Abstract
Purpose
While PSA is the best biomarker for predicting prostate cancer, its predictive performance needs to be improved. Results from the Prostate Cancer Prevention Trial (PCPT) revealed the overall performance measured by the areas under curve (AUC) of the receiver operating characteristic (ROC) at 0.68. The goal of the present study is to assess the ability of genetic variants as a PSA independent method to predict prostate cancer risk.
Experimental Design
We systematically evaluated all prostate cancer risk variants that were identified from genome-wide association studies during the past year in a large population-based prostate cancer case-control study population in Sweden, including 2,893 prostate cancer patients and 1,781 men without prostate cancer.
Results
Twelve SNPs were independently associated with prostate cancer risk in this Swedish study population. Using a cutoff of any 11 risk alleles or family history, the sensitivity and specificity for predicting prostate cancer were 0.25 and 0.86, respectively. The overall predictive performance of prostate cancer using genetic variants, family history, and age, measured by AUC was 0.65 (95% CI: 0.63–0.66), significantly improved over that of family history and age (0.61%, 95% CI: 0.59–0.62), P = 2.3 × 10−10.
Conclusion
The predictive performance for prostate cancer using genetic variants and family history is similar to that of PSA. The utility of genetic testing, alone and in combination with PSA levels, should be evaluated in large studies such as the European Randomized Study for Prostate Cancer trial and PCPT.
Keywords: prostate cancer, prediction, PSA, association
Introduction
PSA level is currently the best available method to predict an individual’s risk of prostate cancer. About three quarters of men age 50 years or older in the United States have been tested for serum PSA levels for early detection of prostate cancer (1). However, the sensitivity and specificity of PSA test are not ideal. Results from the large PCPT trial suggest that 25% to 30% of men with abnormal PSA levels (≥ 4.0 ng/mL) had cancer on prostate biopsy while ~15% of men whose PSA levels were considered to be normal, i.e. < 4.0 ng/mL, also had a positive biopsy for cancer (2). Clearly, supplemental methods are urgently needed to improve the predictive value of PSA. Several other methods have been developed to screen for prostate cancer, including PSA velocity (change of PSA levels over time) (3), the percentage of free PSA among total PSA (4), PCA3 (5,6), and EPCA-2 (7,8). The predictive performance of these methods remains under evaluation.
Recently, several genetic variants in the form of single nucleotide polymorphisms (SNPs) have been implicated in prostate cancer risk by genome-wide association studies (9–17). In contrast to previously reported prostate cancer risk variants, these novel risk variants are common in the general population and can be consistently replicated in multiple study populations (18–26). While each of them are only moderately associated with prostate cancer risk, a strong cumulative effect of the first five discovered risk variants was observed in a large Swedish study (CAPS) (27).
In this study, we sought to systematically evaluate all genetic variants reported to be significantly associated with prostate cancer risk from recent genome-wide association studies, and assess their predictive performance by estimating sensitivity, specificity, and AUC statistics of ROC.
Methods
Study population
The study sample was described in detail elsewhere (27). Briefly, we conducted a large-scale population-based case-control study in Sweden, named CAPS (CAncer Prostate in Sweden) from the National Prostate Cancer Register (28). Prostate cancer patients were identified and recruited from four of the six regional cancer registries in Sweden. The inclusion criterion for case subjects was pathological or cytological verified adenocarcinoma of the prostate, diagnosed between July, 2001 and October, 2003. Among 3,648 identified prostate cancer case subjects, 3,161 (87%) agreed to participate. DNA samples from blood and TNM stage, Gleason grade (biopsy), and PSA levels at diagnosis were available for 2,899 patients (92%). These case subjects were classified as having aggressive (advanced) disease if they met any of the following criteria: T3/4, N+, M+, Gleason score sum ≥ 8, or PSA > 50 ng/ml; otherwise, they were classified as non-aggressive (localized). Control subjects were recruited concurrently with case subjects. They were randomly selected from the Swedish Population Registry, and matched according to the expected age distribution of cases (groups of five-year intervals) and geographical region. A total of 3,153 controls were invited and 2,149 (68%) agreed to participate. DNA samples from blood were available for 1,722 control subjects (80%). Serum PSA level was measured for all control subjects but was not used as an exclusion variable. A history of prostate cancer among first-degree relatives was obtained from a questionnaire for both cases and controls. Table 1 presents the demographic and clinical characteristics of the study subjects. The study received institutional approval at the Karolinska Institutet, Umeå University, and Wake Forest University School of Medicine.
Table 1.
Clinical and demographic characteristics of subjects in CAPS
| Characteristics | # (%) of cases
|
# (%) of controls (N=1,722) | ||
|---|---|---|---|---|
| Aggressive (N=1,231) | Non-aggressive (N=1,619) | All cases (N=2,899) | ||
| Age at enrollment (Year) | ||||
| Mean (sd) | 68.04 (7.32) | 65.14 (6.74) | 66.36 (7.13) | 67.15 (7.39) |
| Age at disgnosis | ||||
| ≤ 65 | 514 (41.75) | 926 (57.19) | 1469 (50.78) | N/A |
| > 65 | 717 (58.25) | 693 (42.81) | 1424 (49.22) | N/A |
| Family History (first-degree relatives) | ||||
| No | 1013 (82.29) | 1295 (79.99) | 2342 (80.95) | 1565 (90.57) |
| Yes | 218 (17.71) | 324 (20.01) | 551 (19.05) | 163 (9.43) |
| Missing data | 0 | 0 | 0 | 0 |
| PSA levels at diagnosis for cases or at enrollment for controls (ng/ml) | ||||
| ≤ 4 | 36 (2.95) | 185 (11.61) | 221 (7.85) | 1438 (83.56) |
| 5–9.99 | 171 (14.00) | 755 (47.39) | 926 (32.91) | 230 (13.36) |
| 10–19.99 | 216 (17.69) | 438 (27.50) | 654 (23.24) | 37 (2.15) |
| 20–49.99 | 252 (20.64) | 215 (13.50) | 467 (16.60) | 13 (0.76) |
| 50–99.99 | 229 (18.76) | 0 | 229 (8.14) | 2 (0.12) |
| ≥ 100 | 317 (25.96) | 0 | 317 (11.27) | 1 (0.06) |
| Missing | 10 | 26 | 85 | 1 |
| T-stage | ||||
| T0 | 2 (0.16) | 7 (0.44) | 9 (0.32) | N/A |
| T1 | 147 (12.07) | 933 (58.24) | 1080 (38.30) | N/A |
| T2 | 242 (19.87) | 662 (41.32) | 904 (32.06) | N/A |
| T3 | 724 (59.44) | 0 | 724 (25.67) | N/A |
| T4 | 103 (8.46) | 0 | 103 (3.65) | N/A |
| TX | 13 | 17 | 79 | N/A |
| N-stage | ||||
| N0 | 222 (70.03) | 302 (100.00) | 524 (84.65) | N/A |
| N1 | 95 (29.97) | 0 | 95 (15.35) | N/A |
| NX | 914 | 1317 | 2280 | N/A |
| M-stage | ||||
| M0 | 589 (68.25) | 655 (100.00) | 1244 (81.95) | N/A |
| M1 | 274 (31.75) | 0 | 274 (18.05) | N/A |
| MX | 368 | 964 | 1381 | N/A |
| Gleason (biopsy) | ||||
| ≤ 4 | 9 (0.83) | 98 (6.32) | 107 (4.06) | N/A |
| 5 | 43 (3.96) | 247 (15.93) | 290 (10.99) | N/A |
| 6 | 153 (14.08) | 832 (53.64) | 985 (37.34) | N/A |
| 7 | 414 (38.09) | 374 (24.11) | 788 (29.87) | N/A |
| 8 | 258 (23.74) | 0 | 258 (9.78) | N/A |
| 9 | 185 (17.02) | 0 | 185 (7.01) | N/A |
| 10 | 25 (2.30) | 0 | 25 | N/A |
| Missing | 144 | 68 | 261 | N/A |
43 patients can not be classifed as aggressive or localized cases because of missing phenotypes
Selection of SNPs for evaluation and SNP genotyping
We selected 19 SNPs implicated in four prostate cancer genome-wide association studies (9–17). These included five SNPs at 8q and 17q (three separate sub-regions at 8q24, and one region each at 17q12 and 17q24.3) that have been previously evaluated in CAPS (27) and 14 SNPs at 2q15, 3p12, 6q25, 7p15, 7q21, 9q33, 10q11, 10q26, 11q13, 19q13, and Xp11 that were recently reported and have yet to be independently confirmed (15–17).
These 19 SNPs were genotyped among case and control subjects using a MassARRAY QGE iPLEX system (Sequenom, Inc. San Diego, CA). The average genotype call rate for these SNPs was 98.3% (98.2% in cases and 98.4% in controls, P > 0.05) and the average concordance rate was 99.8%. The missing data were treated as missing values in the analyses. Each of the SNPs in the autosomal chromosomes was in Hardy-Weinberg equilibrium (P > 0.05) among the control group.
Statistical analyses
Tests for Hardy-Weinberg equilibrium were performed for each SNPseparately among case patients and control subjects using Fisher’s exact test. Pair-wise linkage disequilibrium (LD) was tested for SNPs within the same chromosomal region in control subjects using SAS/Genetics software (Version 9.0).
Allele frequency differences between case patients and control subjects were tested for each SNP using a chi-square test with 1 degree of freedom. Allelic odds ratio (OR) and 95% confidence interval (95% CI) were estimated based on a multiplicative model. For SNPs that were significantly associated with prostate cancer risk from allelic test (nominal P < 0.05), a series of genetic models (additive, dominant or recessive) were performed using unconditional logistic regression with adjustment for age and geographic region. The model that had the highest likelihood was considered as the best-fitting genetic model for the respective SNP.
We then tested the independent association of prostate cancer risk with each of these SNPs by including all the significant SNPs (from single SNP analysis) and family history in a logistic regression model using a backward selection procedure. SNPs that remained significant at P < 0.05 are considered to be independently associated with prostate cancer risk. Multiplicative interactions between these SNPs were tested for each pair of SNPs by including both main effects and an interaction term (product of two main effects) in a logistic regression model.
We calculated sensitivity and specificity for cutoff values of number of genetic risk factors in predicting men with or without prostate cancer. We also evaluated the overall predictive performance of predictive models for prostate cancer by constructing receiver operating characteristic (ROC) curves and calculating the area under the curve (AUC) statistics. The AUC statistics was estimated for several predictive models after fitting a logistic regression using SAS software, including models with 1) age only, 2) age and family history, and 3) age, family history, and genetic variants. The statistical differences in AUC statistics between the models were tested using a non-parametric method (29).
Results
All 19 SNPs tested were in Hardy-Weinberg equilibrium in cases and controls (P > 0.05). The frequencies of the previous reported risk alleles for the 19 SNPs tested were all higher in cases than in controls. For 16 of these SNPs, the differences were statistically significant at a nominal P < 0.05 (Table 2). Various genetic models (additive, dominant, and recessive) were tested for each of these 16 SNPs using a logistic regression analysis with adjustment for age, family history, and geographic region. Results for the genetic model that gave the highest likelihood for each of these SNPs are presented in Table 2. Additive model was the best fitting model for 14 of the 16 SNPs.
Table 2.
Association of prostate cancer risk and 19 SNPs identified from previous genome-wide association studies
| Variable | Chr | Position* | Allele tests
|
Model | P# | Reference publications | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Allele
|
Allele frequency | OR (95% CI) | P* | ||||||||
| Ref | Risk | Cases | Controls | ||||||||
| rs721048 | 2p15 | 63,043,382 | G | A | 0.20 | 0.19 | 1.09 (0.98–1.22) | 0.11 | -- | -- | Gudmundsson |
| rs2660753 | 3p12 | 87,193,364 | C | T | 0.10 | 0.08 | 1.32 (1.13–1.54) | 3.4E-04 | addtive | 3.9E-04 | Eeles |
| rs9364554 | 6q25 | 160,804,075 | C | T | 0.33 | 0.31 | 1.12 (1.02–1.22) | 0.02 | addtive | 0.02 | Eeles |
| rs10486567 | 7p15 | 27,749,803 | T | C | 0.78 | 0.76 | 1.12 (1.01–1.24) | 0.03 | dominant | 5.3E-03 | Thomas |
| rs6465657 | 7q21 | 97,654,263 | T | C | 0.51 | 0.47 | 1.16 (1.06–1.26) | 6.7E-04 | addtive | 8.9E-04 | Eeles |
| rs16901979 | 8q24 (2) | 128,194,098 | C | A | 0.06 | 0.03 | 1.66 (1.34–2.07) | 3.1E-06 | addtive | 4.2E-06 | Gudmundsson |
| rs6983267 | 8q24 (3) | 128,482,487 | T | G | 0.56 | 0.51 | 1.22 (1.12–1.33) | 3.6E-06 | addtive | 3.7E-06 | Yeager, Zheng |
| rs1447295 | 8q24 (1) | 128,554,220 | C | A | 0.17 | 0.14 | 1.21 (1.08–1.36) | 1.6E-03 | dominant | 8.4E-04 | Amundadottir |
| rs1571801 | 9q33 | 121,506,927 | G | T | 0.31 | 0.28 | 1.15 (1.05–1.26) | 3.8E-03 | addtive | 4.1E-03 | Duggan |
| rs7920517 | 10q11 | 51,202,627 | A | G | 0.47 | 0.44 | 1.12 (1.03–1.22) | 9.2E-03 | addtive | 1.0E-02 | Eeles |
| rs10993994 | 10q11 | 51,219,502 | C | T | 0.43 | 0.39 | 1.15 (1.05–1.25) | 1.6E-03 | addtive | 1.7E-03 | Eeles, Thomas |
| rs4962416 | 10q26 | 126,686,862 | A | G | 0.24 | 0.23 | 1.07 (0.97–1.18) | 0.20 | -- | -- | Thomas |
| rs7931342 | 11q13 | 68,751,073 | T | G | 0.51 | 0.47 | 1.13 (1.04–1.24) | 4.0E-03 | addtive | 4.3E-03 | Eeles |
| rs10896449 | 11q13 | 68,751,243 | A | G | 0.49 | 0.46 | 1.14 (1.05–1.25) | 2.1E-03 | addtive | 2.3E-03 | Eeles, Thomas |
| rs4430796 | 17q12 | 33,172,153 | C | T | 0.61 | 0.56 | 1.24 (1.14–1.35) | 8.5E-07 | addtive | 6.7E-07 | Gudmundsson |
| rs1859962 | 17q24.3 | 66,620,348 | T | G | 0.54 | 0.50 | 1.17 (1.08–1.28) | 2.0E-04 | addtive | 2.2E-04 | Gudmundsson |
| rs2735839 | 19q13 | 56,056,435 | A | G | 0.88 | 0.88 | 1.03 (0.91–1.18) | 0.62 | -- | -- | Eeles |
| rs5945572 | Xp11 | 51,062,719 | C | T | 0.42 | 0.38 | 1.19 (1.05–1.35) | 5.0E-03 | -- | 5.0E-03 | Gudmundsson |
| rs5945619 | Xp11 | 51,074,708 | A | G | 0.42 | 0.38 | 1.20 (1.06–1.36) | 3.5E-03 | -- | 3.5E-03 | Eeles |
Build35
P* is based on the allelic test which assumed a multiplicative model
P# is based on Cochran-Armitag trend test for additve models and χ2 test for dominant models
To assess whether these 16 SNPs are independently associated with prostate cancer risk, we tested associations of prostate cancer risk by including all 16 SNPs (assuming the best fitting model for each SNP) in a logistic regression model using a backward selection procedure. Twelve SNPs remained independently associated with prostate cancer risk (P < 0.05) following adjustment for other SNPs as well as age, family history, and geographic region (Table 3). Three SNPs (rs7920517 at 10q11, rs7931342 at 11q13, and rs5945572 at Xp11) were no longer significant, most likely because they are in strong LD with another SNP in the same chromosomal region; rs7920517 is in LD with rs10993994 at 10q11, r2 = 0.76, rs7931342 is in LD with rs10896449 at 11q13, r2 = 0.95, and rs5945572 is in LD with rs5945619 at Xp11, r2 = 0.91. When multiplicative interaction was tested for each possible pair of these 12 SNPs using an interaction term in logistic regression, none was significant at P < 0.05.
Table 3.
Multivariate association of prostate cancer risk with each of the SNPs and family history
| Chr | Position* | model | OR* | 95%CI | P* | |
|---|---|---|---|---|---|---|
| Family history | 2.19 | 1.80–2.67 | 6.5E-15 | |||
| Age | 1.02 | 1.00–1.03 | 6.8E-03 | |||
| Genographic region | 0.46 | 0.38–0.54 | 1.7E-19 | |||
| rs2660753 | 3p12 | 87,193,364 | additive | 1.32 | 1.12–1.55 | 8.0E-04 |
| rs9364554 | 6q25 | 160,804,075 | additive | 1.08 | 0.98–1.19 | 0.11 |
| rs10486567 | 7p15 | 27,749,803 | dominant | 1.39 | 1.04–1.85 | 0.03 |
| rs6465657 | 7q21 | 97,654,263 | additive | 1.14 | 1.04–1.25 | 4.1E-03 |
| rs16901979 | 8q24 (2) | 128,194,098 | additive | 1.65 | 1.32–2.08 | 1.6E-05 |
| rs6983267 | 8q24 (3) | 128,482,487 | additive | 1.22 | 1.12–1.34 | 1.3E-05 |
| rs1447295 | 8q24 (1) | 128,554,220 | dominant | 1.16 | 1.01–1.34 | 0.04 |
| rs1571801 | 9q33 | 121,506,927 | additive | 1.15 | 1.04–1.27 | 0.01 |
| rs10993994A | 10q11 | 51,219,502 | additive | 1.16 | 1.06–1.27 | 1.8E-03 |
| rs10896449B | 11q13 | 68,751,243 | additive | 1.12 | 1.02–1.22 | 0.02 |
| rs4430796 | 17q12 | 33,172,153 | additive | 1.22 | 1.11–1.33 | 3.7E-05 |
| rs1859962 | 17q24.3 | 66,620,348 | additive | 1.17 | 1.07–1.28 | 9.0E-04 |
| rs5945619C | Xp11 | 51,074,708 | dominant | 1.19 | 1.05–1.36 | 8.7E-03 |
Build35
rs79201517 (51,202,627) is in strong LD with rs10993994, r2=0.76)
rs7931342 (68,751,073) is in strong LD with rs10896449, r2=0.95)
rs5945572 (51,062,719) is in strong LD with rs5945619, r2=0.91)
OR* and P* are based on tests assuming an additive model, ajusting for age, geographic region and family history
Among these 12 SNPs, five have been previously confirmed by us and in other study populations (18–27), including three at 8q24 (rs1447295, rs6983267, and rs16901979), one at 17q12 (rs4430796), and one at 17q24.3 (rs1859962). One SNP (rs1571801 at 9q33) was initially discovered by a genome-wide association study of 500 cases and 500 controls of this CAPS study population (14). Another SNP (rs5945619 at Xp11) has already been evaluated in CAPS and was included as a replication study population in the original discovery paper (16). Our results for the remaining five SNPs represent the first independent confirmation of the initial reports (15–17). They are rs2660753 at 3p12, rs10486567 at 7p15, rs6465657 at 7q21, rs10993994 at 10q11, and rs10896449 at 11q13.
To assess the utility of these SNPs and family history in predicting men with and without prostate cancer, we estimated the sensitivity and specificity for predicting prostate cancer using various cutoffs of number of risk alleles and family history (Table 4). We chose to use the risk allele as a unit rather than risk genotype given the consideration that an additive model was the best fitting model for vast majority of these SNPs. The SNP rs1571801 at 9q33 was not included in this analysis because it was originally discovered in this study population and has not been extensively confirmed in other study populations. Among a possible 23 risk factors (22 risk alleles from the 11 SNPs and family history), a cutoff of 11 risk factors provided a sensitivity and specificity (0.25 and 0.86, respectively) that were similar to that of the PSA level cutoff of 4.1 ng/mL (0.21 and 0.94, respectively) estimated from the Prostate Cancer Prevention Trial (PCPT) (2). In our study, about 14% of the controls have 11 or more of these risk factors.
Table 4.
Sensitivity and specificity for prostate cancer using cumulative effect of 12 risk factors (11 SNPs and risk family hisory)
| 11 SNPs &family history | psa | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| all cases vs controls | agg cases vs controls
|
nonaggcases vs controls
|
age ≥ 65 yr
|
age < 65 yr
|
cutoff | Sensitivity | Specificity | ||||||
| 2899 cases and 1722 controls
|
1280 cases and 1722 controls
|
1668 cases and 1722 controls
|
1558 cases and 1017 controls
|
1341 cases and 705 controls
|
|||||||||
| cutoff | sensitivity | specificity | sensitivity | specificity | sensitivity | specificity | sensitivity | specificity | sensitivity | specificity | |||
| 1 | 1.000 | 0.001 | 0.999 | 0.001 | 1.000 | 0.001 | 1.000 | 0.000 | 0.999 | 0.001 | |||
| 2 | 0.998 | 0.003 | 0.996 | 0.003 | 1.000 | 0.003 | 0.999 | 0.003 | 0.997 | 0.004 | |||
| 3 | 0.997 | 0.007 | 0.994 | 0.007 | 0.999 | 0.007 | 0.998 | 0.007 | 0.994 | 0.007 | |||
| 4 | 0.989 | 0.020 | 0.986 | 0.020 | 0.991 | 0.020 | 0.990 | 0.020 | 0.985 | 0.023 | |||
| 5 | 0.976 | 0.048 | 0.972 | 0.048 | 0.978 | 0.048 | 0.974 | 0.044 | 0.968 | 0.074 | |||
| 6 | 0.944 | 0.105 | 0.937 | 0.105 | 0.948 | 0.105 | 0.944 | 0.098 | 0.908 | 0.150 | |||
| 7 | 0.865 | 0.208 | 0.857 | 0.208 | 0.869 | 0.208 | 0.865 | 0.206 | 0.817 | 0.292 | 1.1 | 0.834 | 0.389 |
| 8 | 0.741 | 0.363 | 0.714 | 0.363 | 0.762 | 0.363 | 0.730 | 0.361 | 0.671 | 0.477 | 1.6 | 0.67 | 0.587 |
| 9 | 0.581 | 0.552 | 0.559 | 0.552 | 0.600 | 0.552 | 0.573 | 0.554 | 0.477 | 0.674 | 2.1 | 0.526 | 0.725 |
| 10 | 0.403 | 0.738 | 0.384 | 0.738 | 0.419 | 0.738 | 0.388 | 0.752 | 0.304 | 0.817 | 2.6 | 0.405 | 0.811 |
| 3.1 | 0.322 | 0.867 | |||||||||||
| 11 | 0.245 | 0.860 | 0.234 | 0.860 | 0.255 | 0.860 | 0.235 | 0.869 | 0.161 | 0.928 | 4.1 | 0.205 | 0.938 |
| 12 | 0.129 | 0.946 | 0.120 | 0.946 | 0.137 | 0.946 | 0.122 | 0.947 | 0.069 | 0.980 | |||
| 13 | 0.051 | 0.985 | 0.051 | 0.985 | 0.052 | 0.985 | 0.050 | 0.984 | 0.021 | 0.997 | 6.1 | 0.046 | 0.985 |
| 14 | 0.017 | 0.997 | 0.016 | 0.997 | 0.018 | 0.997 | 0.015 | 0.995 | 0.010 | 1.000 | 8.1 | 0.017 | 0.994 |
| 15 | 0.006 | 0.999 | 0.007 | 0.999 | 0.005 | 0.999 | 0.004 | 0.998 | 0.003 | 1.000 | 10.1 | 0.009 | 0.997 |
| 16 | 0.002 | 1.000 | 0.001 | 1.000 | 0.002 | 1.000 | 0.001 | 1.000 | 0.000 | 1.000 | |||
| 17 | 0.000 | 1.000 | 0.001 | 1.000 | 0.000 | 1.000 | 0.001 | 1.000 | 0.000 | 1.000 | |||
We also calculated sensitivity and specificity of these genetic risk factors to predict specific types of prostate cancer, including aggressive prostate cancer, non-aggressive prostate cancer, early age-diagnosed prostate cancer (< 65 years), and late age-diagnosed prostate cancer (≥ 65 years). No differences in the results were observed for any specific types of cancer (Table 4).
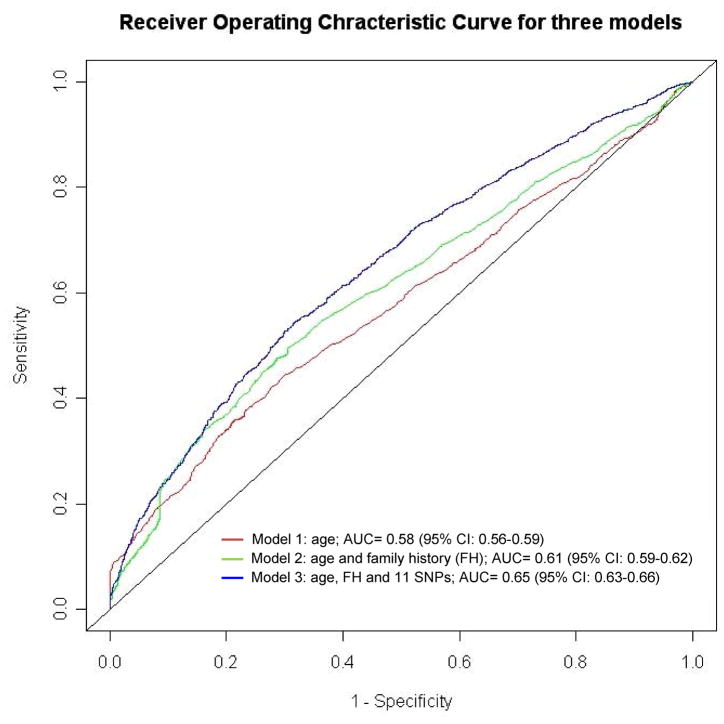
In addition to using a specific cutoff number of risk factors, we also estimated the overall predictive performance of genetic risk factors by estimating the AUC statistic of ROC curves for several nested regression models. The AUC measures the predictive performance of a predictive model where a value of 0.50 represents chance alone. We found AUC gradually improved from 0.58 (95% CI: 0.56–0.59) for Model 1 with age alone, to 0.61 (0.59–0.62) for Model 2 with age and family history, and to 0.65 (0.63–0.66) for Model 3 with age, family history, and the 11 risk SNPs (Fig 1). The differences in AUC were statistically significant between Models 2 and 1 for the predictive effect of additional family history (P = 1.36 × 10−7), and between Models 3 and 2 for the predictive effect of adding the 12 SNPs (P = 2.3 × 10−10). The AUC for Model 3 is again similar to that of PSA levels 0.68 (95% CI: 0.67–0.69) in the PCPT (2). The AUC for Model 3 (with 11 SNPs) was significantly higher than for the model with age, family history and only 5 previously evaluated SNPs (27) (0.63, 95% CI, 0.62–0.65), P = 0.003 (data not shown).
Figure 1.
Receiver operating characteristic (ROC) curve for prostate cancer for three models, including Model 1 with age alone, Model 2 with age and family history, and Model 3 with age, family history, and 11 risk SNPs. The estimate of AUC (area under ROC curve) for each model is presented. The AUC gradually improved from 0.58 (95% CI: 0.56–0.59) for Model 1, to 0.61 (0.59–0.62) for Model 2 with age and family history, and to 0.65 (0.63–0.66) for Model 3 with age, family history, and the 11 risk SNPs. The differences in AUC, using a nonparametric method (29), were statistically significant between Models 2 and 1 for the predictive effect of additional family history (P = 1.36 × 10−7), and between Models 3 and 2 for the predictive effect of adding genetic variants (P = 2.3 × 10−10).
Discussion
In this systematic evaluation of all the prostate cancer risk variants reported from recent genome-wide association studies, we found 12 SNPs are independently associated with prostate cancer risk in a large population-based study in Sweden. Two points are noteworthy: 1) a large number of independent SNPs are associated with prostate cancer risk; and 2) these risk variants were confirmed in this relatively homogeneous Swedish population, although most of them were initially discovered in populations originating from Iceland, the U.S., and UK. These findings demonstrate well the general nature of these associations, the complexity of prostate cancer genetics and the polygenic basis for prostate cancer.
Probably the most important finding of our study is that the predictive performance for prostate cancer using these genetic risk variants, family history and age is similar to that of PSA levels as estimated from the PCPT (2). Genetic risk factors are different from PSA levels in predicting prostate cancer in several important ways: the former predict the likelihood of developing prostate cancer and can be measured at any age while the latter become informative only when prostate cancer (or a cancer associated process) has developed and is generally measured well into adulthood. Genetic markers have several additional advantages in predicting individual prostate cancer risk. They need to be measured only once in a lifetime, while PSA levels and the interpretation of these levels are dynamic and change over time for a variety of reasons, some cancer-associated, and some not. Furthermore, genetic markers can be measured accurately, while there is considerable variation in the measurement of PSA levels among different methods and laboratories.
However, there are several limitations in the current study. First, different from the prospective PCPT study, our case-control study cannot provide an unbiased estimate of positive predictive value (PPV) for prostate cancer because the estimates of PPV will be influenced by the proportions of cases and controls in the study. Second, the control subjects in our study were not extensively examined for prostate cancer status; therefore, misclassification in control subjects is a concern. This may affect estimates of specificity and AUC. Third, although PSA screening in Sweden is not as prevalent as in some other parts of world, a fraction of cases in our study were diagnosed due to elevated PSA levels alone. Therefore, it is inappropriate to assess predictive performance of PSA on prostate cancer risk in our study. This limitation further prevents us to assess the joint predictive performance of genetic risk factors and PSA on prostate cancer risk, a critical question for further study. Finally, the fact that these SNPs do not differentiate between aggressive and non-aggressive disease does not address the problem of over-diagnosis of prostate cancer. This limitation may be addressed when genetic variants that are associated with aggressive prostate cancer are identified in the future.
Nevertheless, the novel findings on the predictive performance of prostate cancer using genetic risk factors in our study, if confirmed in additional independent study populations, represent an important first step and provide a basis for combining genetic variants with PSA in predicting prostate cancer risk. Several study populations where all men in the study were biopsied for prostate cancer diagnosis regardless of PSA levels, such as European Randomized Study for Prostate Cancer Trial and PCPT may help to address this important question.
Statement of Clinical Relevance.
While PSA is the best biomarker for predicting prostate cancer, its predictive performance is not ideal and needs to be improved. Results from our study suggest that the predictive performance for prostate cancer using risk variants and family history is similar to that of PSA. If this finding is confirmed and expanded, these genetic markers might be used to supplement PSA to improve its predictive value. Better prediction of prostate cancer risk could augment efforts to more efficiently diagnose prostate cancer at an early curative stage, and possibly reduce unnecessary biopsies.
Acknowledgments
The authors thank all the study subjects who participated in the CAPS study and urologists who included their patients in the CAPS study. The authors take full responsibility for the study design, data collection, analysis and interpretation of the data, the decision to submit the manuscript for publication, and the writing of the manuscript.
The study is partially supported by National Cancer Institute CA105055, CA106523 and CA95052 to Dr. Xu., Department of Defense grant PC051264 to Dr. Xu, Swedish Cancer Society (Cancerfonden) to Dr. Gronberg and Swedish Academi of Sciences (Vetenskapsrådet) to Dr. Gronberg. The support of William T Gerrard, Mario A Duhon, John and Jennifer Chalsty, and David Koch to W.B.I is gratefully acknowledged.
Footnotes
Competing interest
A patent application has been filed to preserve patent rights for the technology and results related to the first five genetic variants (three at 8q24 and one each at 17q12 and 17q24.3) by the Wake Forest University School of Medicine, Johns Hopkins University School of Medicine, and Dr. Henrik Grönberg at Karolinska Institutet, Stockholm. All authors declare no competing interest.
References
- 1.Weir HK, Thun MJ, Hankey BF, et al. Annual report to the nation on the status of cancer, 1975–2000, featuring the uses of surveillance data for cancer prevention and control. J Natl Cancer Inst. 2003;95:1276–1299. doi: 10.1093/jnci/djg040. [DOI] [PubMed] [Google Scholar]
- 2.Thompson IM, Ankerst DP, Chi C, et al. Operating characteristics of prostate-specific antigen in men with an initial PSA level of 3. 0 ng/ml or lower. JAMA. 2005;294:66–70. doi: 10.1001/jama.294.1.66. [DOI] [PubMed] [Google Scholar]
- 3.Carter HB, Pearson JD, Metter EJ, et al. Longitudinal evaluation of prostate specific antigen levels in men with and without prostate disease. JAMA. 1992;267:2215–2220. [PMC free article] [PubMed] [Google Scholar]
- 4.Catalona WJ, Partin AW, Slawin KM, et al. Use of the percentage of free prostate-specific antigen to enhance differentiation of prostate cancer from benign prostatic disease: a prospective multicenter clinical trial. JAMA. 1998;279:1542–1547. doi: 10.1001/jama.279.19.1542. [DOI] [PubMed] [Google Scholar]
- 5.de Kok JB, Verhaegh GW, Roelofs RW, et al. DD3(PCA3), a very sensitive and specific marker to detect prostate tumors. Cancer Res. 2002;62:2695–2698. [PubMed] [Google Scholar]
- 6.Fradet Y, Saad F, Aprikian A, et al. uPM3, a new molecular urine test for the detection of prostate cancer. Urology. 2004;64:311–315. doi: 10.1016/j.urology.2004.03.052. [DOI] [PubMed] [Google Scholar]
- 7.Leman ES, Cannon GW, Trock BJ, et al. EPCA-2: a highly specific serum marker for prostate cancer. Urology. 2007;69:714–720. doi: 10.1016/j.urology.2007.01.097. [DOI] [PubMed] [Google Scholar]
- 8.Diamandis EP. POINT: EPCA-2: A promising new serum biomarker for prostatic carcinoma? Clin Biochem. 2007;40:1437–1439. doi: 10.1016/j.clinbiochem.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 9.Amundadottir LT, Sulem P, Gudmundsson J, et al. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;38:652–658. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 10.Freedman ML, Haiman CA, Patterson N, et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc Natl Acad Sci USA. 2006;103:14068–14073. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gudmundsson J, Sulem P, Manolescu A, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39:631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 12.Yeager M, Orr N, Hayes RB, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 13.Gudmundsson J, Sulem P, Steinthorsdottir V, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet. 2007;39:977–983. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 14.Duggan D, Zheng SL, Knowlton M, et al. Two genome-wide association studies of aggressive prostate cancer implicate putative prostate tumor suppressor gene DAB2IP. J Natl Cancer Inst. 2007;99:1836–1844. doi: 10.1093/jnci/djm250. [DOI] [PubMed] [Google Scholar]
- 15.Thomas G, Jacobs KB, Yeager M, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 16.Gudmundsson J, Sulem P, Rafnar T, et al. Common sequence variants on 2p15 and Xp11. 22 confer susceptibility to prostate cancer. Nat Genet. 2008;40:281–283. doi: 10.1038/ng.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eeles RA, Kote-Jarai Z, Giles GG, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40:316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 18.Haiman CA, Patterson N, Freedman ML, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet. 2007;39:638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng SL, Sun J, Cheng Y, et al. Association between two unlinked loci at 8q24 and prostate cancer risk among European Americans. JNCI. 2007;99:1525–1533. doi: 10.1093/jnci/djm169. [DOI] [PubMed] [Google Scholar]
- 20.Severi G, Hayes VM, Padilla EJ, et al. The common variant rs1447295 on chromosome 8q24 and prostate cancer risk: results from an Australian population-based case-control study. Cancer Epidemiol Biomarkers Prev. 2007;16:610–612. doi: 10.1158/1055-9965.EPI-06-0872. [DOI] [PubMed] [Google Scholar]
- 21.Wang L, McDonnell SK, Slusser JP, et al. Two common chromosome 8q24 variants are associated with increased risk for prostate cancer. Cancer Res. 2007;67:2944–2950. doi: 10.1158/0008-5472.CAN-06-3186. [DOI] [PubMed] [Google Scholar]
- 22.Schumacher FR, Feigelson HS, Cox DG, et al. A common 8q24 variant in prostate and breast cancer from a large nested case-control study. Cancer Res. 2007;67:2951–2956. doi: 10.1158/0008-5472.CAN-06-3591. [DOI] [PubMed] [Google Scholar]
- 23.Suuriniemi M, Agalliu I, Schaid DJ, et al. Confirmation of a positive association between prostate cancer risk and a locus at chromosome 8q24. Cancer Epidemiol Biomarkers Prev. 2007;16:809–814. doi: 10.1158/1055-9965.EPI-06-1049. [DOI] [PubMed] [Google Scholar]
- 24.Robbins C, Torres JB, Hooker S, et al. Confirmation study of prostate cancer risk variants at 8q24 in African Americans identifies a novel risk locus. Genome Res. 2007;17:1717–1722. doi: 10.1101/gr.6782707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng I, Plummer SJ, Jorgenson E, et al. 8q24 and prostate cancer: association with advanced disease and meta-analysis. Eur J Hum Genet. 2008;16:496–505. doi: 10.1038/sj.ejhg.5201959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun J, Purcell L, Gao Z, et al. Association between sequence variants at 17q12 and 17q24. 3 and prostate cancer risk in European and African Americans. Prostate. 2008;68:691–697. doi: 10.1002/pros.20754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng SL, Sun J, Wiklund F, et al. Cumulative association of five genetic variants with prostate cancer. N Engl J Med. 2008;358:910–919. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]
- 28.Adolfsson J, Garmo H, Varenhorst E, et al. Clinical characteristics and primary treatment of prostate cancer in Sweden between 1996 and 2005. Scand J Urol Nephrol. 2007;12:1–22. doi: 10.1080/00365590701673625. [DOI] [PubMed] [Google Scholar]
- 29.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the Areas Under Two or More Correlated Receiver Operating Characteristic Curves: A Nonparametric Approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]