Abstract
Olfactory receptors, in addition to being involved in first step of the physiological processes that leads to olfaction, occupy an important place in mammalian genomes. ORs constitute super families in these genomes. Elucidating ol-factory receptor function at a molecular level can be aided by a computationally derived structure and an understanding of its interactions with odor molecules. Experimental functional analyses of olfactory receptors in conjunction with computational studies serve to validate findings and generate hypotheses. We present here a review of the research efforts in: creating computational models of olfactory receptors, identifying binding strategies for these receptors with odorant molecules, performing medium to long range simulation studies of odor ligands in the receptor binding region, and identifying amino acid positions within the receptor that are responsible for ligand-binding and olfactory receptor activation. Written as a primer and a teaching tool, this review will help researchers extend the methodologies described herein to other GPCRs.
Keywords: Olfactory receptors, modeling, molecular dynamic simulation, genomics, ORDB, HORDE
INTRODUCTION AND BACKGROUND
An olfactory receptor (OR) interacting with an odorant molecule constitutes the first of a series of steps leading to olfaction. As genomes have been published [1-5], starting with the initial draft of the human genome, efforts have been undertaken to identify their olfactory repertoires [6-9]. As these genomes are inevitably revised, these repertoires are likely to change [3]. Mining olfactory gene sequences from genomes includes performing sequence comparison studies between genes previously identified through experimental cloning studies and genes published in the genomes. Sequence motifs common to olfactory receptors, and presumably responsible for olfactory function (specifically, G-protein coupling or activation) are used to make these comparisons. In other cases, where genome wide sequence similarities exist (for example, the close similarity between the genomes of humans and chimpanzees), orthologs are identified and listed as putative olfactory receptors [10].
Olfactory Receptor Database (ORDB) (http://senselab.-med.yale.edu/ORDB), a resource maintained by the author under the auspices of the SenseLab project, is housed at the Yale University School of Medicine [11, 12]. It is a repository of genomic and proteomic information related to chemosensory receptors. ORDB represents the research efforts of more than 100 laboratories for more than 40 species that include mammals and other vertebrates, as well as invertebrates. In addition to ORs, information related to taste receptors, vomeronasal and pheromone receptors, insect olfactory receptors and fungal receptors are also stored in ORDB. At last count, information for more than 14,500 receptors is housed at ORDB. HORDE (Human Olfactory Receptor Data Exploratorium) database (http://bioportal.weizmann.ac.il/-HORDE/) [1, 13], at the Weizmann Institute of Science in Israel also contains comprehensive listings of olfactory receptors from mammalian genomes. ORs from human, chimpanzee, dog [6, 14], opossum and platypus [15] are currently listed at HORDE. HORDE provides for each receptor: information about its family and subfamily (according to the HORDE classification, adopted by HUGO [Human Genome Organization]) classification, other classifiers such as chromosome cluster, synteny, links to mouse, rat, chimpanzee orthologs, results of SNP (single nucleotide polymorphism) analysis and links to other OR-resources that house this gene.
Certain age-related neurodegenerative disorders are known to find their origins in olfactory dysfunction. Unlike eyesight and hearing, the sense of smell does not degenerate with age. At a young age, olfactory dysfunction might be a prognosticator for: Alzheimer's disease, [16] Parkinson's disease, [17] HIV, [18] motor neuron disease, [19] schizophrenia, [20] and anorexia [21]. A glimpse into OR function at a molecular level would go a long way towards identifying specific markers and diagnostic tests for these conditions.
Structurally and functionally, ORs belong to the family of membrane receptors known as GPCRs (GTP-binding Protein Coupled Receptors). These are ubiquitous proteins that traduce cell membranes, variably serving as a conduits from outside the cell to within or activating signal transduction (or other intracellular) processes [22-25]. ORs are embedded in the mucus membrane of the nasal cavity, in the cilia of olfac-tory neurons. Odor molecules when inhaled interact with, and activate, ORs. OR activation is possibly the result of a structural change [26]. The ensuing signal transduction steps are converted into electrical signals leading to an olfactory response.
Experimentally elucidating a structural change in an OR at the molecular level is currently not possible. No experimentally derived structure from nuclear magnetic resonance (NMR) or x-ray diffraction studies exists for ol-factory receptors. GPCRs—from the two structures that have been determined by x-ray diffraction methods, rhodopsin [27] and the recently published structure of beta-adrenergic receptor [28, 29]—are presumed to possess seven transmembrane (TM) helical domains, an extra cellular N-terminus, an intra-cellular C-terminus, three extracellular and three intracellular loops that connect TM helices. The rhodop sin structure [30] has thus far been used as the model structure for all computational models of GPCRs, including ORs. It is likely that future models of GPCRs will incorporate structural features of the beta adrenergic receptor.
Experimental research in identifying functionality in ORs involves associating receptors with the odors that activate them. To do this, large panels of odors are tested, often unsuccessfully. Interactions between ORs and odors are confounded: more than one odor might activate an OR; on the other hand, one odor might also be able to bind more than one receptor [31]. ORDB, which lists more than 14,500 chemosensory receptors, includes fewer than 50 odorant molecules, which have been experimentally shown to bind and activate olfactory receptors, work related to several of these odorants having been published in the a single paper [32].
Presented here is an overview of certain key aspects of the computational biology of olfactory receptors, namely: OR modeling, docking of odors, and simulation of ligands’ dynamic behavior, in the OR binding region. Bioinformatics efforts in identifying residues that are complicit in odorant-binding are also presented.
COMPUTATIONAL MODELING OF ORs
The following steps are involved in creating an OR model. Some variations in the details of the protocols exist, but the general principles are the same.
Identifying the Transmembrane Regions
Different methods have been used to identify the helical TM domains of an OR. They are identified through secondary structure prediction methods that use hydrophobicity profiles because of the hydropathic nature of helices [33]. Hidden Markov Models (HMM) [34] have also been used to identify TMs. There are several online resources that identify transmembrane helical domains in a protein; the only requirement is to enter the protein sequence into a text-box. TMHMM (TransMembrane [through] Hidden Markov Models) [35, 36] and HMMTOP (Hidden Markov Models [based] Topology) [37, 38] are two (of several) preferred [39] such programs. These resources provide information that not only identifies the helical regions, but also the location of the loops—extra-cellular or intracellular.
Creating the Transmembrane Scaffold
As mentioned in the Introduction, for want of an experimentally derived structure of ORs, modelers have used the structure of rhodopsins as a template structure. The first model of an OR, rat OR I7 was created from an electron diffraction structure of rhodopsin with a resolution of over 6 Å by Singer [33]. Singer created canonical helices and manually positioned these over the helical electron densities of the experimental rhodopsin. Canonical helices are idealized and might be missing features that might be complicit in receptor-ligand binding. A similar starting step is still used in the Membstruk (membrane structure) OR (or GPCR) creation-protocol which uses a low resolution structure of rhodopsin [40]. Criticisms have arisen as to the use of these low resolution structures, especially since a high resolution (2.2 Å) experimental structure is available at the Protein Data Bank (ID: 1u19) [30]. Lai et al. have shown [41], that the low resolution structure created by Singer [33] is well matched with the high resolution structure of bovine rhodopsins. The only discrepancy is with the TM 1, which is slightly shifted. Also, the higher resolution mostly defines the side chains of residues at the atomic level, while the positioning of the TM helices is resolved at a lower level.
Sequence similarity between ORs and rhodopsin is at best, approximately, 40 percent. OR models use the rhodopsin structure as the template during homology modeling using structure similarity not sequence similarity. To these are matched the TM regions of olfactory receptors. Homology modeling programs such as Modeler [42, 43] are used for template-matching followed by structure normalization.
There are critical structural differences between the rhodopsin and OR sequence [44]. In rhodopsin, the helical regions are significantly longer than those for ORs; and the loops are shorter than OR loop regions. On the other hand, the predicted TM regions for ORs are relatively shorter with significantly longer loops. The shorter OR helices are positioned, manually or through homology modeling, towards the center of the rhodopsins helix [33]. The seventh TM in rhodopsins has a significant kink which forces a portion of this helix at almost right angles to the central axis of the seventh TM. There are several molecular viewers at PDB, which one can use to observe the kink in TM 7 of rhodopsin (PDB ID: 1U19). This kinked region is ignored in the modeling of ORs.
Because of the lack of homology between rhodopsin and ORs, homology modeling can only be the first step, merely to establish helical position and some side-chain geometry. Homology modeling forces the geometry of the target molecule to mimic the geometry of the template molecule. For ORs, this might introduce some rhodopsins-specific structural artifacts into the OR model, which might not be appropriate given the lack of sequence homology between ORs and rhodopsin. To alleviate this problem, the helical geometries are minimized by performing molecular dynamic simulations in the aqueous medium to relax away any rhodopsin specific feature such as kinks.
Preservation of the Hydrophobic Binding Pocket: Rotating of the Helices in the TM Scaffold
Another reason to not rely solely on the results of homology modeling to create the seven TM scaffold is that the helical assembly has to recognize the biochemical imperative of the role of the GPCR structure: to stabilize within the lipid bi-layer while retaining the hydrophilic nature of the interior of the TM bundle. In Membstruk, the helices are manually positioned such that the hydrophobic moment is pointed away from the binding region into the bi-layer. This is followed by rigorous energy optimization while each helix is rotated in turn by small angle increments until the minimal energy for the entire helical assembly is achieved. PER-SCAN [45] (not currently available) was used to position helices during creation of the first OR model [33]. The hydrophobic moments were identified using the software and the helices were then manually positioned based on these moments. PERSCAN was successfully used to predict the hydrophobic moments in the TM helical positions of bacteriorhodopsin [45]. Bacteriorhodopsin bears structural similarities with a GPCR (PDB ID: 1S52) but does not have the sequence homology to be classed as one.
Another approach in positioning TM helices in ORs is illustrated in Equation (1). Each helix is represented by an idealized helical wheel, where amino acids occupy every 20° on the wheel. The effective hydrophobicity, Θθ at each 20° interval on this wheel is calculated using the following expression:
| (1) |
Θθ is computed by summing the arc contributions to the hydrophobicity moment on that residue from all other points along the helical wheel for a given TM. μθ is the hydrophobicity for a residue. Equation (1) allows the hydrophobicity to be defined along an arc of a circle (or a cumulative hydro-phobic effect at a specific angle contributed from hydrophobicities at all other angles) as opposed to the hydrophobicity at a specific residue. In a recent publication [46], effective hydrophobicities for human olfactory receptor OR17-210 [47] were determined using equation (1); for μ values, the consensus hydrophobicity values as devised by David Eisenberg [48] were used. Other hydrophobicity scales (which are μ values in equation (1)) such as those by Kyte and Doolittle [49] or Segrest and co-workers [50] can also be used.
Assigning Loops to the OR Model
In the first model of the OR [33], loops were not built into the computational model. For that particular binding experiment, loops were not considered necessary. There is no experimental evidence from mutational analysis that specifies residues (or certain positions) in the loops that might impact binding. Loops are difficult to model because of their lack of defined structure. Certain constraints such as disulfide bridges may be modeled into the loop structure. Three critical factors have to be considered in modeling loops: 1) That the loops in olfactory receptors are longer than rhodopsin loops, therefore using homology modeling will result in template-specific artifacts (the appearance of the loops being stretched); 2) The dynamic nature of loops, though bound within the crystallographic unit cell, cannot be effectively modeled. 3) Loops have to be abstracted out before the helical scaffold is built, to allow the rotation of each transmembrane helix.
After the TM helices have been individually optimized and rotated, the TM scaffold is put together and loops are built, using ab initio methods. One end of a loop is anchored to the helix and the other loop is allowed to optimize such that it attaches to the assigned helix terminus after a rigorous process of optimization. The end result of this process is accepted as a loop orientation for this model. Since this is an in silico optimization, one cannot be assured that the loop configuration simulates an in vivo environment. Prior to loop optimization, the distances of cysteines from each other and the likelihood to form disulfide linkages are first assessed. The Membstruk protocol [40] uses the WHAT IF modeling and visualization software [51] (http://swift.cmbi.ru.nl/-servers/html/) to add loops. The Modeler program also has a module that can be used to attach loops to the secondary structural elements of a protein and optimize their energies [42].
Overall Structural Optimization
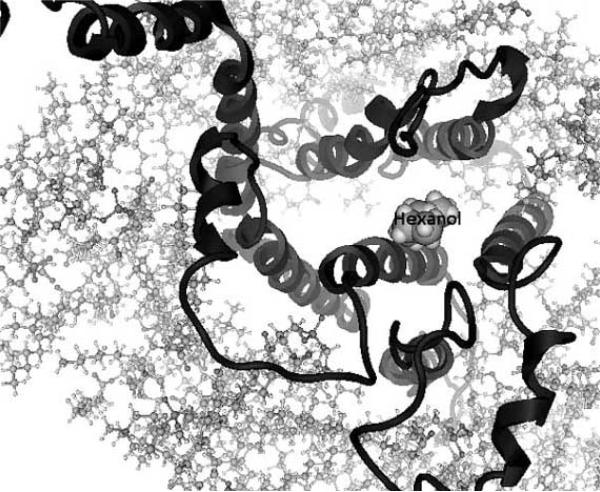
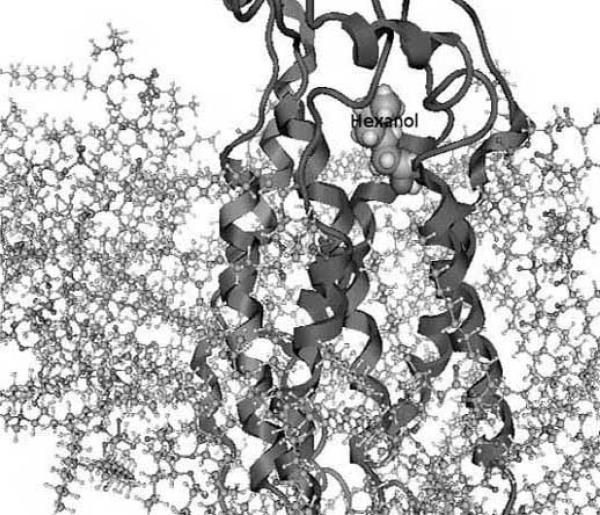
Once the transmembrane scaffold is built, the helices are rotated for hydrophobicity-based positioning and the loops and termini are added, the overall structure is optimized to an energy minimum. One can also preserve the helical super-structure by performing energy minimizations while fixing the positions of the alpha Carbon atoms (only of the helices) of the protein backbone while allowing the side chains and helices to optimize [41]. The Membstruk protocol allows the positional parameters of the entire protein to optimize. Here, the surrounding lipid bi-layer is simulated using 52 molecules of dilauroylphosphatidyl choline lipid [40]. Figs. (1a) and (1b) shows the model of the S25 mouse olfactory receptor [31] with a docked hexanol molecule surrounded by the lipid layer, as created by Membstruk. Depending on whether the molecules simulating the lipid layer are allowed to optimize, it is possible to identify the structural differences between an activated and de-activated protein following ligand docking, by letting every atom of the protein optimize independently.
Fig.(1a).
Figure shows a top-down view of the S25 mouse olfactory receptor as described in Malnic et al. [31] with hexanol (labeled)docked in its binding region. The lipid layer surrounding the protein (helices and loops shown) is created using 52 molecules of dilauroyl phosphatidyl choline lipid. These are the results of the modeling and docking studies using the Membstruk [40] and Hierdock [61] protocols. Figure courtesy of Dr. Wely Floriano, Department of Chemistry, California Polytechnic and State University, Pomona.
Fig.(1b).
Figure shows a side-on view of the S25 mouse olfactory receptor as described in Malnic et al. [31] with hexanol (labeled) docked in its binding region. The lipid layer surrounding the protein (helices and loops shown) is created using 52 molecules of dilauroylphosphatidyl choline lipid. These are the results of the modeling and docking studies using the Membstruk [40] and Hierdock [61] protocols. Figure courtesy of Dr. Wely Floriano, Department of Chemistry, California Polytechnic and State University, Pomona.
LIGAND DOCKING
Ligand docking is carried out to identify the binding characteristics of a ligand with an OR. There is sufficient evidence from experimental results (and supported by computational studies), that more than one aspect of the OR binding pocket might be responsible for interaction and activation [31]. For example, rat OR I7 experimentally binds several eight carbon, straight chain aldehyde molecules, and to a somewhat lesser extent seven and nine carbon straight chain aldehydes [33, 52, 53]. Much like the aldehyde group of retinal that binds a key Lysine residue forming a Schiff base [54] in vertebrate rhodopsins, docking studies of rat OR 17 and octanal have shown electrostatic interactions with Lysine 164 on TM4 of the receptor [33]. However, longer than nine-carbon atom chains aldehydes did not show binding with rat I7. This can be attributed to steric factors that preclude the odorant molecule from fitting into the receptor binding region in a configuration that allows the electrostatic interaction with the lysine. Interestingly enough, smaller carbon chain aldehydes, derivatives of butanal, pentanal and hexanal do not activate the receptor [33, 55]. One might conclude that, in addition to the primary electrostatic interaction, additional Van der Waals forces (given the hydrophobic nature of the binding pocket) also are involved in stabilizing the receptor during activation. This is supported by another functional assessment study for ORs, which showed that site-directed mutagenesis of a key residue that interacts with a ligand via electrostatic interactions results in lowering of activation levels, but not completely eliminating them [56].
The program DOCK is often used for ligand binding studies [57]. The program first determines charges on the protein atoms. Void volumes are then calculated for the entire protein for areas wherein a ligand might fit. Regions that are biochemically not feasible (regions on the intracellular side of the receptor or regions on the outside of the receptor where the lipid bi-layer is likely to be) are rejected. Computational binding studies have shown that extra-cellular side of the protein, bound by TMs 3, 4, 5 and 6 is where binding likely takes place [33, 58, 59]. DOCK then determines intermolecular binding by generating force fields and interaction parameters, which are used to test several hundred ligand configurations over a range of distances, angles and torsion angles. These configurations are then listed in increasing energy of the OR-odor docked system. DOCK also allows the scaling (up or down) of electrostatic parameters. This helps position a ligand in a configuration that is favorable to electrostatic interactions, especially if such an interaction has been observed experimentally. This methodology has been used to confirm the docking of the ligand in the I7 binding pocket, forming a non-bonded, electrostatic interaction with the Lys164 of the OR [41].
The Hierdock protocol has been used to dock ligands in the binding pocket of olfactory and other GPCRs at the California Institute of Technology [32, 60]. Hierdock is often used in conjunction with, and subsequent to, Membstruk. Hierdock involves a progression of steps that increases in granularity of docking, discarding ligand configurations that do not meet the criteria established by this granularity at each step. The overall binding energy of a ligand is calculated as a difference between the potential energy of ligand in the protein and the ligand in the solvent. Five of the lower energy structures are then further refined. The side-chains are replaced by accessing a high resolution (~1.0 A) rotamer library of theoretically-derived side chains. This is perhaps because the protein model created in Membstruk is based on a low resolution electron diffraction structure of rhodopsin (~7.5 A), which is not sufficient to resolve side chain configurations. The strategy to replace the residues is sound because it removes any rotameric features that are specific to rhodopsin residues. The binding energy is then recalculated. This docking procedure has been successfully used to identify putative binding site for several ORs and GPCRs, and compare well with the results of experimental binding studies [58, 59].
While other docking procedures are also employed, which vary in certain steps or the docking programs used, [44] this section provides the reader with a general idea of the steps involved in OR docking with odorants.
Simulation Studies
Lai et al. have used molecular dynamics studies to simulate the interactions between ORs and ligands [41]. There are several advantages to using simulation studies. Receptor ligand interactions that are not observed from static docking become evident. Long-term interactions can serve to substantiate or repudiate inferences from static docking. Simulation studies also show if short-term interactions are sustained over longer time periods. Static docking enables researchers to identify the ligand positions in a single configuration, whereas simulation studies can be used to trace a path of the ligand within the receptor's binding region. This transit pathway of a ligand can allow researchers to predict whether the ligand remains spatially stable over a longer time period. Simulation studies also show how key amino acids in the receptor interact with the ligand; whether these interactions are sustained or whether, over time, other amino acids interact with the odorant molecule. More importantly, one can discern through these studies if intra-receptor amino acid interactions facilitate odorant binding.
Lai et al. published the first simulation studies of olfac-tory receptors and ten odorant ligands [41] using the first computationally created OR model [33]. The experimental binding studies for these ligands had been previously published [52]. Each ligand was docked in the binding pocket of the OR in the method described above. This was followed by the dynamic simulation of the system for up to 200 picoseconds. Ligand conformational energies were minimized using the Discover module in InsightII (Acceryls, Inc.). Following docking, the highest scoring (lowest energy) docked configuration was selected for molecular dynamics simulation studies. For every ligand tested, the lowest energy-docking configuration tethered to the Lys164 on TM4 (the importance of the interaction of this residue with the ligand ahs been mentioned earlier in this review [33]).
In each docked system, Ligand-OR system energies were first minimized. In order to preserve the integrity of the helical regions, the positional parameters of alpha-carbon atoms in the helical domains were restrained. Introduction of a simulated bi-layer [40] results in a more realistic representation of the membrane OR protein system. The lack of a simulated bi-layer however, does not influence the nature of docking (restricted to the binding pocket) nor the OR–ligand binding energies—the latter are used to establish levels of OR activation [61].
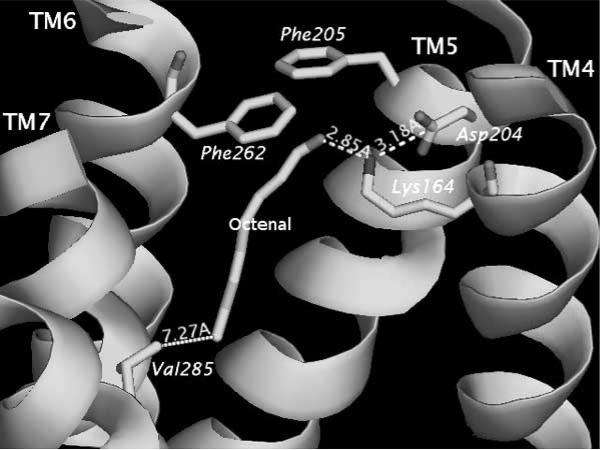
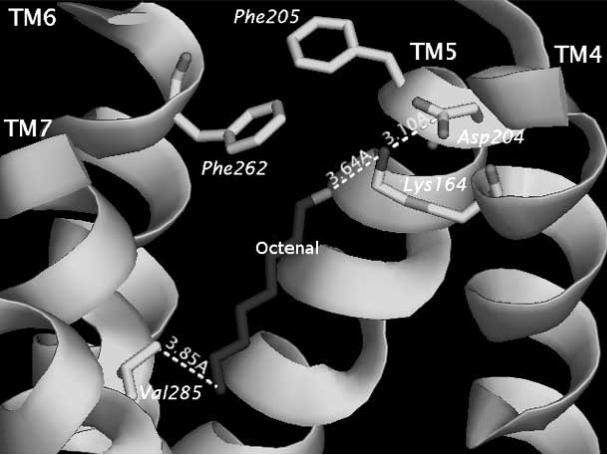
The results of Lai et al. [41] studies can be found on line at http://senselab.med.yale.edu/OR-Model. Links to pdb-formatted files containing positional parameters for the ten OR-odor systems can be found at this web resource. Each ligand–OR simulation was performed between 100–200 picoseconds (ps) at 300 K with 100 femtoseconds (fs) equilibration time. Trajectories were stored every 10 fs. Specific interaction parameters between OR and ligand: distance, angles and energies of interactions were traced and graphed. The graphs show how specific distances changes are correlated with other distance or angular changes within the receptor or between the receptor and the ligand. While it can be difficult to quantify the correlation between behavior of the ligand in the OR binding pocket, Lai et al. [41] established that ligands in the experimental study [52] that showed good activation strengths also showed definitive interactions with specific residues in the OR binding region, Figs. (2a) and (2b) show the minimum and maximum distances (as the simulation proceeds) between the octenal ligand and its primary electrostatic tether with Lys164, which is on the fourth TM.
Fig. (2a).
Figure shows the ligand trans-2-octenal (labeled) in the minimized docked position in the binding region of rat OR I7. The key distances show a strong electrostatic interaction between the ligand carbonyl group and the Lys164. The positions of F205 and F262 are also shown. Simulations showed that the phenylalanines did not impact the transit pathway of the ligand through the receptor.
Fig. (2b).
This figure shows the maximum distance between the ligand trans-2-octenal (labeled) and the Lys164 (the position electrostatic interaction). The movie of this simulation traces the pathway for the ligand. The distance with Val285 which was more than 7Å in Fig. (2a) shows that the ligand has moved back significantly.
The OR-Model web page also lists figures that illustrate the distances between the specific amino acids (implicated in binding) and the ligands, along with movie files of the ligand's pathway within the binding region of the receptor.
Studies to Identify Residues Implicated in Ligand Binding
Ligand binding with olfactory receptors is consequential to our understanding of how ORs might be associated with activating or inhibiting ligands. Simulation studies of rat OR17 with eight carbon-chain aldehydes showed that the Lys164 and the carbonyl group of the aldehyde formed a primary electrostatic tether. A similar interaction is observed between the aldehyde group of retinal and a lysine in rhodopsin [62]. But, as has been mentioned previously, the lack of olfactory activity following interactions with smaller aldehydes indicate that binding (and subsequent activation) is also supported by weaker Van der Waal interactions involving other olfactory receptors [33, 52, 56]. The Goddard group at CalTech, in numerous papers, has identified residues which are within hydrogen bonding distance from the ligand, following static docking [32, 40, 58, 59, 63-66]. These studies predicted that residues, localized on TMs 3 to 7 were potentially responsible for binding. Singer et al. performed correlated mutation analyses to identify residues that might be responsible for binding [55, 67]. Eight such residues were identified, including residues on TM6, which were possible identified with forming hydrogen bonds or with interacting with hydroxyl groups (phenols, alcohols, etc.).
One of the notions that have been advanced is the variability of residues in specific positions in ORs as a way to identify those that are responsible for binding. Conserved regions would be responsible for stabilizing the structure, G-protein binding, interactions with the surrounding lipid bi-layer. Since there are thousands of odorant ligands, it is assumed that any variability in binding would be affected by variability in the primary structure of the receptor. It was initially predicted that TMs 3 to 6 had the most variable receptors. Man et al. performed a comprehensive, comparative study of the orthologs and paralogs of mouse and human ORs [68]. Conserved residue pairs were first identified from orthologous sequences that had more than 77% sequence similarity. From 218 orthologous sequences, 146 positions were identified. Variable positions among paralogs were identified for sequences. 22 positions were identified that were statistically significant as potential binding sites. These were largely distributed over TMs 3, 4, 5, 6 and 7. TM3 had the most residue positions identified. Two positions were also identified on the extra-cellular loop between TMs 4 and 5. There is however, no structural evidence for the involvement of loops in odor binding.
DISCUSSION AND CONCLUSION
Identifying ligands that will potentially bind receptors has strong implications for introducing a paradigm for olfac-tory receptor function. This combined with modeling, docking and simulation studies will no doubt go a long way towards helping us understand the mechanism of interactions between olfactory receptors and odorant molecules, the first step in the process that leads to olfaction.
One has to be aware of the pitfalls of relying too much on the results from computational modeling. Ideally, a computational model is used to generate a hypothesis about the structure of the OR, the conditions leading to its activation, and residues involved in odor-binding. Experimentalists would then confirm or refute the inferences drawn from computational studies, helping fine tune the computational efforts.
There is folly in solely relying on the crystallographically determined structure of rhodopsin as a template for an OR structure. There is not enough homology between rhodopsin and ORs; loops in rhodopsin are shorter than those for ORs; the helices in rhodopsin are longer than those for typical ORs; there are structural features in rhodopsin such as kinks or an additional intracellular helix that is not a TM. There is no evidence that these features might be reproduced in rhodopsin.
Researchers have to create protocols that best represent the positions of the helices, the hydrophobicity of the binding pocket, the arrangement of loops, and the overall helical superstructure in as scientifically rigorous a manner as possible. Simulation studies [41] were carried out in vacuo, which is not an adequate representation of a biological process. The process of rotating the helices to meet the electronic character of the binding pocket also depends upon semi empirical methods such as in equation 1, or ab initio methods of energy minimizing such as that used in Membstruk.
There is some speculation that loops are involved in binding. Certainly the work of Man et al, point to two positions on the extra-cellular loop 2 as possible binding residues [69]. But because there is no way to adequately model loops, a computational model will not correctly show the impact of loop position on binding.
Despite this, computational models afford olfactory receptor researchers a view into the interactions between ORs and odorant molecules.
ACKNOWLEDGEMENTS
This work was generously supported in part by grant 2 P01 DC 004732-05 from the National Institute for Deafness and Communicative Disorders, National Institutes of Health. The author is also grateful to Dr. Wely Floriano, of the Department of Chemistry, California State Polytechnic University, Pomona for providing Figs. (1a) and (1b). The author is also supported by a Faculty Development Grant at the University of Alabama at Birmingham.
REFERENCES
- 1.Glusman G, Yanai I, Rubin I, et al. The complete human olfactory subgenome. Genome Res. 2001;11(5):685–702. doi: 10.1101/gr.171001. [DOI] [PubMed] [Google Scholar]
- 2.Niimura Y, Nei M. Comparative evolutionary analysis of olfactory receptor gene clusters between humans and mice. Gene. 2005;346:13–21. doi: 10.1016/j.gene.2004.09.025. [DOI] [PubMed] [Google Scholar]
- 3.Niimura Y, Nei M. Extensive gains and losses of olfactory receptor genes in Mammalian evolution. PLoS ONE. 2007;2:e708. doi: 10.1371/journal.pone.0000708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zozulya S, Echeverri F, Nguyen T. The human olfactory receptor repertoire. Genome Biol. 2001;2(6):RESEARCH0018. doi: 10.1186/gb-2001-2-6-research0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malnic B, Godfrey PA, Buck LB. The human olfactory receptor gene family. Proc Natl Acad Sci USA. 2004;101(8):2584–9. doi: 10.1073/pnas.0307882100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quignon P, Giraud M, Rimbault M, et al. The dog and rat olfactory receptor repertoires. Genome Biol. 2005;6(10):R83. doi: 10.1186/gb-2005-6-10-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Young JM, Shykind BM, Lane RP, et al. Odorant receptor expressed sequence tags demonstrate olfactory expression of over 400 genes, extensive alternate splicing and unequal expression levels. Genome Biol. 2003;4(11):R71. doi: 10.1186/gb-2003-4-11-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang X, Firestein S. The olfactory receptor gene superfamily of the mouse. Nat Neurosci. 2002;5(2):124–33. doi: 10.1038/nn800. [DOI] [PubMed] [Google Scholar]
- 9.Godfrey PA, Malnic B, Buck LB. The mouse olfactory receptor gene family. Proc Natl Acad Sci USA. 2004;101(7):2156–61. doi: 10.1073/pnas.0308051100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilad Y, Man O, Glusman G. A comparison of the human and chimpanzee olfactory receptor gene repertoires. Genome Res. 2005;15(2):224–30. doi: 10.1101/gr.2846405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crasto C, Marenco L, Miller P, et al. Olfactory Receptor Database: a metadata-driven automated population from sources of gene and protein sequences. Nucleic Acids Res. 2002;30(1):354–60. doi: 10.1093/nar/30.1.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crasto C, Singer MS, Shepherd GM. The olfactory receptor family album. Genome Biol. 2001;2(10):REVIEWS1027. doi: 10.1186/gb-2001-2-10-reviews1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olender T, Feldmesser E, Atarot T, et al. The olfactory receptor universe--from whole genome analysis to structure and evolution. Genet Mol Res. 2004;3(4):545–53. [PubMed] [Google Scholar]
- 14.Olender T, Fuchs T, Linhart C, et al. The canine olfactory subgenome. Genomics. 2004;83(3):361–72. doi: 10.1016/j.ygeno.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 15.Aloni R, Olender T, Lancet D. Ancient genomic architecture for mammalian olfactory receptor clusters. Genome Biol. 2006;7(10):R88. doi: 10.1186/gb-2006-7-10-r88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mesholam RI, Moberg PJ, Mahr RN, et al. Olfaction in neurode-generative disease: a meta-analysis of olfactory functioning in Alzheimer's and Parkinson's diseases. Arch Neurol. 1998;55(1):84–90. doi: 10.1001/archneur.55.1.84. [DOI] [PubMed] [Google Scholar]
- 17.Murofushi T, Mizuno M, Osanai R, et al. Olfactory dysfunction in Parkinson's disease. ORL J Otorhinolaryngol Relat Spec. 1991;53(3):143–6. doi: 10.1159/000276208. [DOI] [PubMed] [Google Scholar]
- 18.Westervelt HJ, McCaffrey RJ, Cousins JP, et al. Longitudinal analysis of olfactory deficits in HIV infection. Arch Clin Neuropsychol. 1997;12(6):557–65. [PubMed] [Google Scholar]
- 19.Elian M. Olfactory impairment in motor neuron disease: a pilot study. J Neurol Neurosurg Psychiatry. 1991;54(10):927–8. doi: 10.1136/jnnp.54.10.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moberg PJ, Doty RL, Mahr RN, et al. Olfactory identification in elderly schizophrenia and Alzheimer's disease. Neurobiol Aging. 1997;18(2):163–7. doi: 10.1016/s0197-4580(97)00015-8. [DOI] [PubMed] [Google Scholar]
- 21.Fedoroff IC, Stoner SA, Andersen AE, et al. Olfactory dysfunction in anorexia and bulimia nervosa. Int J Eat Disord. 1995;18(1):71–7. doi: 10.1002/1098-108x(199507)18:1<71::aid-eat2260180108>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 22.Abaffy T, Malhotra A, Luetje CW. The molecular basis for ligand specificity in a mouse olfactory receptor: a network of functionally important residues. J Biol Chem. 2007;282(2):1216–24. doi: 10.1074/jbc.M609355200. [DOI] [PubMed] [Google Scholar]
- 23.Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65(1):175–87. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- 24.Mombaerts P. Molecular biology of odorant receptors in vertebrates. Annu Rev Neurosci. 1999;22:487–509. doi: 10.1146/annurev.neuro.22.1.487. [DOI] [PubMed] [Google Scholar]
- 25.Mombaerts P. Odorant receptor gene choice in olfactory sensory neurons: the one receptor-one neuron hypothesis revisited. Curr Opin Neurobiol. 2004;14(1):31–6. doi: 10.1016/j.conb.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 26.Wang J, Luthey-Schulten ZA, Suslick KS. Is the olfactory receptor a metalloprotein? Proc Natl Acad Sci USA. 2003;100(6):3035–9. doi: 10.1073/pnas.262792899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katada S, Touhara K. [A molecular basis for odorant recognition: olfactory receptor pharmacology]. Nippon Yakurigaku Zasshi. 2004;124(4):201–9. doi: 10.1254/fpj.124.201. [DOI] [PubMed] [Google Scholar]
- 28.Cherezov V, Rosenbaum DM, Hanson MA, et al. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318(5854):1258–65. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rasmussen SG, Choi HJ, Rosenbaum DM, et al. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450(7168):383–7. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 30.Okada T. X-ray crystallographic studies for ligand-protein interaction changes in rhodopsin. Biochem Soc Trans. 2004;32(Pt 5):738–41. doi: 10.1042/BST0320738. [DOI] [PubMed] [Google Scholar]
- 31.Malnic B, Hirono J, Sato T, et al. Combinatorial receptor codes for odors. Cell. 1999;96(5):713–23. doi: 10.1016/s0092-8674(00)80581-4. [DOI] [PubMed] [Google Scholar]
- 32.Floriano WB, Vaidehi N, Goddard WA, 3rd, et al. Molecular mechanisms underlying differential odor responses of a mouse ol-factory receptor. Proc Natl Acad Sci USA. 2000;97(20):10712–6. doi: 10.1073/pnas.97.20.10712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singer MS. Analysis of the molecular basis for octanal interactions in the expressed rat 17 olfactory receptor. Chem Senses. 2000;25(2):155–65. doi: 10.1093/chemse/25.2.155. [DOI] [PubMed] [Google Scholar]
- 34.Schuster-Bockler B, Bateman A. An introduction to hidden Markov models. Curr Protoc Bioinformatics. 2007 doi: 10.1002/0471250953.bia03as18. Appendix 3(Appendix 3A. [DOI] [PubMed] [Google Scholar]
- 35.Krogh A, Larsson B, von Heijne G, et al. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305(3):567–80. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 36.Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–82. [PubMed] [Google Scholar]
- 37.Ikeda M, Arai M, Lao DM, et al. Transmembrane topology prediction methods: a re-assessment and improvement by a consensus method using a dataset of experimentally-characterized transmembrane topologies. In Silico Biol. 2002;2(1):19–33. [PubMed] [Google Scholar]
- 38.Tusnady GE, Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17(9):849–50. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- 39.Moller S, Croning MD, Apweiler R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics. 2001;17(7):646–53. doi: 10.1093/bioinformatics/17.7.646. [DOI] [PubMed] [Google Scholar]
- 40.Vaidehi N, Floriano WB, Trabanino R, et al. Prediction of structure and function of G protein-coupled receptors. Proc Natl Acad Sci USA. 2002;99(20):12622–7. doi: 10.1073/pnas.122357199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lai PC, Singer MS, Crasto CJ. Structural activation pathways from dynamic olfactory receptor-odorant interactions. Chem Senses. 2005;30(9):781–92. doi: 10.1093/chemse/bji070. [DOI] [PubMed] [Google Scholar]
- 42.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234(3):779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 43.Eswar N, Marti-Renom MA, Webb B, et al. Comparative Protein Structure Modeling Using MODELLER. In: Coligan JE, et al., editors. Current Protocols in Bioinformatics. John Wiley & Sons; New York: 2006. pp. 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Katada S, Hirokawa T, Oka Y, et al. Structural basis for a broad but selective ligand spectrum of a mouse olfactory receptor: mapping the odorant-binding site. J Neurosci. 2005;25(7):1806–15. doi: 10.1523/JNEUROSCI.4723-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Donnelly D, Overington JP, Blundell TL. The prediction and orientation of alpha-helices from sequence alignments: the combined use of environment-dependent substitution tables, Fourier transform methods and helix capping rules. Protein Eng. 1994;7(5):645–53. doi: 10.1093/protein/7.5.645. [DOI] [PubMed] [Google Scholar]
- 46.Lai PC, Bahl G, Gremigni M, et al. An olfactory receptor pseudogene whose function emerged in humans: a case study in the evolution of structure-function in GPCRs. J Struct Funct Genomics. 2008 doi: 10.1007/s10969-008-9043-x. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matarazzo V, Clot-Faybesse O, Marcet B, et al. Functional characterization of two human olfactory receptors expressed in the baculovirus Sf9 insect cell system. Chem Senses. 2005;30(3):195–207. doi: 10.1093/chemse/bji015. [DOI] [PubMed] [Google Scholar]
- 48.Eisenberg D. Three-dimensional structure of membrane and surface proteins. Annu Rev Biochem. 1984;53:595–623. doi: 10.1146/annurev.bi.53.070184.003115. [DOI] [PubMed] [Google Scholar]
- 49.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157(1):105–32. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 50.Segrest JP, Jackson RL, Morrisett JD, et al. A molecular theory of lipid-protein interactions in the plasma lipoproteins. FEBS Lett. 1974;38(3):247–58. doi: 10.1016/0014-5793(74)80064-5. [DOI] [PubMed] [Google Scholar]
- 51.Vriend G. WHAT IF: a molecular modeling and drug design program. J Mol Graph. 1990;8(1):52–6. 29. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 52.Araneda RC, Kini AD, Firestein S. The molecular receptive range of an odorant receptor. Nat Neurosci. 2000;3(12):1248–55. doi: 10.1038/81774. [DOI] [PubMed] [Google Scholar]
- 53.Singer MS, Oliveira L, Vriend G, et al. Potential ligand-binding residues in rat olfactory receptors identified by correlated mutation analysis. Receptors Channels. 1995;3(2):89–95. [PubMed] [Google Scholar]
- 54.Schertler GF. Signal transduction: the rhodopsin story continued. Nature. 2008;453(7193):292–3. doi: 10.1038/453292a. [DOI] [PubMed] [Google Scholar]
- 55.Singer MS, Hughes TE, Shepherd GM, et al. Identification of ol-factory receptor mRNA sequences from the rat olfactory bulb glomerular layer. Neuroreport. 1998;9(16):3745–8. doi: 10.1097/00001756-199811160-00032. [DOI] [PubMed] [Google Scholar]
- 56.Shirokova E, Schmiedeberg K, Bedner P, et al. Identification of specific ligands for orphan olfactory receptors. G protein-dependent agonism and antagonism of odorants. J Biol Chem. 2005;280(12):11807–15. doi: 10.1074/jbc.M411508200. [DOI] [PubMed] [Google Scholar]
- 57.Kuntz ID, Blaney JM, Oatley SJ, et al. A geometric approach to macromolecule-ligand interactions. J Mol Biol. 1982;161(2):269–88. doi: 10.1016/0022-2836(82)90153-x. [DOI] [PubMed] [Google Scholar]
- 58.Freddolino PL, Kalani MY, Vaidehi N, et al. Predicted 3D structure for the human beta 2 adrenergic receptor and its binding site for agonists and antagonists. Proc Natl Acad Sci USA. 2004;101(9):2736–41. doi: 10.1073/pnas.0308751101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hall SE, Floriano WB, Vaidehi N, et al. Predicted 3-D structures for mouse I7 and rat I7 olfactory receptors and comparison of predicted odor recognition profiles with experiment. Chem Senses. 2004;29(7):595–616. doi: 10.1093/chemse/bjh063. [DOI] [PubMed] [Google Scholar]
- 60.Floriano WB, Vaidehi N, Zamanakos G, et al. HierVLS hierarchical docking protocol for virtual ligand screening of large-molecule databases. J Med Chem. 2004;47(1):56–71. doi: 10.1021/jm030271v. [DOI] [PubMed] [Google Scholar]
- 61.Floriano WB, Vaidehi N, Goddard WA., 3rd Making sense of olfaction through predictions of the 3-D structure and function of olfactory receptors. Chem Senses. 2004;29(4):269–90. doi: 10.1093/chemse/bjh030. [DOI] [PubMed] [Google Scholar]
- 62.Janz JM, Farrens DL. Role of the retinal hydrogen bond network in rhodopsin Schiff base stability and hydrolysis. J Biol Chem. 2004;279(53):55886–94. doi: 10.1074/jbc.M408766200. [DOI] [PubMed] [Google Scholar]
- 63.Hummel P, Vaidehi N, Floriano WB, et al. Test of the Binding Threshold Hypothesis for olfactory receptors: explanation of the differential binding of ketones to the mouse and human orthologs of olfactory receptor 912-93. Protein Sci. 2005;14(3):703–10. doi: 10.1110/ps.041119705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kalani MY, Vaidehi N, Hall SE, et al. The predicted 3D structure of the human D2 dopamine receptor and the binding site and binding affinities for agonists and antagonists. Proc Natl Acad Sci USA. 2004;101(11):3815–20. doi: 10.1073/pnas.0400100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Singer MS, Shepherd GM, Greer CA. Olfactory receptors guide axons. Nature. 1995;377(6544):19–20. doi: 10.1038/377019b0. [DOI] [PubMed] [Google Scholar]
- 66.Trabanino RJ, Hall SE, Vaidehi N, et al. First principles predictions of the structure and function of g-protein-coupled receptors: validation for bovine rhodopsin. Biophys J. 2004;86(4):1904–21. doi: 10.1016/S0006-3495(04)74256-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Singer MS, Weisinger-Lewin Y, Lancet D, et al. Positive selection moments identify potential functional residues in human olfactory receptors. Receptors Channels. 1996;4(3):141–7. [PubMed] [Google Scholar]
- 68.Man O, Gilad Y, Lancet D. Prediction of the odorant binding site of olfactory receptor proteins by human-mouse comparisons. Protein Sci. 2004;13(1):240–54. doi: 10.1110/ps.03296404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Man O, Willhite DC, Crasto CJ, et al. A framework for exploring functional variability in olfactory receptor genes. PLoS ONE. 2007;2:e682. doi: 10.1371/journal.pone.0000682. [DOI] [PMC free article] [PubMed] [Google Scholar]