009; Bryant et al., 2010), both known regulators of cortical epithelial polarity. Additionally, FIP5 is known to bind Kinesin II (Schonteich et al., 2008), providing a link to a potential mechanism for the transport and targeting of FIP5-positive vesicles. Although the full picture of the interactions involved in lumen morphogenesis has yet to be completed, emerging data suggest that Rab11 and its binding proteins play a key role in integrating apical transport and epithelial PM polarization during tissue morphogenesis.
Figure 9.
Proposed model of the roles of FIP5 and SNX18 in apical lumen formation and endosomal scission.
Materials and methods
Antibodies
Rabbit polyclonal anti-SNX18 antibodies were prepared as described previously (Prekeris et al., 2000) using recombinant purified human full-length SNX18. Antibodies were affinity purified using recombinant SNX18 conjugated to Affigel (Bio-Rad Laboratories) and eluted with 0.1 M glycine buffer, pH 2.5. Rabbit anti-FIP5, anti-FIP1, and anti-FIP3 polyclonal antibodies have been described previously (Prekeris et al., 2000; Peden et al., 2004; Wilson et al., 2005). Mouse monoclonal anti-TfR, anti-Ezrin, and anti–e-Cadherin antibodies were obtained from BD. Rabbit anti-GFP was obtained from Invitrogen. 10 nm gold-conjugated anti–rabbit IgG was purchased from British BioCell International. Mouse monoclonal anti-Occludin antibody was obtained from Invitrogen. Mouse monoclonal anti-gp135 antibody was a gift from C. Yeaman (University of Iowa, Iowa City, IA) and G. Ojakian (State University of New York Downstate Medical Center, Brooklyn, NY). Rabbit polyclonal anti-cingulin antibody was provided by S. Citi (University of Geneva, Geneva, Switzerland). Anti-endolyn antibodies were a gift from O. Weisz (University of Maryland, College Park, MD). Fluorescein-labeled anti–rabbit IgG antibody, Texas red–labeled anti-mouse IgG antibody, and goat anti–mouse AffiniPure F(ab’)2 fragments were obtained from Jackson ImmunoResearch Laboratories, Inc. Cell-permeant Hoechst DNA stain and Tf conjugated to Alexa Fluor 594 were obtained from Invitrogen.
Expression constructs and protein purification
SNX18, all SNX18 truncation mutants, and SNX9 (provided by D. Yarar, Whitehead Institute, Massachusetts Institute of Technology, Cambridge, MA) were expressed as GST fusion proteins using the pGEX-KG plasmid (GE Healthcare). GST fusion constructs were expressed and purified using the BL21-(FE3)-RIPL Escherichia coli strain as described previously (Junutula et al., 2004). In brief, E. coli were lysed using a French press and then incubated with glutathione agarose beads (Sigma-Aldrich). Beads were then washed and either eluted with 25 mM glutathione or cleaved with thrombin (GE Healthcare).
Full-length human FIP3 and FIP5 were tagged with an N-terminal 6His tag followed by a tobacco etch virus cleavage site by subcloning into the baculovirus transfer vector, pVL1392. Cotransfection and amplification of recombinant baculovirus was conducted using BacPAK transfection reagents (BD) according to the manufacturer’s instructions. In brief, 106 Sf9 cells were seeded into a 6-well plate, and the Bacfectin–DNA mixture was added drop-wise. After 5 d, the P1 viral stock was harvested and further amplified to P2 and P3 stages. For protein production, 1L of Sf9 cells at 2 million cells/ml were infected with 2 ml of P3 viral stock (approximate MOI of 0.5) and harvested after 65 h. Cells were lysed in 50 mM Tris buffer, pH 7.5, containing 300 mM NaCl, and the cleared lysate was loaded onto a Ni-NTA column. Eluted 6His-FIP3 was dialyzed overnight against buffer (50 mM Tris, pH 7.5, 300 mM NaCl, and 5 mM BME) and frozen in liquid nitrogen. Yields were typically 3–5 mg/liter with an estimated purity of >75%.
Generation and purification of adenovirus expression constructs
shRNA-resistant FIP5-GFP and myc-SNX18 adenoviral constructs and recombinant adenovirus were generated using the AdEasy system (He et al., 1998). In brief, each gene was cloned into pShuttle-CMV, and the resultant clones were linearized with PmeI and used to transform E. coli BJ5183 cells carrying the viral DNA plasmid pAdEasy-1. Recombinant plasmids were digested with PacI to expose the inverted terminal repeats, and 8 µg of each construct was used to transfect, by calcium phosphate coprecipitation, 6-cm dishes of 50% confluent HEK 293 cells modified to express adenovirus preterminal protein, DNA polymerase, and DNA-binding protein. The medium was aspirated and replaced after 24 h, and the cells were incubated for 10 d, until many plaques had formed. Virus was released by repeated freeze/thaw cycles and amplified by the addition of the adenoviral vector to fifty 10 cm dishes of HEK 293 cells, which were incubated for 48 h. Virus harvesting and purification were conducted as described previously (Orlicky and Schaack, 2001). In brief, the cells were harvested by centrifugation, and the virus was released by three freeze/thaw cycles followed by centrifugation to pellet the cell debris. Two rounds of virus back-extraction were performed on the cell pellet. The supernatants were combined and purified via centrifugation on a cesium chloride step gradient of 1 ml of 1.4 g/c2 and 2 ml of 1.25 g/c2 CsCl in PBS using an SW41 rotor centrifuged at 36,000 rpm. The virus banded at the interface of the CsCl steps, and was collected by side puncture with a syringe. The virus was next mixed with 1.35 g/c2 CsCl in PBS and centrifuged overnight at 65,000 rpm in an NVT100 rotor, and again collected by syringe side puncture. The resulting purified virion-containing solution was dialyzed four times for 2 h each at 4° against a modified previously published buffer containing 10 mM Tris, 10 mM His, 75 mM NaCl, 1 mM MgCl2, 100 µM EDTA, 0.5% vol/vol EtOH, pH 7.4, and 50% vol/vol glycerol (Evans et al., 2004).
Virus particle concentrations were determined by OD260 spectrophotometry, with one OD260 unit equal to 1012 particles. 30 plaque forming units/cell were used for each experiment, as this was the amount of virus determined to result in the production of exogenous FIP5 or SNX18 equal to the amount of endogenous protein in dox− cells (as determined by immunoblotting).
Glutathione bead pull-down assays
In glutathione bead pull-down assays, glutathione beads (50 µl) were coated with 10 µg of GST fusion protein or GST alone and incubated with varying amounts of soluble protein in a final volume of 0.5 ml of reaction buffer (50 mM Hepes, pH 7.4, 150 mM NaCl, 5 mM MgCl2, 0.1% Triton X-100, 0.1% bovine serum albumin, and 1 mM phenylmethylsulfonyl fluoride). Samples were incubated at 22°C for 1 h on a nutator with constant rotation. The samples were pelleted at 2,000 g for 3 min and washed three times with 1 ml of reaction buffer. Bound proteins were eluted with 1% SDS, analyzed by SDS-PAGE, and either stained with Coomassie blue or immunoblotted.
Liposome preparation and tubulation analysis
All lipids were porcine brain lipid extract purchased from Avanti Polar Lipids, Inc. Liposomes were made from 90% PC, 5% PS, and 5% PI(4,5)P2. Lipids were mixed, dried under a nitrogen stream, and lyophilized in vacuum for 2 h. Dried lipids were rehydrated in a solution of 25 mM Hepes, pH 7.5, 25 mM KCl, 2.5 mM magnesium acetate, and 150 mM potassium glutamate at room temperature for 1 h, after which the liposomes were sonicated for 30 min. GST, GST-SNX9, 6His-FIP3, GST-SNX18, or 6His-FIP5 proteins were incubated with liposomes for 30 min at 37°C. Samples were immediately processed and analyzed via transmission electron microscopy. For electron microscopy analysis, samples were prepared using a modified previously published procedure (Farsad et al., 2001). In brief, 10 µl aliquots of liposome/protein incubations were adsorbed for 5 min onto 250 µM hexagonal mesh copper grids (Electron Microscopy Sciences) and coated with 0.5% formvar solution in ethylene dichloride (Electron Microscopy Sciences) and carbon coated using an Auto 306 Vacuum Coater (Edwards). Grids were stained with 1–2% uranyl acetate, blotted, washed with water, allowed to air dry, and imaged using a Techna G2 12 transmission electron microscope (FEI Company) at 80 kV with Digital Micrograph 3.7.1 GMS1.2 imaging software (Gatan, Inc.).
ITC
ITC experiments were performed using a VP-ITC calorimeter (Microcal LLC) as recommended by the manufacturer. GST, GST-SNX18, or GST-SNX9 were loaded in the sample cell (in PBS) and titrated with either 6His-FIP5 or 6His-FIP3 proteins in the same buffer (10 µl injections up to a total of 30 injections). The titrations were performed while samples were stirred at 300 rpm at 25°C or at the indicated temperature. An interval of 4 min was allowed between each injection for the baseline to stabilize. The blank ITC titration was performed against buffer by injecting 6His-FIP5. The blank subtraction was done for all data used for analysis. The data were fitted via the one-set-of-sites model to calculate the binding constant (K) using Origin software (Microcal, LLC).
Cell culture and immunofluorescence microscopy
HeLa cells and MDCK-MIIR cells stably expressing human TfR and pIgA-R were cultured in DME with 4.5 g/liter glucose, 5.84 g/liter l-glutamine, and 10% heat-inactivated tet-free FBS (Takara Bio Inc.), and supplemented with 100 IU/ml penicillin and 100 µg/ml streptomycin.
For 2D filter cell culture assays, MDCK-MIIR cells were plated on collagen-coated 0.4 µm pore-size Transwell filters (Corning) and allowed to polarize for 4 d. Cells were then fixed with 4% paraformaldehyde for 15 min and permeabilized for 10 min in PBS containing 0.4% saponin, and nonspecific sites were blocked with PBS containing 0.2% BSA and 1% FBS. Cells were incubated with specific antibodies, washed in PBS, and mounted in VectaShield (Vector Laboratories). Cells were imaged with an inverted Axiovert 200M deconvolution microscope (Carl Zeiss) with a 63× oil immersion lens and QE charge-coupled device camera (Sensicam). Image processing was performed using 3D rendering and exploration software (Intelligent Imaging Innovations).
3D cell culture assays for cyst formation were conducted according to a previously described method (Vieira et al., 2006). In brief, actively dividing cells were mixed with growth factor reduced Matrigel (BD) and plated in 12-µl drops on 8-well slides. The Matrigel–cell mixture was allowed to harden for 30 min at 37°C, and 400 µl of medium was added. The cells were incubated for the indicated period of time and the media was changed every other day. 3D cell cultures were stained according to a modified previously published protocol (Debnath et al., 2003). In brief, 3D cultures were fixed with 3% paraformaldehyde for 20 min, permeabilized with PBS and 0.5% Triton X-100 for 10 min, and quenched three times for 15 min each wash with a glycine/PBS solution (130 mM NaCl, 7 mM Na2HPO4, 3.5 mM NaH2PO4, and 100 mM glycine). Cells were incubated in primary block (10% FBS, 130 mM NaCl, 7 mM Na2HPO4, 3.5 mM NaH2PO4, 7.7 mM NaN3, 0.1% BSA, 0.2% Triton X-100, and 0.05% Tween-20) for 4 h, followed by incubation in secondary block (primary block with 20 µg/ml goat anti–mouse F(ab’)2 fragments) for 1 h. After washing, cells were left overnight in primary block with primary antibody and Hoerscht nuclear stain. Cells were then washed and incubated for 1 h with secondary antibody in primary block. Cells were washed, dried for 1 h, and mounted with VectaShield.
RNA interference
In HeLa cells, SNX18 was depleted with siRNA designed based on human SNX18 sequences (SNX18-1, 5′-AATGTCAGACAGACGCGAAAA-3′; SNX18-2, 5′-AAGCACCTGACCTATGAGAAC-3′). siRNAs were cotransfected into HeLa cells using Lipofectamine2000 (Invitrogen). Transfected cells were incubated for either 48 or 74 h and analyzed for SNX18 expression by Western blotting. Remaining cells were used for flow cytometry studies.
To create tet-inducible MDCK shRNA cells lines, SNX18 and FIP5 shRNAs were designed using canine sequences (FIP5, 5′-GATGAAGGGCAAGAAGAAG-3′; SNX18, 5′-ATCACAGATGCAGCATTC-3′). shRNAs were then cloned into the pHUSH retroviral expression vector (provided by D. Davis, Genentech, South San Francisco, CA). Stable, clonal cell lines were then selected using 1 µg/ml of puromycin and grown using regular MDCK media supplemented with tet-free FBS (Takara Bio Inc.). To knock down either SNX18 or FIP5, stable cells lines were incubated in the presence of 1 µg/ml of dox for 72 h.
Analysis of Tf-biotin uptake and recycling in MDCK cells
MDCK cells expressing the human Tf receptor were grown on collagen-coated 10 cm filters with a 0.4 µm pore size (10 mm filters; Corning). 50 µg/ml of biotin-labeled human Tf (Sigma-Aldrich) was added to the basolateral chamber and allowed to internalize for 1 h at 37°C. The filters were then washed three times and the medium was replaced with serum-supplemented medium containing 50 µg/ml unlabeled human Tf, and cells were incubated for either 0 or 60 min at 37°C. The medium was collected at these time points and incubated for 30 min at room temperature with streptavidin-conjugated agarose resin (Thermo Fisher Scientific). The beads were washed five times with PBS, and the biotin-Tf was eluted by heating to 90°C for 10 min in the presence of 1% SDS. Samples were separated by SDS-PAGE, followed by Western blot analysis using streptavidin conjugated to IRDye800 (LI-COR Biosciences). Immunoblots were scanned and quantified using the Li-COR Odyssey Infrared Imager.
Analysis of Tf uptake in HeLa cells
For uptake assays, mock or SNX18 siRNA-treated HeLa cells were incubated with 20 µg/ml Tf-Alexa647 at 37°C for the indicated period of time. Cells were immediately pelleted, washed, and fixed for 20 min on ice in PBS with 4% paraformaldehyde. Cells were sedimented and resuspended in PBS, and the amount of internalized Tf was determined by flow cytometric analysis. Flow cytometry analyses were performed using a Cytomics FC 500 flow cytometer (Beckman Coulter) equipped with 488-nm and 647-nm lasers.
Immunoprecipitation and proteomics
HeLa cell lysates from twenty 10-cm plates were harvested in the presence of 1% Triton X-100. Cell lysates were then incubated overnight with either 50 µg/ml lysate of affinity-purified rabbit anti-FIP5 antibody or 50 µg/ml lysate of purified nonspecific rabbit IgG, followed by the addition of 100 µl of protein A–Sepharose per 1 ml of lysate. Beads were then washed, eluted with 1% SDS, and separated on a 7–14% gradient acrylamide gel. The resulting gel was Coomassie stained and bands present only in the anti-FIP5 immunoprecipitate were isolated. Gel bands were cut into three equal sections and in-gel digested using three proteases. Peptides were extracted from the gel pieces with 0.1% trifluoroacetic acid/60% acetonitrile and lyophilized. Dried peptide samples from each gel band were rehydrated and loaded onto a microcapillary column (100 µm–inner diameter fused silica) packed with 15 cm Aqua C18 reverse-phase material, then placed in-line with an LTQ linear trap mass spectrometer. Peptides were eluted with a 2 h mobile gradient of acetonitrile/0.1% formic acid. Tandem mass spectra were analyzed via Sequest using a human/mouse/rat database concatenated to a randomized human/mouse/rat database. DTASelect was used to reassemble identified peptides into proteins. Identified proteins were filtered at <5% false discovery rate.
Lipid binding assays
In lipid binding assays, PC/PS (90%:5% at a final concentration of 0.5 mg/ml) liposomes with or without phosphatidylinositides (at 5%) were incubated for 30 min at room temperature with 5 µM of recombinant purified SNX18 or 6His-FIP5. Liposomes were then sedimented by centrifugation for 60 min at 50,000 rpm in a TLA100 rotor. Liposomes were resuspended in PBS with 1% SDS. Samples were then separated on SDS/PAGE gels, and the amount of bound protein was determined by Coomassie staining.
Electron microscopy analysis
For analysis of Tf localization, HeLa cells were transfected with or without GFP-FIP5-F1 and incubated with Tf-HRP 1:40 in L15 for 45 min at 37°C, 48 h after transfection. The cells were then fixed with paraformaldehyde and glutaraldehyde before performing the DAB reaction, dehydration, and embedding, essentially as described by Stinchcombe et al. (1995). For immunolabeling, HeLa cells previously transfected with or without GFP-FIP5-F1 (24 h) were loaded with Tf-HRP 1:40 in L15 for 45 min at 37°C, then chilled on ice. DAB labeling was performed using 3% DAB (TAAB) and 0.003% H2O2 (Sigma-Aldrich) in Tris/HCl 0.05 M, pH 7.6, for 30 min in the dark, followed by digitonin permeabilization, paraformaldehyde fixation, and incubation with rabbit anti-GFP antibody (1:1500) and 10 nm gold-conjugated anti–rabbit IgG antibody, essentially as described by Futter et al. (1998). Ultrathin sections (60-70 nm thick) were cut using a microtome (Ultracut E; Reichert-Jung), stained with lead citrate, and viewed using a CM12 transmission electron microscope (Phillips).
Online supplemental material
Figs. S1 and S5 show data characterizing MDCK cells expressing tet-inducible FIP5 shRNA (Fig. S1) and SNX18 shRNA (Fig. S5). Fig. S2 shows the images characterizing early stages (2–4 cells) of epithelial lumen formation. Fig. S3 contains additional EM images of cells expressing FIP5 siRNA, as well as characterization of new anti-SNX18 and anti-SNX9 antibodies. Fig. S4 shows subcellular localization of endogenous SNX18 as well as GFP-tagged SNX18 and SNX9 in HeLa and MDCK cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201011112/DC1.
Supplementary Material
Acknowledgments
We thank Dr. Brian Doctor (University of Colorado Denver) for critical reading of the manuscript. We are appreciative of Dr. Charles Yeaman and Dr. George Ojakian for anti-gp135 antibodies. We are also grateful to Dr. Defne Yarar for SNX9 and SNX18 cDNAs, and Dr. David Davis for the pHUSH expression vector. Finally, we thank Dr. O. Weisz for anti-endolyn antibodies and Dr. Sandra Citi for anti-cingulin antibodies.
This work was supported by a grant from the National Institutes of Health-NIDDK (DK064380 to R. Prekeris).
Footnotes
Abbreviations used in this paper:
- AMIS
- apical membrane initiation site
- dox
- doxycycline
- FIP
- Rab11 family interacting protein
- LC
- low complexity
- PC
- 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine
- PI
- phosphatidylinositide
- PI(4,5)P2
- l-α-phosphatidyinositol-4,5-bisphosphate
- pIgA-R
- polymeric IgA receptor
- PM
- plasma membrane
- PS
- 1,2-dipalmitoyl-glycero-3-phosphoserine
- shRNA
- short hairpin RNA
- SNX
- sorting nexin
- tet
- tetracycline
- Tf
- transferrin
- TfR
- transferrin receptor
References
- Bryant D.M., Datta A., Rodríguez-Fraticelli A.E., Peränen J., Martín-Belmonte F., Mostov K.E. 2010. A molecular network for de novo generation of the apical surface and lumen. Nat. Cell Biol. 12:1035–1045 10.1038/ncb2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis G.E., Bayless K.J. 2003. An integrin and Rho GTPase-dependent pinocytic vacuole mechanism controls capillary lumen formation in collagen and fibrin matrices. Microcirculation. 10:27–44 [DOI] [PubMed] [Google Scholar]
- Debnath J., Muthuswamy S.K., Brugge J.S. 2003. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 30:256–268 10.1016/S1046-2023(03)00032-X [DOI] [PubMed] [Google Scholar]
- Desclozeaux M., Venturato J., Wylie F.G., Kay J.G., Joseph S.R., Le H.T., Stow J.L. 2008. Active Rab11 and functional recycling endosome are required for E-cadherin trafficking and lumen formation during epithelial morphogenesis. Am. J. Physiol. Cell Physiol. 295:C545–C556 10.1152/ajpcell.00097.2008 [DOI] [PubMed] [Google Scholar]
- Di Paolo G., De Camilli P. 2006. Phosphoinositides in cell regulation and membrane dynamics. Nature. 443:651–657 10.1038/nature05185 [DOI] [PubMed] [Google Scholar]
- Eathiraj S., Mishra A., Prekeris R., Lambright D.G. 2006. Structural basis for Rab11-mediated recruitment of FIP3 to recycling endosomes. J. Mol. Biol. 364:121–135 10.1016/j.jmb.2006.08.064 [DOI] [PubMed] [Google Scholar]
- Evans R.K., Nawrocki D.K., Isopi L.A., Williams D.M., Casimiro D.R., Chin S., Chen M., Zhu D.M., Shiver J.W., Volkin D.B. 2004. Development of stable liquid formulations for adenovirus-based vaccines. J. Pharm. Sci. 93:2458–2475 10.1002/jps.20157 [DOI] [PubMed] [Google Scholar]
- Farsad K., Ringstad N., Takei K., Floyd S.R., Rose K., De Camilli P. 2001. Generation of high curvature membranes mediated by direct endophilin bilayer interactions. J. Cell Biol. 155:193–200 10.1083/jcb.200107075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futter C.E., Gibson A., Allchin E.H., Maxwell S., Ruddock L.J., Odorizzi G., Domingo D., Trowbridge I.S., Hopkins C.R. 1998. In polarized MDCK cells basolateral vesicles arise from clathrin-γ-adaptin-coated domains on endosomal tubules. J. Cell Biol. 141:611–623 10.1083/jcb.141.3.611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray D.C., Hoeflich K.P., Peng L., Gu Z., Gogineni A., Murray L.J., Eby M., Kljavin N., Seshagiri S., Cole M.J., Davis D.P. 2007. pHUSH: a single vector system for conditional gene expression. BMC Biotechnol. 7:61 10.1186/1472-6750-7-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hales C.M., Griner R., Hobdy-Henderson K.C., Dorn M.C., Hardy D., Kumar R., Navarre J., Chan E.K., Lapierre L.A., Goldenring J.R. 2001. Identification and characterization of a family of Rab11-interacting proteins. J. Biol. Chem. 276:39067–39075 10.1074/jbc.M104831200 [DOI] [PubMed] [Google Scholar]
- Håberg K., Lundmark R., Carlsson S.R. 2008. SNX18 is an SNX9 paralog that acts as a membrane tubulator in AP-1-positive endosomal trafficking. J. Cell Sci. 121:1495–1505 10.1242/jcs.028530 [DOI] [PubMed] [Google Scholar]
- He T.C., Zhou S., da Costa L.T., Yu J., Kinzler K.W., Vogelstein B. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA. 95:2509–2514 10.1073/pnas.95.5.2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagoe W.N., Lindsay A.J., Read R.J., McCoy A.J., McCaffrey M.W., Khan A.R. 2006. Crystal structure of rab11 in complex with rab11 family interacting protein 2. Structure. 14:1273–1283 10.1016/j.str.2006.06.010 [DOI] [PubMed] [Google Scholar]
- Junutula J.R., Schonteich E., Wilson G.M., Peden A.A., Scheller R.H., Prekeris R. 2004. Molecular characterization of Rab11 interactions with members of the family of Rab11-interacting proteins. J. Biol. Chem. 279:33430–33437 10.1074/jbc.M404633200 [DOI] [PubMed] [Google Scholar]
- Kamei M., Saunders W.B., Bayless K.J., Dye L., Davis G.E., Weinstein B.M. 2006. Endothelial tubes assemble from intracellular vacuoles in vivo. Nature. 442:453–456 10.1038/nature04923 [DOI] [PubMed] [Google Scholar]
- Knödler A., Feng S., Zhang J., Zhang X., Das A., Peränen J., Guo W. 2010. Coordination of Rab8 and Rab11 in primary ciliogenesis. Proc. Natl. Acad. Sci. USA. 107:6346–6351 10.1073/pnas.1002401107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay A.J., McCaffrey M.W. 2004. The C2 domains of the class I Rab11 family of interacting proteins target recycling vesicles to the plasma membrane. J. Cell Sci. 117:4365–4375 10.1242/jcs.01280 [DOI] [PubMed] [Google Scholar]
- López-Novoa J.M., Nieto M.A. 2009. Inflammation and EMT: an alliance towards organ fibrosis and cancer progression. EMBO Mol Med. 1:303–314 10.1002/emmm.200900043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubarsky B., Krasnow M.A. 2003. Tube morphogenesis: making and shaping biological tubes. Cell. 112:19–28 10.1016/S0092-8674(02)01283-7 [DOI] [PubMed] [Google Scholar]
- Martin-Belmonte F., Mostov K. 2008. Regulation of cell polarity during epithelial morphogenesis. Curr. Opin. Cell Biol. 20:227–234 10.1016/j.ceb.2008.01.001 [DOI] [PubMed] [Google Scholar]
- McCaffrey L.M., Macara I.G. 2009. The Par3/aPKC interaction is essential for end bud remodeling and progenitor differentiation during mammary gland morphogenesis. Genes Dev. 23:1450–1460 10.1101/gad.1795909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers J.M., Prekeris R. 2002. Formation of mutually exclusive Rab11 complexes with members of the family of Rab11-interacting proteins regulates Rab11 endocytic targeting and function. J. Biol. Chem. 277:49003–49010 10.1074/jbc.M205728200 [DOI] [PubMed] [Google Scholar]
- Orlicky D.J., Schaack J. 2001. Adenovirus transduction of 3T3-L1 cells. J. Lipid Res. 42:460–466 [PubMed] [Google Scholar]
- Park J., Kim Y., Lee S., Park J.J., Park Z.Y., Sun W., Kim H., Chang S. 2010. SNX18 shares a redundant role with SNX9 and modulates endocytic trafficking at the plasma membrane. J. Cell Sci. 123:1742–1750 10.1242/jcs.064170 [DOI] [PubMed] [Google Scholar]
- Peden A.A., Schonteich E., Chun J., Junutula J.R., Scheller R.H., Prekeris R. 2004. The RCP-Rab11 complex regulates endocytic protein sorting. Mol. Biol. Cell. 15:3530–3541 10.1091/mbc.E03-12-0918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prekeris R. 2003. Rabs, Rips, FIPs, and endocytic membrane traffic. ScientificWorldJournal. 3:870–880 10.1100/tsw.2003.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prekeris R., Klumperman J., Scheller R.H. 2000. A Rab11/Rip11 protein complex regulates apical membrane trafficking via recycling endosomes. Mol. Cell. 6:1437–1448 10.1016/S1097-2765(00)00140-4 [DOI] [PubMed] [Google Scholar]
- Pylypenko O., Lundmark R., Rasmuson E., Carlsson S.R., Rak A. 2007. The PX-BAR membrane-remodeling unit of sorting nexin 9. EMBO J. 26:4788–4800 10.1038/sj.emboj.7601889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlüter M.A., Pfarr C.S., Pieczynski J., Whiteman E.L., Hurd T.W., Fan S., Liu C.J., Margolis B. 2009. Trafficking of Crumbs3 during cytokinesis is crucial for lumen formation. Mol. Biol. Cell. 20:4652–4663 10.1091/mbc.E09-02-0137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonteich E., Wilson G.M., Burden J., Hopkins C.R., Anderson K., Goldenring J.R., Prekeris R. 2008. The Rip11/Rab11-FIP5 and kinesin II complex regulates endocytic protein recycling. J. Cell Sci. 121:3824–3833 10.1242/jcs.032441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin N., Ahn N., Chang-Ileto B., Park J., Takei K., Ahn S.G., Kim S.A., Di Paolo G., Chang S. 2008. SNX9 regulates tubular invagination of the plasma membrane through interaction with actin cytoskeleton and dynamin 2. J. Cell Sci. 121:1252–1263 10.1242/jcs.016709 [DOI] [PubMed] [Google Scholar]
- Stinchcombe J.C., Nomoto H., Cutler D.F., Hopkins C.R. 1995. Anterograde and retrograde traffic between the rough endoplasmic reticulum and the Golgi complex. J. Cell Biol. 131:1387–1401 10.1083/jcb.131.6.1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strizzi L., Hardy K.M., Seftor E.A., Costa F.F., Kirschmann D.A., Seftor R.E., Postovit L.M., Hendrix M.J. 2009. Development and cancer: at the crossroads of Nodal and Notch signaling. Cancer Res. 69:7131–7134 10.1158/0008-5472.CAN-09-1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarbutton E., Peden A.A., Junutula J.R., Prekeris R. 2005. Class I FIPs, Rab11-binding proteins that regulate endocytic sorting and recycling. Methods Enzymol. 403:512–525 10.1016/S0076-6879(05)03045-4 [DOI] [PubMed] [Google Scholar]
- Townsend T.A., Wrana J.L., Davis G.E., Barnett J.V. 2008. Transforming growth factor-beta-stimulated endocardial cell transformation is dependent on Par6c regulation of RhoA. J. Biol. Chem. 283:13834–13841 10.1074/jbc.M710607200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega-Salas D.E., Salas P.J., Rodriguez-Boulan E. 1987. Modulation of the expression of an apical plasma membrane protein of Madin-Darby canine kidney epithelial cells: cell-cell interactions control the appearance of a novel intracellular storage compartment. J. Cell Biol. 104:1249–1259 10.1083/jcb.104.5.1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira O.V., Gaus K., Verkade P., Fullekrug J., Vaz W.L., Simons K. 2006. FAPP2, cilium formation, and compartmentalization of the apical membrane in polarized Madin-Darby canine kidney (MDCK) cells. Proc. Natl. Acad. Sci. USA. 103:18556–18561 10.1073/pnas.0608291103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Fang W.H., Krupinski J., Kumar S., Slevin M., Kumar P. 2008a. Pax genes in embryogenesis and oncogenesis. J. Cell. Mol. Med. 12:2281–2294 10.1111/j.1582-4934.2008.00427.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Kaan H.Y., Hooda R.N., Goh S.L., Sondermann H. 2008b. Structure and plasticity of endophilin and sorting nexin 9. Structure. 16:1574–1587 10.1016/j.str.2008.07.016 [DOI] [PubMed] [Google Scholar]
- Wilson G.M., Fielding A.B., Simon G.C., Yu X., Andrews P.D., Hames R.S., Frey A.M., Peden A.A., Gould G.W., Prekeris R. 2005. The FIP3-Rab11 protein complex regulates recycling endosome targeting to the cleavage furrow during late cytokinesis. Mol. Biol. Cell. 16:849–860 10.1091/mbc.E04-10-0927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Mehta S.Q., Pichaud F., Bellen H.J., Quiocho F.A. 2005. Sec15 interacts with Rab11 via a novel domain and affects Rab11 localization in vivo. Nat. Struct. Mol. Biol. 12:879–885 10.1038/nsmb987 [DOI] [PubMed] [Google Scholar]
- Zurzolo C., Polistina C., Saini M., Gentile R., Aloj L., Migliaccio G., Bonatti S., Nitsch L. 1992. Opposite polarity of virus budding and of viral envelope glycoprotein distribution in epithelial cells derived from different tissues. J. Cell Biol. 117:551–564 10.1083/jcb.117.3.551 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.