Abstract
Early land plant mitochondrial genomes captured important changes of mitochondrial genome evolution when plants colonized land. The chondromes of seed plants show several derived characteristics, e.g., large genome size variation, rapid intra-genomic rearrangement, abundant introns, and highly variable levels of RNA editing. On the other hand, the chondromes of charophytic algae are still largely ancestral in these aspects, resembling those of early eukaryotes. When the transition happened has been a long-standing question in studies of mitochondrial genome evolution. Here we report complete mitochondrial genome sequences from an early-diverging liverwort, Treubia lacunosa, and a late-evolving moss, Anomodon rugelii. The two genomes, 151,983 and 104,239 base pairs in size respectively, contain standard sets of protein coding genes for respiration and protein synthesis, as well as nearly full sets of rRNA and tRNA genes found in the chondromes of the liverworts Marchantia polymorpha and Pleurozia purpurea and the moss Physcomitrella patens. The gene orders of these two chondromes are identical to those of the other liverworts and moss. Their intron contents, with all cis-spliced group I or group II introns, are also similar to those in the previously sequenced liverwort and moss chondromes. These five chondromes plus the two from the hornworts Phaeoceros laevis and Megaceros aenigmaticus for the first time allowed comprehensive comparative analyses of structure and organization of mitochondrial genomes both within and across the three major lineages of bryophytes. These analyses led to the conclusion that the mitochondrial genome experienced dynamic evolution in genome size, gene content, intron acquisition, gene order, and RNA editing during the origins of land plants and their major clades. However, evolution of this organellar genome has remained rather conservative since the origin and initial radiation of early land plants, except within vascular plants.
Introduction
Among major lineages of eukaryotes, land plants (embryophytes) have mitochondrial genomes that exhibit several derived features setting them apart from chondromes of other eukaryotes: large and highly variable genome sizes, frequent intra-genomic rearrangements, rich and varied intron contents, highly variable RNA editing levels, and incorporation of foreign DNAs [1], [2], [3], [4], [5], [6]. When and how these evolutionary novelties arose in land plants, which represent a clade spanning at least 475 million years of evolution [7], have remained largely unknown until recently. Over the last twenty years, chondromes from representatives of all major lineages of early land plants and some charophytic algae have been sequenced. These include four charophytes (Mesostigma viride [8], Chlorokybus atmophyticus [9], Chaetosphaeridium globosum [10], and Chara vulgaris [11]), two liverworts (Marchantia polymorpha [12] and Pleurozia purpurea [13]), one moss (Physcomitrella patens [14]), two hornworts (Megaceros aenigmaticus [15] (despite its change to Nothoceros aenigmaticus [16], we will use the original name to be consistent with literature) and Phaeoceros laevis [17]), and two lycophytes (Isoetes engelmanii [18] and Selaginella moellendorffii [19]). These data show that the mitochondrial genomes experienced dynamic evolution during the origin and early evolution of land plants but have remained rather stable within liverworts and hornworts [6]. Further, these data and those from seed plant chondromes [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30] suggest that the extremely fluid mitochondrial genomes first found in flowering plants [31], [32], [33] are restricted to vascular plants.
More specifically, these sequenced chondromes indicate that the mitochondrial genome size increased significantly during the origin of land plants and had remained relatively constant in bryophytes, which represent the first stage of land plant evolution [34], [35]. The wide genome size variation is seen so far mostly in angiosperms [26]. Frequent intra-genomic rearrangements did not occur until the emergence of vascular plants. Intron contents, while being relatively stable within most of the major lineages of land plants (liverworts, hornworts, and vascular plants), differ significantly among these lineages. RNA editing clearly occurs in liverworts, the most basal lineage of land plants [34], [36], [37], but only two lycophytes, Isoetes and Selaginella, have been found to show extremely high levels of editing so far [19], [38]. Finally, no foreign DNA has been seen in any of the bryophyte chondromes, and the first land plant that has sequences of both chloroplast and nuclear origins in the chondrome is the lycophyte Isoetes [18].
To determine the mode of mitochondrial genome evolution across the entire diversity of bryophytes, we sequenced chondromes of the moss Anomodon rugelii and the liverwort Treubia lacunosa. Anomodon rugelii is the second moss, after Physcomitrella patens, that is sequenced for the chondrome. The two species span much of the clade true mosses, which represents 94% of the moss species diversity [39], [40], [41]. Treubia lacunosa represents one of the two liverwort families (Treubiaceae and Haplomitraceae) in the clade that is sister to all other liverworts [40], [42]. Hence, these two chondromes from critically positioned taxa add significantly to the data that already exist and allow a comprehensive examination of mitochondrial genome organization and evolution in early land plants.
Results and Discussion
General Features of the Treubia and Anomodon Mitochondrial Genomes
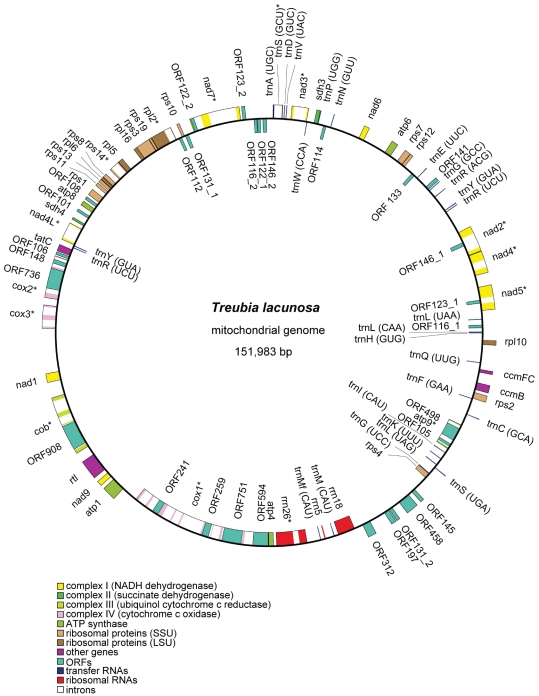
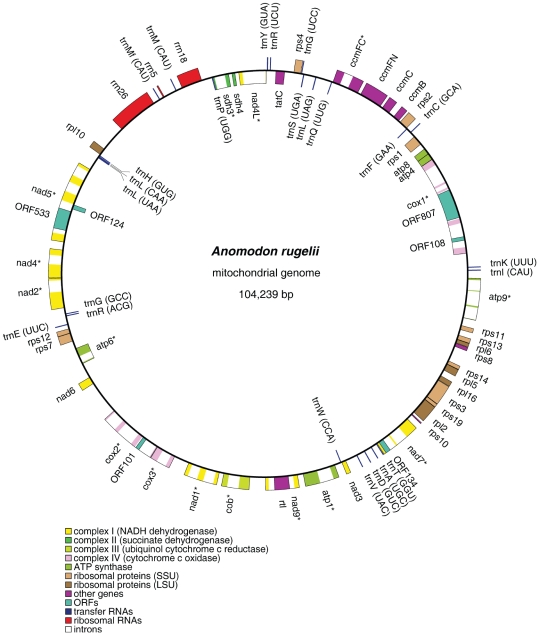
Both Treubia and Anomodon chondromes are assembled as single circular molecules (Figs. 1 and 2, deposited in GenBank under accessions JF973315 and JF973314). Their sizes are 151,983 and 104,239 base pairs (bp) respectively, with AT contents of 56.6% and 58.8%. The percentages of the various sequence elements (genes, exons, introns, and intergenic spacers) in the genome are shown in Table 1.
Figure 1. The gene map of Treubia lacunosa mitochondrial genome.
Genes (exons indicated as closed boxes) shown on the outside of the circle are transcribed clockwise, whereas those on the inside are transcribed counter-clockwise. Genes with group I or II introns (open boxes) are labeled with asterisks. Pseudogenes are indicated with the prefix “ψ”.
Figure 2. The gene map of Anomodon rugelii mitochondrial genome.
Genes (exons indicated as closed boxes) shown on the outside of the circle are transcribed clockwise, whereas those on the inside are transcribed counter-clockwise. Genes with group I or II introns (open boxes) are labeled with asterisks. Pseudogenes are indicated with the prefix “ψ”.
Table 1. Genome sizes and proportions of the various types of sequence in the mitochondrial genomes of Chara vulgaris and seven bryophytes.
| Species | Genome size (bp) | AT (%) | Genes (%) | Exons (%) | Introns (%) | Intergenic spacers (%) |
| Chara vulgaris | 67,737 | 59.1 | 91 | 52 | 39 | 9 |
| Treubia lacunosa | 151,983 | 56.6 | 53 | 26 | 27 | 47 |
| Marchantia polymorpha | 186,609 | 57.6 | 51 | 23 | 28 | 49 |
| Pleurozia purpurea | 168,526 | 54.6 | 52 | 29 | 23 | 48 |
| Physcomitrella patens | 105,340 | 59.4 | 65 | 37 | 28 | 35 |
| Anomodon rugelii | 104,239 | 58.8 | 70 | 39 | 31 | 30 |
| Phaeoceros laevis | 209,482 | 55.4 | 47 | 11 | 36 | 53 |
| Megaceros aenigmaticus | 184,908 | 54.0 | 50 | 16 | 34 | 50 |
Because this is the first time that mitochondrial genome sequences from more than one species in each of the three bryophyte clades are available, and similar data have also become available from related charophytes and lycophytes recently, we will provide some comparative analyses across major lineages of early land plants on all aspects of the genomes discussed.
Comparison of the seven bryophyte chondromes with the Chara chondrome shows that the genome size increased roughly by three times during the origin of land plants and has been relatively stable afterwards, especially within liverworts, mosses, and hornworts (Table 1). The genome size variation seems to be more correlated with the change in proportion of intergenic spacers. The AT content in mitochondrial genomes does not show much variation in Chara and the bryophytes.
RNA editing seems to occur in both Treubia and Anomodon chondromes but at very low levels, as use of the standard genetic code allowed annotation of almost all protein-coding genes but a few that required reconstitution of start or stop codons and removal of internal stop codons (Table 2). The low level of editing in the Treubia chondrome is interesting, as its close relative Haplomitrium likely has a significantly higher level of editing, at least in nad1 and nad7 [43], [44]. Given that both taxa are relic members of an ancient lineage, such unequal levels of editing in the two taxa are difficult to explain by any known mechanisms, adding another example to the previously observed phenomenon of highly lineage-specific occurrence of RNA editing in land plant organellar genomes [45]. The low level of editing in the Anomodon chondrome parallels the situation in the Physcomitrella chondrome, where only 11 editing events occur in the entire genome [46]. However, some mosses such as Takakia may have high levels of editing according to a comparative analysis of nad1 sequences [43].
Table 2. Start and stop codons altered by putative RNA editing in coding sequences of Treubia lacunosa and Anomodon rugelii mitochondrial genomes1.
| Species | Gene | Start codon created | No. of stop codons removed | Stop codon created |
| Treubia lacunosa | sdh3 | 1 UAA -> CAA | ||
| Anomodon rugelii | atp1 | ACG -> AUG | CAA -> UAA | |
| cox3 | CAA -> UAA | |||
| ccmC | CAA -> UAA | |||
| ccmFN | CAA -> UAA | |||
| tatC | ACG -> AUG |
GTG is the start codon for rpl16 in the Treubia chondrome, and is also the start codon for nad9 and rpl16 in the Anomodon chondrome.
No foreign DNA was detected in either Treubia or Anomodon mitochondrial genome. This result is consistent with those of several previous studies, which found no DNA of chloroplast or nuclear origin in any of the five bryophyte chondromes [12], [13], [14], [15], [17].
Gene Contents
The Treubia and Anomodon mitochondrial genomes contain standard sets of protein-coding genes involved in respiration and protein synthesis as found in the chondromes of the liverworts Marchantia and Pleurozia and the moss Physcomitrella (Table S1 in File S1). The ribosomal and transfer RNA genes in these two chondromes are also similar to those in the liverwort and moss mitochondrial genomes sequenced before. In fact, gene content of the Anomodon chondrome is identical to that of the Physcomitrella chondrome, even for pseudogenes.
There are several aspects that deserve special attention. First, in the Treubia chondrome the genes involved in cytochrome c biogenesis are either pseudogenized (ccmB and ccmFC) or lost (ccmC and ccmFN). This is the first liverwort that has no functional mitochondrial gene encoding for this enzyme complex. Previously, two hornworts, Megaceros and Phaeoceros, were known to have no functional mitochondrial gene for cytochrome c biogenesis [15], [17] (Table S1 in File S1). Second, nad7 seems to be a functional gene in Treubia, similar to the situation in Haplomitrium [44]. In all other liverworts that have been investigated, this gene is a pseudogene [44], with a functional copy residing in the nuclear genome [47]. Third, trnTggu is absent in Treubia, as in Apotreubia, the other genus of Treubiaceae, which was reported previously in a survey of trnA-trnT-nad7 gene cluster in a wide variety of liverworts and mosses [48]. This gene is present on the same strand as trnA and nad7 in Chara, Blasia (the sister group of complex thalloid liverworts to which Marchantia belongs), 10 diverse mosses (including Physcomitrella and Anomodon) [48], and two hornworts (in which nad7 has been lost) [15], [17] (Fig. 3). However, in Marchantia and four other complex thalloid liverworts this gene is located on the opposite strand in comparison to trnA and nad7; further, it is absent in 10 other complex thalloid liverworts apparently due to a loss in their common ancestor [48]. Fourth, rpl10, previously known as an open reading frame (ORF) in all five sequenced bryophyte chondromes [12], [13], [14], [15], [17] and recently characterized as a functional gene encoding ribosomal protein 10 in the large subunit [49], [50], is present in both Treubia and Anomodon. Fifth, rtl, which codes for a reverse transcriptase, is a functional mitochondrial gene in Treubia as in Marchantia. In Pleurozia, Physcomitrella, and Anomodon, however, it is a pseudogene. In the two mosses and Chara, rtl is located inside a group II intron, nad9i283 and nad3i211 respectively, whereas in the three liverworts it is a free-standing gene positioned between cob and nad9, likely originating from a group II intron-derived reverse transcriptase gene. Finally, trnRucu and trnYgua both have a duplicated copy in the Treubia chondrome. The two genes are next to each other (Figs. 1 and 3) and the duplication involved both genes, which likely occurred in the ancestor of all liverworts as the same gene arrangement is found in all three sequenced liverwort chondromes. What is intriguing is that in Marchantia, one copy of trnRucu appears to have given rise to trnRucg, a tRNA gene that was lost in the mitochondrial genome of the common ancestor of all green plants (Chlorophyta sensu stricto and Streptophyta), but has been re-created from a duplicated copy of either trnRucu in Marchantia and Chlorokybus or trnRacg in Nephroselmis and Mesostigma by simply modifying the anticodon and a few other nucleotides. The detailed evidence on this finding was presented in a previous study by us when reporting the Pleurozia mitochondrial genome [13].
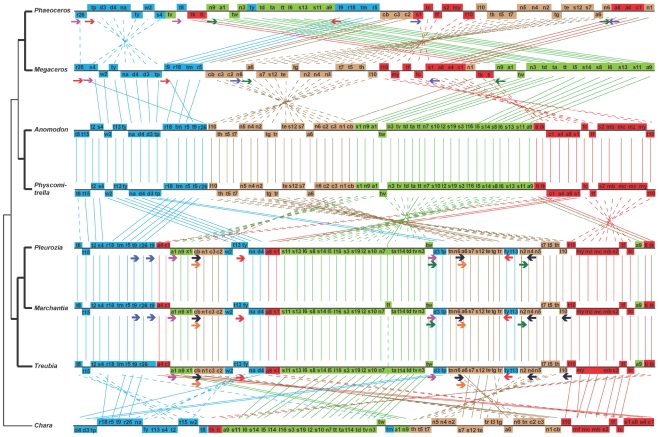
Figure 3. Gene order comparison among mitochondrial genomes of Chara vulgaris, Treubia lacunosa, Marchantia polymorpha, Pleurozia purpurea, Physcomitrella patens, Anomodon rugelii, Phaeoceros laevis, and Megaceros aenigmaticus.
Species are arranged according to the organismal phylogeny [39], [40], [42], [81] except that positions of the two hornworts are reversed as the Megaceros gene order more likely represents the ancestral condition according to a parsimony criterion (a supplementary figure (Fig. S1) is presented in which the two hornworts are placed in their correct organismal phylogeny positions). Solid lines connect orthologous genes between species with the same orientation, whereas dashed lines connect those with the reversed orientation. Repeat sequences are color-coded: in liverworts, RepA – black, RepB – green, RepB2 – purple, RepC – red, RepD – blue, and RepE – orange; in hornworts, RepA – red, RepB – blue (responsible for the inversion between Megaceros and Phaeoceros), RepC – green (this class of repeats was not annotated in either hornwort due to their length of <100 bp, inverted in Megaceros but direct in Phaeoceros), and RepD – purple.
There are 20 ORFs longer than 100 codons located in intergenic spacers of the Treubia chondrome (Fig. 1), no such ORF was detected in the Anomodon chondrome (Fig. 2). In Treubia, the five relatively large ORFs, ORF145, ORF458, ORF131-2, ORF197, and ORF312, are located in the long intergenic spacer between rrn18 and rps4, and may represent unidentified genes. Nevertheless, none of these ORFs has a homolog in Marchantia or Pleurozia and hence they may simply represent chance occurrences of reading frames. Lack of ORFs in the Anomodon chondrome is probably due to the overall genome size economy in the moss mitochondrion (Table 1).
Comparison of mitochondrial gene contents among three bryophyte lineages shows that liverworts and mosses are rather similar in this regard, with only minor difference in the tRNA gene complement (Table S1 in File S1). These two early lineages of land plants have mitochondrial gene contents that can rival their close charophytic alga relatives. On the other hand, hornworts have lost or are in the process of losing many genes for both respiration and protein synthesis, approaching the condition of two lycophytes sequenced so far, Isoetes and Selaginella (Table S1 in File S1).
Two recent studies have reported pseudogene pieces in intergenic spacers [48], [51]. Systematic surveys of these gene pieces are carried out here for all genes in all spacers of the seven bryophyte chondromes. The three liverwort chondromes harbor a large number of gene pieces in 30 spacers (Table S2 in File S1). The two hornwort chondromes contain only nad6 pseudogene pieces in the nad4-nad5 and rrn18-trnMfcau spacers (Table S2 in File S1). No such gene piece was found in any spacer in the two moss chondromes, almost certainly caused by the overall genome compactness in the moss mitochondria. These gene pieces are likely the results of retroposition as they lack any intron in cases of intron-containing genes (at least one intron, cobi783, has been retroposed separately from exons into three spacers (see below)). Most of these pieces are rather short, accounting for only a small portion of the gene. The only exceptions are the pieces for cob in liverworts and nad6 in hornworts, which account for over 80% of the gene length in both cases. In addition, some of these pieces were probably retroposed into the spacers in the common ancestor of all liverworts as they are present in the three species, and others were resulted from more recent retroposition events in more restricted scopes of taxa. Finally, some genes or portions of a gene seem to be favored targets of retroposition, as their pieces appear in more than one spacer, e.g., atp8, ccmFC, cob, cox2, nad2, rpl2, rps7, and rtl (Table S2 in File S1). No piece of ribosomal RNA or tRNA gene was detected in any spacer. At least one group II intron, cobi783, was retroposed (see below), but no systematic survey was conducted for introns because sequence divergence varies significantly in different domains of an intron, which makes BLAST searches difficult.
Gene Orders and Repeat Sequences
The gene order in the Treubia chondrome is identical to those in the Marchantia and Pleurozia chondromes (Fig. 3). Likewise, the Anomodon and Physcomitrella mitochondrial genomes have identical gene orders. Since the three liverworts span the entire liverwort clade [40], [42], the gene order of these three species likely represents the ancestral condition of all liverworts. The two mosses cover only the diversity of the true mosses. Several other major lineages that represent relic and ancient mosses have not been sampled, i.e., Takakiales, Sphagnales, Andreaeales, Tetraphidales, Polytrichales, Buxbaumiales, and Diphysciales [39], [40]. Hence, there may not be sufficient data to infer gene order of the ancestral moss mitochondrial genome. Nevertheless, the gene orders of the two moss chondromes are not very different from those in the liverwort and hornwort chondromes (Fig. 3), which suggests that they may not be too different from the ancestral condition of the moss mitochondrial genome.
Four classes of long repeat sequences longer than 100 bp are present in the Treubia chondrome (Table 3). The classes A and C, both inverted repeats, have been characterized in Pleurozia before [13] and are also present in Marchantia. Two new classes, B2 and E, both direct repeats, are found in Treubia and Marchantia; they are present in Pleurozia but were not characterized previously because of their short length (<100 bp). The classes B and D found in Pleurozia and Marchantia are absent in Treubia, and probably arose after Treubiaceae/Haplomitraceae had diverged. Five of these classes (B, B2, C, D and E) have two copies each in the genome and their locations are listed in Table 3. The class A, however, has four copies in Treubia and Marchantia and three copies in Pleurozia. Two of these copies are closely related, sharing a long stretch of identical sequences, and are located in a group II intron (cobi783) and a spacer between nad4 and nad5. They are present in all three species at the same locations. The additional more divergent copies are located in the atp6-nad6 and trnQuug-rpl10 spacers (the latter copy has been lost in Pleurozia) (Fig. 3).
Table 3. Repeat sequences in the mitochondrial genomes of Treubia lacunosa, Marchantia polymorpha, and Pleurozia purpurea 1.
| Treubia lacunosa | Marchantia polymorpha | Pleurozia purpurea | |||||||||
| Name | Location | Length (bp) | Direction | Name | Location | Length (bp) | Direction | Name | Location | Length (bp) | Direction |
| RepA_Tl2 | within cobi783, between nad4 & nad5 | 788 | inverted | RepA_Mp | within cobi783, between nad4 & nad5 | 428 | inverted | RepA_Pp | within cobi783, between nad4 & nad5 | 446 | inverted |
| --- | --- | --- | --- | RepB_Mp | between sdh3 & trnWcca, part nad2 | 134 | direct | RepB_Pp | between sdh3 & trnWcca, part nad2 | 187 | direct |
| RepB2_Tl | between sdh3 & trnWcca, part atp1 | 208 | direct | RepB2_Mp | between sdh3 & trnWcca, part atp1 | 169 | direct | RepB2_Pp | between sdh3 & trnWcca, part atp1 | 65 | direct |
| RepC_Tl3 | between nad4L & tatC, between nad2 & trnRacg | 486 | inverted | RepC_Mp | between nad4L & tatC, between nad2 & trnRacg | 154 | inverted | RepC_Pp | between nad4L & tatC, between nad2 & trnRacg | 630 | inverted |
| --- | --- | --- | --- | RepD_Mp | trnMfcau region, trnMfcau region | 191 | direct | RepD_Pp | trnMfcau region, trnMfcau region | 270 | direct |
| RepE_Tl | between atp6 & nad6, part cob | 239 | direct | RepE_Mp | between atp6 & nad6, part cob | 140 | direct | RepE_Pp | between atp6 & nad6, part cob | 73 | direct |
Because Genbank accepts only identical sequences as the definition of repeat sequences, the repeat length is only for the longest piece in the repeat region that has identical sequence with another piece elsewhere in the genome. There may be sequences up- and/or downstream of the piece that are parts of the repeat, which are not included for length calculation here because of nucleotide variation. Some repeats <100 bp are included here because their homologs (orthologs) in other species exceed 100 bp. All repeats have only two copies in the genome excep RepA, which has one or two additional more divergent copies in Treubia and Marchantia respectively (between atp6 & nad6 (Marchantia only), and between trnQuug & trnHgug).
Additional more divergent copies of RepA_Tl are located in the spacers between atp6 & nad6 and between trnQuug & rpl10.
RepC includes trnRucu and trnYgua in all three species.
One intriguing aspect about these long repeats is that all six classes involve duplication of genes or introns in intergenic spacers, probably via retroposition. Recently, it has been reported in the hornwort Megaceros chondrome that all three classes of long repeats involve duplication of genes or an intron [15]. Out of ten classes of repeats now known in the liverwort and hornwort chondromes, only one class (D in Phaeoceros) does not involve duplication of any gene or intron [17]. There is no homology between the repeat sequences in the liverworts and the hornworts.
No long repeat sequence was found in the Anomodon chondrome. Short (<100 bp) repeat sequences were identified in both Treubia and Anomodon chondromes. In the former, they are of diverse sequence compositions, whereas in the latter they are mostly domains V and VI or other elements of closely related group II introns (data not shown). Only one family of microsatellite sequence, AT dinucleotides reiterating 10 times in the second exon of cob, was found in the Anomodon chondrome. No such sequence was detected in the Treubia chondrome.
Though there are no or only a few changes in mitochondrial gene order within each of the three bryophyte lineages as shown by the seven sequenced chondromes, a minimum of 17 and 6 changes (inversions and translocations) need to be invoked to explain the gene order differences among liverworts, mosses, and hornworts (Fig. 3). How these genomic rearrangements happened are difficult historical events to reconstruct. Repeat sequences have been suggested to serve as sites of homologous recombination, resulting gene order changes in organellar genomes [52], [53], [54]. A pair of long repeat sequences were recently identified in the Megaceros and Phaeoceros chondromes that were located outside a region that had been inverted [17]. The repeat sequences detected in the liverwort chondromes in this and an earlier study [13] may have been responsible for mitochondrial gene order changes between liverworts and other early land plants. One class, repeat A, is particularly noteworthy (Table 3). A major block of genes, including many encoding ribosomal proteins, respiratory proteins, and some tRNAs, are located on opposite strands between Chara-liverworts and mosses-hornworts (Fig. 3). The former condition can be traced back to Cyanidioschyzon merolae (a unicellular red alga) [55] and Nephroselmis olivacea (a unicellular prasinophyceae green alga) [56] (also see Fig. 2 in [15]), whereas the latter condition extends to Huperzia squarrosa (a basal lycophyte and an early vascular plant; Y. Liu, B. Wang, P. Cui, L. Li, J.-Y. Xue, J. Yu, & Y.-L. Qiu, unpublished data). It is thus possible that the Chara-liverworts gene arrangement represents the ancestral condition and that of the mosses and hornworts is an evolutionarily derived condition. This change was perhaps caused by a major inversion within the mitochondrial genome of the common ancestor of mosses-hornworts-vascular plants after liverworts had diverged. The class A repeats, likely present in the chondrome of that common ancestor and retained in the modern liverwort chondromes, probably served as sites of homologous recombination. The positions of the two members, one located in the second intron in cob and the other in the spacer between trnQuug and rpl10, match roughly but rather nicely with the boundary points defining the region that has undergone inversion (Fig. 3). Two facts are consistent with this hypothesis. One is that the trnQuug-rpl10 copy shows a higher level of divergence to the cob intron copy than the nad4-nad5 spacer copy, indicating its more ancient history. Additionally, most members of this class are present in the three diverse liverworts, again indicating their ancient history. One might argue that absence of this class of repeats in the moss and hornwort chondromes does not support the hypothesis. However, this evidence can be used to explain evolutionary fixation of rearranged gene orders in the chondromes of mosses, hornworts, and basal vascular plants, which indeed have not reverted back to the Chara-liverwort condition. Further, the moss chondromes are rather economical in size (Table 1), and repeat sequences might have been purged from the genome shortly after the origin of mosses. It would be desirable to sequence chondromes of some basal moss lineages such as Takakia and Polytrichum to see if they have larger genomes and harbor long repeat sequences.
Evolution of gene order in bryophyte chondromes is overall quite conservative, both within and among major lineages. The extent of conservation is especially striking when compared with rapidly rearranged mitochondrial genomes of vascular plants [18], [19], [21], [23], [25], [26], [28]. A total of 19, 18 and 7 inversions and translocations (plus some deletions) can explain gene order differences of chondromes between Chara and Treubia, Pleurozia and Physcomitrella, and Anomodon and Megaceros respectively (Fig. 3). These are very modest number of changes for the lineages that have existed for at least 375 million years, the age of liverworts inferred from a well-preserved fossil [57]. In sharp contrast, two cytotypes of maize differ by 16 rearrangements in their chondromes [23]. If any adaptive explanation is sought, which has been explored in several recent studies of molecular evolution in organellar genomes [45], [58], [59], one wonders what are the reasons behind such high levels of conservation. One factor may be that organellar genomes, like their ancestral bacterial genomes, have polycistronic operons [60], [61], [62], [63], [64]. Some gene clusters, which likely represent polycistronic operons, have widespread phylogenetic distribution in green algae and early land plants, with some even found in Reclinomonas americana (a basal eukaryote) [65] or Cyanidioschyzon merolae [55]. These include rRNA genes, ribosomal protein genes, nad2-nad4-nad5, ccm genes, nad4L-sdh genes, and cox2-cox3-cob-nad6 [15]. Expression of these genes has probably exerted strong functional selection pressure on tight linkage and proper arrangement order of these genes in certain regions of the mitochondrial genome. Further, the products of these genes are assembled into protein/enzyme complexes involved in respiration and protein synthesis together with those encoded by nuclear genes, which were originally mitochondrial genes that migrated to the nucleus during post-endosymbiosis evolution [66]. This process would add another layer of functional constraint on inheritance and expression of the organellar genes. Recently, some nuclear genes encoding proteins that suppress recombination in mitochondria and chloroplasts in the plant cell have been characterized in Physcomitrella and some angiosperms [54], [67], [68]. Thus, conservative gene order evolution in organellar genomes seems to be resulted from both historical/genome structural reasons and functional non-autonomy of these tiny genomes. Nevertheless, these reasons are insufficient to explain the extreme level of conservation in the bryophyte mitochondrial genomes, because the same organellar genome is radically recombinogenic in vascular plants. Hence, additional, likely organismal, explanations need to be sought to account for such drastically different levels of gene order conservation in bryophyte and vascular plant chondromes.
Intron Contents
The intron content of the Treubia chondrome is similar to those in the Marchantia and Pleurozia chondromes. Both group I and group II introns are present and they are all cis-spliced (Table S3 in File S1). However, Treubia does not have either of the two group II introns (atp1i989 and atp1i1050) found in atp1 of Marchantia and Pleurozia. Likewise, it lacks one of the two group II introns in nad4L (nad4Li283) of the other two liverworts. The sole group II intron in Marchantia rrn18 (rrn18i1065) is also lacking in Treubia, as in Pleurozia.
The intron content in the Anomodon chondrome is exactly the same as that in the Physcomitrella chondrome, again all being cis-spliced, whether they are group I or group II (Table S3 in File S1).
With intron contents determined from the seven completely sequenced bryophyte chondromes, it is clear that the largely unique intron content in each of the three bryophyte lineages is shared by most members of the lineage and can be attributed to independent gains of these mobile genetic elements in the common ancestors of liverworts, mosses, and hornworts respectively (Table S3 in File S1). Secondary losses and later acquisitions seem to have happened, but only sporadically. This pattern of intron distribution is consistent with the rather conservative mode of gene order and gene content evolution in liverworts, mosses, and hornworts.
Dynamic and Conservative Evolution of Mitochondrial Genomes in Early Land Plants
The data from the Treubia and Anomodon chondromes reinforce the conclusion of several recent studies that mitochondrial genomes in early land plants show a mixed mode of dynamic and conservative evolution [13], [14], [15], [17]. During the origin of land plants, changes in the following aspects were quite dynamic: genome size increase, gene order change, gene structural alteration (ccmF fractured into ccmFC and ccmFN), and appearance of RNA editing machinery. The dynamic mode of evolution continued as major clades of land plants appeared: genome size increase or decrease, massive waves of intron acquisition, extensive gene losses in hornworts, generation of repeat sequences, and major inversions and translocations resulting gene order change. On the other hand, several aspects of the genome were rather conservative during this phase of plant evolution, in particular in comparison with vascular plant chondromes. These include: overall genome size, genome structure (lack of recombination that generated subgenomic circles), sequence composition (AT%), gene order, and genome content (lack of foreign DNA). Within liverworts, mosses, and hornworts, conservative evolution of mitochondrial genomes was even more extreme. Not only gene and intron contents varied little among diverse species within each of the three major clades of bryophytes, pseudogene gene contents and retroposed pseudogene pieces were highly similar as well. Gene orders were also identical among different species of liverworts and mosses respectively, and those of two hornworts were also rather similar. Before these bryophyte chondromes were sequenced, no prediction could be made about what they might look like based on what was known of mitochondrial genomes in angiosperms and pteridophytes [25], [28], [31], [33], [69].
With at least two species sequenced for their chondromes in each of the three bryophyte lineages, most features of this organellar genome seem to follow some patterns within the lineage. However, RNA editing, which is correlated to changes in protein hydrophobicity and molecular size in land plant organellar genomes [45], remains unpredictable, as its abundance can vary dramatically in closely related taxa such as Treubia and Haplomitrium. In lycophytes, similarly highly disparate occurrence of RNA editing has been observed in Isoetes [38] and Selaginella [19] versus Huperzia (Y. Liu, B. Wang, P. Cui, L. Li, J.-Y. Xue, J. Yu, & Y.-L. Qiu, unpublished data). Underlying causes of this enigmatic molecular evolutionary phenomenon need to be pursued in future studies.
Finally, what may be speculated about is that an evolutionarily derived type of mitochondrial genome expression system seems to have evolved in vascular plants, as bryophytes still have the typical and ancestral type of mitochondrial genomes. Notably, the genes in bryophyte chondromes remain organized into long polycistronic operons as in green and red algae as well as the basal eukaryote Reclinomonas [8], [10], [11], [56], [65], despite many changes that took place during the origin of land plants and emergence of liverworts, mosses, and hornworts. Frequent intragenomic rearrangements in vascular plant chondromes have broken most of these operons and many genes become free-standing as in the nuclear genome, which may be caused by mutations of nuclear genes identified recently that suppress recombination in organellar genomes [54], [67], [68]. During the bryophyte-vascular plant transition, the dominant generation in the plant life cycle changed from a haploid gametophyte to a diploid sporophyte, plant size increased by several orders of magnitude, and the physical environment changed correspondingly [35], [70], [71], [72], [73]. The change revealed in mitochondria, the powerhouse of the plant cell, is correlated to this major change in evolution of land plants and the environment on earth; whether there is any causative relationship between them is currently unknown.
Materials and Methods
Approximately 10 g of fresh tissues of Treubia lacunosa (Col.) Prosk. and Anomodon rugelii (C.M.) Keissl. were collected in the field from New Zealand and Michigan, USA, respectively. The Treubia material was collected with a permit (Number BP-17540-FLO) to Dr. Matt von Konrat at Field Museum (USA) issued and coordinated by Paul Cashmore of the New Zealand Department of Conservation. The Anomodon material was collected on public land, requiring no permit because it is a common weedy species. The material was brought to the lab for cleaning under a dissecting scope. For Treubia, a voucher specimen numbered John J. Engel & Matt von Konrat 28345 was deposited at Field Museum. For Anomodon, a voucher specimen numbered Qiu 06002 was deposited at the University Herbarium in the University of Michigan, Ann Arbor.
Total cellular DNA was extracted with the CTAB method [74], and purified with phenol extraction to remove proteins. A fosmid library was constructed using the CopyControlTM kit (EPICENTRE Biotechnologies, Madison, Wisconsin, USA) from the total cellular DNA fragments of 35–45 kb size-selected by agarose gel electrophoresis. No restriction enzyme digestion or mechanical shearing was used before electrophoresis. Clones containing mitochondrial DNA fragments were identified through Southern hybridizations using the HRP chemiluminescent blotting kit (KPL, Inc., Gaithersburg, Maryland, USA), with major mitochondrial genes as probes. The probes were made by amplification from total cellular DNA of Treubia or Anomodon. The inserts were sequenced through primer-walking on an ABI 3100 genetic analyzer (Applied Biosystems, Foster City, California, USA). Sequences were assembled using Sequencher (Gene Codes Corp., Ann Arbor, Michigan, USA).
The mitochondrial genomes were annotated in six steps. First, genes for known mitochondrial proteins and rRNAs were identified by Basic Local Alignment Search Tool (BLAST) searches [75] (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi) of the non-redundant database at the National Center for Biotechnology Information (NCBI). The exact gene and exon/intron boundaries were predicted by alignment of orthologous genes from annotated plant mitochondrial genomes available at the organelle genomic biology website at NCBI (http://www.ncbi.nlm.nih.gov/genomes/ORGANELLES/organelles.html). Putative RNA editing sites were inferred to create proper start and stop codons as well as to remove internal stop codons. Second, genes for hypothetical proteins were identified using the web-based tool Open Reading Frames Finder (ORF-finder; http://www.ncbi.nlm.nih.gov/gorf/gorf.html) with the standard genetic code. Third, genes for tRNAs were found using tRNAscan-SE [76] (http://lowelab.ucsc.edu/tRNAscan-SE/). Fourth, repeated sequences were searched using REPuter [77] (http://bibiserv.techfak.uni-bielefeld.de/reputer/) or BLAST. Fifth, microsatellite sequences were screened using msatcommander 0.8.2 with default settings [78]. Finally, pseudogene pieces in intergenic spacers were identified by BLASTing gene sequences against spacer sequences, and those longer than 50 bp were recorded in this study.
The annotated GenBank files of the mitochondrial genomes of Treubia and Anomodon were used to draw gene maps by using OrganellarGenomeDRAW tool (OGDRAW) [79]. The maps were then examined for further comparison of gene order and content. When sequence homology in some parts of certain genes or intergenic spacers was uncertain, the sequences were aligned using CLUSTAL_X [80], with visual examination followed.
Supporting Information
Gene order comparison among mitochondrial genomes of Chara vulgaris, Treubia lacunosa, Marchantia polymorpha, Pleurozia purpurea, Physcomitrella patens, Anomodon rugelii, Phaeoceros laevis, and Megaceros aenigmaticus. Species are arranged according to the organismal phylogeny [39], [40], [42], [81]. Solid lines connect orthologous genes between species with the same orientation, whereas dashed lines connect those with the reversed orientation. Repeat sequences are color-coded: in liverworts, RepA – black, RepB – green, RepB2 – purple, RepC – red, RepD – blue, and RepE – orange; in hornworts, RepA – red, RepB – blue (responsible for the inversion between Megaceros and Phaeoceros), RepC – green (this class of repeats was not annotated in either hornwort due to their length of <100 bp, inverted in Megaceros but direct in Phaeoceros), and RepD – purple.
(EPS)
Contains Table S1. Gene contents in mitochondrial genomes of selected charophyte and land plants1. Table S2. Pseudogene pieces in intergenic spacers of mitochondrial genomes of Treubia lacunosa, Marchantia polymorpha, Pleurozia purpurea, Phaeoceros laevis, and Megaceros aenigmaticus 1. Table S3. Intron contents in mitochondrial genomes of selected charophyte and land plants.1
(DOCX)
Acknowledgments
We thank Matt von Konrat and John Engel for help with obtaining plant material, and Volker Knoop and Jianzhi Zhang for discussion.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by NSF grants DEB 0531689 and 0332298 to YLQ. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Schuster W, Brennicke A. The plant mitochondrial genome - physical structure, information content, RNA editing, and gene migration to the nucleus. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:61–78. [Google Scholar]
- 2.Gray MW, Burger G, Lang BF. Mitochondrial evolution. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- 3.Palmer JD, Adams KL, Cho YR, Parkinson CL, Qiu YL, et al. Dynamic evolution of plant mitochondrial genomes: Mobile genes and introns and highly variable mutation rates. Proc Natl Acad Sci, USA. 2000;97:6960–6966. doi: 10.1073/pnas.97.13.6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knoop V. The mitochondrial DNA of land plants: peculiarities in phylogenetic perspective. Curr Genet. 2004;46:123–139. doi: 10.1007/s00294-004-0522-8. [DOI] [PubMed] [Google Scholar]
- 5.Knoop V, Volkmar U, Hecht J, Grewe F. Mitochondrial genome evolution in the plant lineage. In: Kempken F, editor. Plant Mitochondria. Dordrecht: Springer; 2010. pp. 3–29. [Google Scholar]
- 6.Liu Y, Xue J-Y, Wang B, Li L, Qiu Y-L. Conservative and dynamic evolution of mitochondrial genomes in early land plants. In: Bock R, Knoop V, editors. Genomics of Chloroplasts and Mitochondria. Dordrecht Kluwer; 2011. in press. [Google Scholar]
- 7.Wellman CH, Osterloff PL, Mohiuddin U. Fragments of the earliest land plants. Nature. 2003;425:282–285. doi: 10.1038/nature01884. [DOI] [PubMed] [Google Scholar]
- 8.Turmel M, Otis C, Lemieux C. The complete mitochondrial DNA sequence of Mesostigma viride identifies this green alga as the earliest green plant divergence and predicts a highly compact mitochondrial genome in the ancestor of all green plants. Mol Biol Evol. 2002;19:24–38. doi: 10.1093/oxfordjournals.molbev.a003979. [DOI] [PubMed] [Google Scholar]
- 9.Turmel M, Otis C, Lemieux C. An unexpectedly large and loosely packed mitochondrial genome in the charophycean green alga Chlorokybus atmophyticus. BMC Genomics. 2007;8:137. doi: 10.1186/1471-2164-8-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turmel M, Otis C, Lemieux C. The chloroplast and mitochondrial genome sequences of the charophyte Chaetosphaeridium globosum: Insights into the timing of the events that restructured organelle DNAs within the green algal lineage that led to land plants. Proc Natl Acad Sci, USA. 2002;99:11275–11280. doi: 10.1073/pnas.162203299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turmel M, Otis C, Lemieux C. The mitochondrial genome of Chara vulgaris: Insights into the mitochondrial DNA architecture of the last common ancestor of green algae and land plants. Plant Cell. 2003;15:1888–1903. doi: 10.1105/tpc.013169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oda K, Yamato K, Ohta E, Nakamura Y, Takemura M, et al. Gene organization deduced from the complete sequence of liverwort Marchantia polymorpha mitochondrial DNA - A primitive form of plant mitochondrial genome. J Mol Biol. 1992;223:1–7. doi: 10.1016/0022-2836(92)90708-r. [DOI] [PubMed] [Google Scholar]
- 13.Wang B, Xue J-Y, Li L, Liu L, Qiu Y-L. The complete mitochondrial genome sequence of the liverwort Pleurozia purpurea reveals extremely conservative mitochondrial genome evolution in liverworts. Curr Genet. 2009;55:601–609. doi: 10.1007/s00294-009-0273-7. [DOI] [PubMed] [Google Scholar]
- 14.Terasawa K, Odahara M, Kabeya Y, Kikugawa T, Sekine Y, et al. The mitochondrial genome of the moss Physcomitrella patens sheds new light on mitochondrial evolution in land plants. Mol Biol Evol. 2007;24:699–709. doi: 10.1093/molbev/msl198. [DOI] [PubMed] [Google Scholar]
- 15.Li L, Wang B, Liu Y, Qiu Y-L. The complete mitochondrial genome sequence of the hornwort Megaceros aenigmaticus shows a mixed mode of conservative yet dynamic evolution in early land plant mitochondrial genomes. J Mol Evol. 2009;68:665–678. doi: 10.1007/s00239-009-9240-7. [DOI] [PubMed] [Google Scholar]
- 16.Villarreal JC, Goffinet B, Duff RJ, Cargill DC. Phylogenetic delineation of Nothoceros and Megaceros (Dendrocerotaceae). The Bryologist. 2010;113:106–113. [Google Scholar]
- 17.Xue J-Y, Liu Y, Li L, Wang B, Qiu Y-L. The complete mitochondrial genome sequence of the hornwort Phaeoceros laevis: retention of many ancient pseudogenes and conservative evolution of mitochondrial genomes in hornworts. Curr Genet. 2010;56:53–61. doi: 10.1007/s00294-009-0279-1. [DOI] [PubMed] [Google Scholar]
- 18.Grewe F, Viehoever P, Weisshaar B, Knoop V. A trans-splicing group I intron and tRNA-hyperediting in the mitochondrial genome of the lycophyte Isoetes engelmannii . Nucleic Acids Res. 2009;15:5093–5104. doi: 10.1093/nar/gkp532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hecht J, Grewe F, Knoop V. Extreme RNA editing in coding islands and abundant microsatellites in repeat sequences of Selaginella moellendorffii mitochondria: the root of frequent plant mtDNA recombination in early tracheophytes. Genome Biol Evol. 2011;3:344–358. doi: 10.1093/gbe/evr027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kubo T, Nishizawa S, Sugawara A, Itchoda N, Estiati A, et al. The complete nucleotide sequence of the mitochondrial genome of sugar beet (Beta vulgaris L.) reveals a novel gene for tRNA(Cys)(GCA). Nucleic Acids Res. 2000;28:2571–2576. doi: 10.1093/nar/28.13.2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaw SM, Shih ACC, Wang D, Wu YW, Liu SM, et al. The mitochondrial genome of the gymnosperm Cycas taitungensis contains a novel family of short interspersed elements, Bpu sequences, and abundant RNA editing sites. Mol Biol Evol. 2008;25:603–615. doi: 10.1093/molbev/msn009. [DOI] [PubMed] [Google Scholar]
- 22.Notsu Y, Masood S, Nishikawa T, Kubo N, Akiduki G, et al. The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol Genet Genomics. 2002;268:434–445. doi: 10.1007/s00438-002-0767-1. [DOI] [PubMed] [Google Scholar]
- 23.Allen JO, Fauron CM, Minx P, Roark L, Oddiraju S, et al. Comparisons among two fertile and three male-sterile mitochondrial genomes of maize. Genetics. 2007;177:1173–1192. doi: 10.1534/genetics.107.073312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clifton SW, Minx P, Fauron CMR, Gibson M, Allen JO, et al. Sequence and comparative analysis of the maize NB mitochondrial genome. Plant Physiol. 2004;136:3486–3503. doi: 10.1104/pp.104.044602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Unseld M, Marienfeld JR, Brandt P, Brennicke A. The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat Genet. 1997;15:57–61. doi: 10.1038/ng0197-57. [DOI] [PubMed] [Google Scholar]
- 26.Alverson AJ, Wei X, Rice DW, Stern DB, Barry K, et al. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). Mol Biol Evol. 2010;27:1436–1448. doi: 10.1093/molbev/msq029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alverson AJ, Zhuo S, Rice DW, Sloan DB, Palmer JD. The mitochondrial genome of the legume Vigna radiata and the analysis of recombination across short mitochondrial repeats. PLoS One. 2011;6:e16404. doi: 10.1371/journal.pone.0016404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Handa H. The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana. Nucleic Acids Res. 2003;31:5907–5916. doi: 10.1093/nar/gkg795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goremykin VV, Salamini F, Velasco R, Viola R. Mitochondrial DNA of Vitis vinifera and the issue of rampant horizontal gene transfer. Mol Biol Evol. 2009;26:99–110. doi: 10.1093/molbev/msn226. [DOI] [PubMed] [Google Scholar]
- 30.Sugiyama Y, Watase Y, Nagase M, Makita N, Yagura S, et al. The complete nucleotide sequence and multipartite organization of the tobacco mitochondrial genome: comparative analysis of mitochondrial genomes in higher plants. Mol Genet Genomics. 2005;272:603–615. doi: 10.1007/s00438-004-1075-8. [DOI] [PubMed] [Google Scholar]
- 31.Palmer JD, Herbon LA. Plant mitochondrial DNA evolves rapidly in structure, but slowly in sequence. J Mol Evol. 1988;28:87–97. doi: 10.1007/BF02143500. [DOI] [PubMed] [Google Scholar]
- 32.Lonsdale DM, Brears T, Hodge TP, Melville SE, Rottmann WH. The plant mitochondrial genome: homologous recombination as a mechanism for generating heterogeneity. Philos Trans R Soc London Ser B. 1988;319:149–163. [Google Scholar]
- 33.Ward B, Anderson R, Bendich A. The size of the mitochondrial genome is large and variable in a family of plants. Cell. 1981;25:793–803. doi: 10.1016/0092-8674(81)90187-2. [DOI] [PubMed] [Google Scholar]
- 34.Kenrick P, Crane PR. Washington, D. C: Smithsonian Institution Press; 1997. The Origin and Early Diversification of Land Plants: A Cladistic Study.441 [Google Scholar]
- 35.Qiu Y-L. Phylogeny and evolution of charophytic algae and land plants. J Syst Evol. 2008;46:287–306. [Google Scholar]
- 36.Mishler BD, Churchill SP. A cladistic approach to the phylogeny of the bryophytes. Brittonia. 1984;36:406–424. [Google Scholar]
- 37.Qiu Y-L, Cho YR, Cox JC, Palmer JD. The gain of three mitochondrial introns identifies liverworts as the earliest land plants. Nature. 1998;394:671–674. doi: 10.1038/29286. [DOI] [PubMed] [Google Scholar]
- 38.Grewe F, Herres S, Viehoever P, Polsakiewicz M, Weisshaar B, et al. A unique transcriptome: 1728 positions of RNA editing alter 1406 codon identites in mitochondrial mRNAs of the lycophyte Isoetes engelmannii Nucleic Acids Res in press. 2010. [DOI] [PMC free article] [PubMed]
- 39.Goffinet B, Cox CJ, Shaw AJ, Hedderson TAJ. The bryophyta (mosses): Systematic and evolutionary inferences from an rps4 gene (cpDNA) phylogeny. Ann Bot. 2001;87:191–208. doi: 10.1006/anbo.2000.1318. [DOI] [PubMed] [Google Scholar]
- 40.Qiu Y-L, Li LB, Wang B, Chen ZD, Knoop V, et al. The deepest divergences in land plants inferred from phylogenomic evidence. Proc Natl Acad Sci, USA. 2006;103:15511–15516. doi: 10.1073/pnas.0603335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crosby MR, Magill RE, Allen B, H S. St. Louis, MO: Missouri Botanical Garden, Tropicos Database; 1999. A Checklist of the Mosses.320 [Google Scholar]
- 42.Forrest LL, Davis EC, Long DG, Crandall-Stotler BJ, Clark A, et al. Unraveling the evolutionary history of the liverworts (Marchantiophyta): multiple taxa, genomes and analyses. The Bryologist. 2006;109:303–334. [Google Scholar]
- 43.Dombrovska O, Qiu Y-L. Distribution of introns in the mitochondrial gene nad1 in land plants: phylogenetic and molecular evolutionary implications. Mol Phylogen Evol. 2004;32:246–263. doi: 10.1016/j.ympev.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 44.Groth-Malonek M, Wahrmund U, Polsakiewicz M, Knoop V. Evolution of a pseudogene: Exclusive survival of a functional mitochondrial nad7 gene supports Haplomitrium as the earliest liverwort lineage and proposes a secondary loss of RNA editing in Marchantiidae. Mol Biol Evol. 2007;24:1068–1074. doi: 10.1093/molbev/msm026. [DOI] [PubMed] [Google Scholar]
- 45.Jobson RW, Qiu Y-L. Did RNA editing in plant organellar genomes originate under natural selection or through genetic drift? Biol Direct. 2008;3:43. doi: 10.1186/1745-6150-3-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rüdinger M, Funk HT, Rensing SA, Maier UG, Knoop V. RNA editing: 11 sites only in the Physcomitrella patens mitochondrial transcriptome and a universal nomenclature proposal. Mol Genet Genomics. 2009;281:473–481. doi: 10.1007/s00438-009-0424-z. [DOI] [PubMed] [Google Scholar]
- 47.Kobayashi Y, Knoop V, Fukuzawa H, Brennicke A, Ohyama K. Interorganellar gene transfer in bryophytes: the functional nad7 gene is nuclear encoded in Marchantia polymorpha. Mol Gen Genet. 1997;256:589–592. doi: 10.1007/pl00008616. [DOI] [PubMed] [Google Scholar]
- 48.Wahrmund U, Groth-Malonek M, Knoop V. Tracing plant mitochondrial DNA evolution: rearrangements of the ancient mitochondrial gene cluster trnA- trnT- nad7 in liverwort phylogeny. J Mol Evol. 2008;66:621–629. doi: 10.1007/s00239-008-9114-4. [DOI] [PubMed] [Google Scholar]
- 49.Mower JP, Bonen L. Ribosomal protein L10 is encoded in the mitochondrial genome of many land plants and green algae. BMC Evol Biol. 2009;9:265. doi: 10.1186/1471-2148-9-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kubo N, Arimura S-I. Discovery of a functional rpl10 gene in diverse plant mitochondrial genomes and its functional replacement by a nuclear gene for chloroplast RPL10 in two lineages of angiosperms. DNA Res. 2010;17:1–9. doi: 10.1093/dnares/dsp024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Groth-Malonek M, Rein T, Wilson R, Groth H, Heinrichs J, et al. Different fates of two mitochondrial gene spacers in early land plant evolution. Int J Plant Sci. 2007;168:709–717. [Google Scholar]
- 52.Palmer JD. Comparative organization of chloroplast genomes. Annu Rev Genet. 1985;19:325–354. doi: 10.1146/annurev.ge.19.120185.001545. [DOI] [PubMed] [Google Scholar]
- 53.Small I, Suffolk R, Leaver CJ. Evolution of plant mitochondrial genomes via substoichiometric intermediate. Cell. 1989;58:69–76. doi: 10.1016/0092-8674(89)90403-0. [DOI] [PubMed] [Google Scholar]
- 54.Maréchal A, Brisson N. Recombination and the maintenance of plant organelle genome stability. New Phytol. 2010;186:299–317. doi: 10.1111/j.1469-8137.2010.03195.x. [DOI] [PubMed] [Google Scholar]
- 55.Ohta N, Sato N, Kuroiwa T. Structure and organization of the mitochondrial genome of the unicellular red alga Cyanidioschyzon merolae deduced from the complete nucleotide sequence. Nucleic Acids Res. 1998;26:5190–5198. doi: 10.1093/nar/26.22.5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Turmel M, Lemieux C, Burger G, Lang BF, Otis C, et al. The complete mitochondrial DNA sequences of Nephroselmis olivacea and Pedinomonas minor: Two radically different evolutionary patterns within green algae. Plant Cell. 1999;11:1717–1729. doi: 10.1105/tpc.11.9.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hernick LV, Landing E, Bartowski KE. Earth’s oldest liverworts – Metzgeriothallus sharonae sp. nov. from the Middle Devonian (Giventian) of eastern New York, USA. Rev Palaeobot Palynol. 2008;148:154–162. [Google Scholar]
- 58.Jobson RW, Qiu Y-L. Amino acid compositional shifts during streptophyte transitions to terrestrial habitats. J Mol Evol. 2011;72:204–214. doi: 10.1007/s00239-010-9416-1. [DOI] [PubMed] [Google Scholar]
- 59.Wang B, Liu J, Jin L, Feng X-Y, Chen J-Q. Complex mutation and weak selection together determined the codon usage bias in bryophyte mitochondrial genomes. J Integ Plant Biol. 2010;52:1100–1108. doi: 10.1111/j.1744-7909.2010.00998.x. [DOI] [PubMed] [Google Scholar]
- 60.Kozak M. Comparison of initiation of protein synthesis in prokaryotes, eukaryotes, and organelles. Microbiol Rev. 1983;47:1–45. doi: 10.1128/mr.47.1.1-45.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bland MM, Levins I, S C, Matzinger DF. The tobacco mitochondrial ATPase subunit 9 gene is closely linked to an open reading frame for a ribosomal protein. Mol Gen Genet. 1986;204:8–16. doi: 10.1007/BF00330180. [DOI] [PubMed] [Google Scholar]
- 62.Hoffmann M, Dombrovski S, Guha C, Binder S. Co-transcription of the rpl5-rps4-cob gene cluster in pea mitochondria. Mol Gen Genet. 1999;261:537–545. doi: 10.1007/s004380050998. [DOI] [PubMed] [Google Scholar]
- 63.Kanno A, Hirai A. A transcription map of the chloroplast genome from rice (Oryza sativa). Curr Genet. 1993;23:166–174. doi: 10.1007/BF00352017. [DOI] [PubMed] [Google Scholar]
- 64.Gagliardi D, Gualberto JM. Gene expression in higher plant mitochondria. In: Day DA, Harvey Millar A, Whelan J, editors. Plant Mitochondria: From Genome to Function. Dordrecht: Kluwer Academic Publishers; 2004. pp. 55–81. [Google Scholar]
- 65.Lang BF, Burger G, Okelly CJ, Cedergren R, Golding GB, et al. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature. 1997;387:493–497. doi: 10.1038/387493a0. [DOI] [PubMed] [Google Scholar]
- 66.Nugent JM, Palmer JD. RNA-mediated transfer of the gene coxII from the mitochondrion to the nucleus during flowering plant evolution. Cell. 1991;66:473–481. doi: 10.1016/0092-8674(81)90011-8. [DOI] [PubMed] [Google Scholar]
- 67.Odahara M, Kuroiwa H, Kuroiwa T, Sekine Y. Suppression of repeat-mediated gross mitochondrial genome rearrangements by RecA in the moss Physcomitrella patens. Plant Cell. 2009;21:1182–1194. doi: 10.1105/tpc.108.064709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shedge V, Arrieta-Montiel M, Christensen AC, Mackenzie SA. Plant mitochondrial recombination surveillance requires unusual RecA and MutS homologs. Plant Cell. 2007;19:1251–1264. doi: 10.1105/tpc.106.048355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Palmer JD, Soltis D, Soltis P. Large size and complex structure of mitochondrial DNA in two nonflowering land plants. Curr Genet. 1992;21:125–129. doi: 10.1007/BF00318471. [DOI] [PubMed] [Google Scholar]
- 70.Berner RA. The effect of the rise of land plants on atmospheric CO2 during the Paleozoic. In: Gensel PG, Edwards D, editors. Plants Invade the Land: Evolutionary & Environmental Perspectives. New York: Columbia University Press; 2001. pp. 173–178. [Google Scholar]
- 71.Schwartzman DW, Volk T. Biotic enhancement of weathering and the habitability of Earth. Nature. 1989;340:457–460. [Google Scholar]
- 72.Mora CI, Driese SG, Colarusso LA. Middle to late Paleozoic atmospheric CO2 levels from soil carbonate and organic matter. Science. 1996;271:1105–1107. [Google Scholar]
- 73.Qiu Y-L, Taylor AB, McManus HA. Evolution of life cycle in land plants. J Syst Evol. 2011. in press.
- 74.Doyle JJ, Doyle JS. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bulletin. 1987;19:11–15. [Google Scholar]
- 75.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 76.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, et al. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Faircloth BC. MSATCOMMANDER: detection of microsatellite repeat arrays and automated, locus-specific primer design. Mol Ecol Resour. 2008;8:92–94. doi: 10.1111/j.1471-8286.2007.01884.x. [DOI] [PubMed] [Google Scholar]
- 79.Lohse M, Drechsel O, Bock R. OrganellarGenomeDRAW (OGDRAW) - a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 2007;52:267–274. doi: 10.1007/s00294-007-0161-y. [DOI] [PubMed] [Google Scholar]
- 80.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Duff RJ, Cargill DC, Villarreal JCC, Renzaglia KS. Phylogenetic relationships of the hornworts based on rbcL sequence data: novel relationships and new insights. In: Goffinet B, Hollowell V, Magill R, editors. Molecular Systematics of Bryophytes. St. Louis: Missouri Botanical Garden Press; 2004. pp. 41–58. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene order comparison among mitochondrial genomes of Chara vulgaris, Treubia lacunosa, Marchantia polymorpha, Pleurozia purpurea, Physcomitrella patens, Anomodon rugelii, Phaeoceros laevis, and Megaceros aenigmaticus. Species are arranged according to the organismal phylogeny [39], [40], [42], [81]. Solid lines connect orthologous genes between species with the same orientation, whereas dashed lines connect those with the reversed orientation. Repeat sequences are color-coded: in liverworts, RepA – black, RepB – green, RepB2 – purple, RepC – red, RepD – blue, and RepE – orange; in hornworts, RepA – red, RepB – blue (responsible for the inversion between Megaceros and Phaeoceros), RepC – green (this class of repeats was not annotated in either hornwort due to their length of <100 bp, inverted in Megaceros but direct in Phaeoceros), and RepD – purple.
(EPS)
Contains Table S1. Gene contents in mitochondrial genomes of selected charophyte and land plants1. Table S2. Pseudogene pieces in intergenic spacers of mitochondrial genomes of Treubia lacunosa, Marchantia polymorpha, Pleurozia purpurea, Phaeoceros laevis, and Megaceros aenigmaticus 1. Table S3. Intron contents in mitochondrial genomes of selected charophyte and land plants.1
(DOCX)