Abstract
Conjugation is an important mode of horizontal gene transfer in bacteria, enhancing the spread of antibiotic resistance. In clinical settings, biofilms are likely locations for antibiotic resistance transfer events involving nosocomial pathogens such as Enterococcus faecalis. Here we demonstrate that growth in biofilms alters the induction of conjugation by a sex pheromone in E. faecalis. Mathematical modeling suggested that a higher plasmid copy number in biofilm cells would enhance a switch-like behavior in the pheromone response of donor cells with a delayed, but increased response to the mating signal. Alterations in plasmid copy number, and a bimodal response to induction of conjugation in populations of plasmid-containing donor cells were both observed in biofilms, consistent with the predictions of the model. The pheromone system may have evolved such that donor cells in biofilms are only induced to transfer when they are in extremely close proximity to potential recipients in the biofilm community. These results may have important implications for development of chemotherapeutic agents to block resistance transfer and treat biofilm-related clinical infections.
Introduction
In microbial biofilms, cell-cell communication and coordinated multicellular interactions are of paramount importance. For example, quorum sensing and related forms of intercellular signaling play critical roles in biofilm development and dispersal (Boles & Horswill, 2008). In addition to communication by extracellular signals, the biofilm environment may contribute to microbial evolution by serving as an important niche for horizontal genetic transfer by transformation or conjugation (Nguyen et al., 2010). Moreover, expression of the gene products involved in these processes have been linked to enhanced biofilm formation (Ghigo, 2001, Chuang-Smith et al., 2010). Small molecule inhibitors of conjugation show promise as compounds for drug development (Lujan et al., 2007).
In spite of the plethora of research linking biofilm development to intercellular signaling and genetic transfer, there is limited information about the molecular aspects of cell-cell signaling in biofilms. The inherent heterogeneity of the biofilm environment likely has important effects on both signal production and response. Furthermore, we know relatively little about the molecular interactions between signal molecules and the biofilm matrix and the resulting effects of these interactions on transmission of signals. With regard to gene transfer, transformation in biofilms has been examined (Li et al., 2001) and there is considerable overlap between regulation of competence and biofilm development (Cvitkovitch, 2001). Numerous studies have described the regulation of conjugation in broth cultures but few studies have examined conjugation in a biofilm setting. It has been shown that the conjugative F pilus promotes biofilm formation (Ghigo, 2001) and has also been demonstrated that pCF10-encoded PrgB can increase biofilm formation (Chuang-Smith et al., 2010). Conjugative transfer frequencies have also been measured in biofilms (Molin & Tolker-Nielsen, 2003) but controlled studies of specific effects of biofilm growth on conjugative gene transfer are limited.
In the genus Enterococcus, the most efficient conjugation systems are plasmid-borne and their induction in donor cells is mediated by peptide sex pheromones excreted by plasmid-free recipient cells. In the case of the E. faecalis pCF10 system, the signaling molecule is the heptapeptide cCF10 (LVTLVFV)(Mori et al., 1988). Since production of cCF10 is encoded within the core genome, the plasmid encodes functions to prevent self-induction of donor cells by endogenous pheromone. These include a membrane protein PrgY which acts to reduce pheromone production by donor cells (Chandler et al., 2005a) and the peptide iCF10 (AITLIFI), which acts as a competitive inhibitor of cCF10 (Nakayama et al., 1994). Both iCF10 and cCF10 are first secreted, then imported into donor cells where they bind to the same site in PrgX, the master regulator controlling initiation of transcription of the transfer genes. Although both peptides bind to the same site, they cause different structural changes in PrgX, ultimately stabilizing (iCF10) or destabilizing (cCF10) a transcription-repressing complex of PrgX and its target sites on pCF10 (Kozlowicz et al., 2004).
The direct effect of pheromone is to increase transcription initiation from the promoter for the prgQ operon, which encodes iCF10 and positive regulatory RNAs at its 5’end, and the molecular machinery for conjugation in downstream regions (Figure 1A). Even in the absence of exogenous pheromone, prgQ transcription occurs, but it terminates upstream of the conjugation genes, producing a 380 nt RNA called QS (Chung & Dunny, 1995). Antisense interactions of nascent Q transcripts with a small, complementary RNA called Anti-Q shifts the secondary structure of Q to a terminator (Bae et al., 2004, Johnson et al., 2010). In uninduced cells there is adequate Anti-Q to effectively block formation of any transcripts longer than QS, whereas in the presence of pheromone, the increased production of Q transcripts titrates all of the Anti-Q and leads to longer transcripts (QL). Some of these transcripts extend into the genes for conjugation such as prgB, whose product promotes donor-recipient aggregation. Interestingly the transcription unit encoding PrgX and Anti-Q is convergent with prgQ, with 220 bp of overlap. This unique arrangement means that several levels of reciprocal post-transcriptional regulation operate in this system. These mechanisms serve to amplify the primary effect of pheromone, and our recent studies suggested that these multiple layers of regulation confer properties of a bistable genetic switch to the system (Chatterjee et al., 2011). From the perspective of a donor cell, this peptide signaling system may be viewed as a form of telesensing (Roux et al., 2009), where donor cells secrete a mixture of the two peptides, with iCF10 present in 50-100 fold excess (Nakayama et al., 1994), and monitor their relative concentrations over time. If the ratio changes in favor of cCF10, as is the case when recipients are in close proximity, the conjugation system is activated.
Figure 1. Biofilm cells containing the fluorescent reporter are viable, growing, and express GFP in response to cCF10.
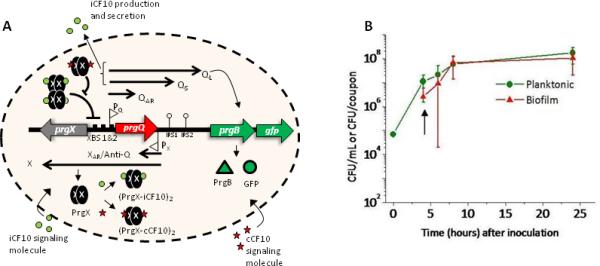
A. The gene regulatory network of prgQ-prgX genetic switch controlling onset of conjugation in pCF10+ donor cells including the position where gfp was fused in-frame to prgB. B. Growth of biofilm and planktonic cells in the CDC Biofilm Reactor. After 4 hours of batch culture growth, the reactor was operated as a continuous flow system at a rate of 8 mL/min (arrow). The viable populations of biofilm and planktonic bacteria were enumerated by plate counts as described in Materials and Methods. Error bars show standard deviation using n≥3 separate experiments.
We previously showed that an in vivo biofilm, namely an endocarditis vegetation, could serve as niche for high-frequency transfer of pCF10 (Hirt et al., 2002, Hirt et al., 2005). We became interested in a systematic analysis of peptide signaling and plasmid transfer in biofilms using the pCF10 system as a model. To initiate these studies we generated a pheromone-inducible fluorescent reporter construct and compared pheromone responses in planktonic versus biofilm cultures. A gfp reporter was trascriptionally fused in-frame to prgB, the gene encoding Aggregation Substance (Asc10) in the pCF10 conjugative plasmid (Figure 1A). As Asc10 production facilitates conjugative transfer of pCF10, the expression of GFP is used as a reporter of conjugation readiness. Using this reporter strain, we found a remarkable difference in the population dynamics of the pheromone response under different growth conditions, consistent with the hypothesis that biofilm growth results in formation of distinct cell types that impact the behavior and regulation of pCF10 transfer.
Results
Cells carrying the GFP reporter are inducible with cCF10 and are growing and viable in both biofilms and liquid culture
Because the experiments described below analyzed the pheromone response of cells carrying the GFP reporter plasmid in biofilm cells versus planktonic cells, we carried out initial experiments to assess the growth of planktonic and biofilm populations grown in the CDC biofilm reactor (CBR) system. In the 4-8 hour time frame, the population densities of both the biofilm and planktonic cells increased to similar extents and then leveled off (Figure 1B). Determination of precise generation times for the two populations in this experiment was difficult because the planktonic cells were continually diluted after the initial 4h of static incubation, and because both growth and adherence could contribute to the population of biofilm cells. Nonetheless, the parallel increases in both populations suggested that they were both actively growing at similar rates, especially for the first 8-10 hours of the experiments. In this and future experiments, planktonic cells refer to those cells growing in the liquid phase of the CBR system (unless otherwise stated) although similar results were obtained with planktonic cells from overnight batch cultures.
Cells containing the pCF10-GFP fusion expressed GFP in response to addition of exogenous cCF10 in liquid culture as well as in biofilms grown in the CBR. When induced with low levels of pheromone for a short period of time, many but not all of the induced cells were “turned on”, or expressed GFP (Figure 2A). Adding higher levels of pheromone for the same amount of time increased the proportion of GFP+ cells (Figure 2B). Under the low induction conditions, the majority of planktonic cells expressed GFP but were less bright than GFP-expressing biofilm cells exposed to the same pheromone concentration (compare Figure 2A and Figure S1A) and the GFP expression level increased with increased pheromone. GFP expression in planktonic cells following low levels of induction is difficult to distinguish from background but could be detected with careful microscopic observation (Figure S1A).
Figure 2. Biofilm cells expressing GFP following pheromone induction.
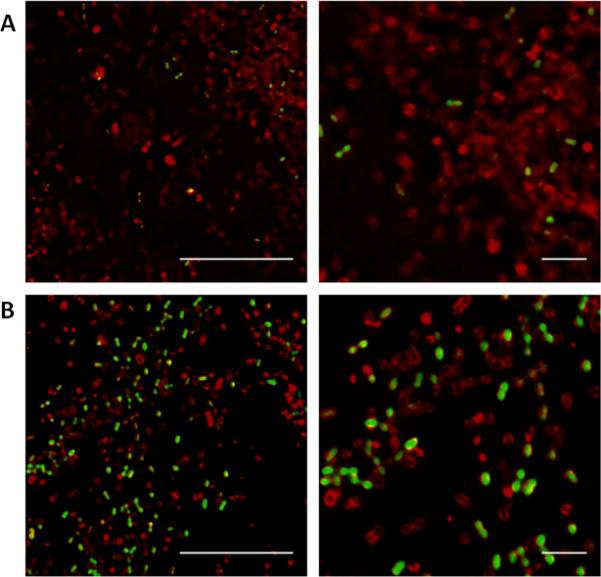
A. Cells induced for 30 minutes with 1ng/mL cCF10. B. Cells induced for 30 minutes with 10ng/mL cCF10. Right panels: Higher zoom of left panels. Green = GFP expression, Red = cell envelope stain. Scale bars: 10 μm (left) 2 μm (right). Images were taken using a 60x objective.
The pattern of conjugation induction differs between cells growing in liquid culture and those grown in a biofilm
The induction pattern of planktonic cells was quantified using a range of cCF10 concentrations with an induction time of 60 minutes. Flow cytometric analysis of planktonic cell induction patterns are shown in Figure 3A. As cells were induced with higher concentrations of cCF10, the entire planktonic population shifted to higher levels of GFP expression, apparently homogeneously (i.e. the detection system of the flow cytometer was not able to resolve two distinct populations). The histograms are representative of results obtained with pheromone titrations ranging from 0.1-10 ng/mL cCF10. A time dependent unimodal increase in GFP expression was observed when identical levels of cCF10 were used to induce planktonic cells for time periods ranging from 0-120 minutes (Figure S2). When cells growing in the biofilm state were similarly induced, the induction pattern was markedly different (Figure 3B). At low concentrations of pheromone, a portion of the cells expressed GFP and formed a small subpopulation distinct from the larger population. As inducer concentrations increased, the subpopulation expressing GFP increased and the proportion of cells not expressing GFP concurrently decreased. This suggests that the cells that were non-responsive at low concentrations of pheromone exposure have the potential to respond to higher pheromone concentrations; cumulative data suggest that at saturating induction levels, >95% of biofilm and planktonic cells express gfp. A bimodal pattern of pheromone response of biofilm cells was also observed when the time course of the response was examined (Figure S2B). We subjected pheromone-treated biofilms to propidium iodide staining to assess the viability of the GFP –positive and –negative populations, and found very low numbers of potential non-viable cells in either population (<2% in several different experiments, as illustrated in Fig 3D). These data rule out cell death as a reason for the lack of pheromone response in the GFP-negative cells.
Figure 3. Growth in a biofilm alters the induction pattern of pCF10 conjugation.
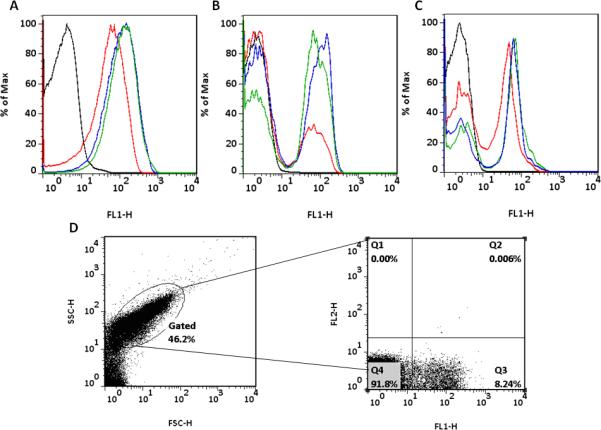
A. Cells grown in a liquid culture were induced for 60 minutes with various concentrations of cCF10. Induced populations shifted to higher GFP expression in a unimodal pattern. B. Coupons containing biofilm cells were induced for 60 minutes with various concentrations of cCF10. Induced cells expressed GFP in a bimodal population distribution. C. Cells grown in a biofilm for 24 hours and then dispersed prior to a 60 minute induction with various concentrations of cCF10 behave as attached biofilm cells showing bimodal response to induction. Horizontal axis = GFP (FL1) expression, Vertical axis = % of maximum cell number. D. Flow cytometry analysis of 24 hour biofilms following induction with 1 ng/mL of cCF10 for 60 minutes. Left panel demonstrates the populations gated to remove debris following biofilm dispersal based on size (FSC) and granularity (SSC). Right panel indicating propidium iodide (PI) staining (FL2) on the y-axis and GFP expression (FL1) on the x-axis. Less than 2% of the sorted cells stained with PI; similar numbers were seen with uninduced cells.
The biofilms used for the induction experiments shown in Figure 3 were grown for 24h, which produced sufficient numbers of bacterial cells for analysis from a relatively small number of coupons. However, by substantially increasing the number of coupons, we were able to do similar induction experiments with 4 hour biofilms, and obtained essentially identical results. This suggests that differentiation of the biofilm cells into distinct sub-populations occurs early in development, while the adherent bacteria are actively growing (Figure 1B). Furthermore, we have carried out numerous experiments involving induction of planktonic cells (including the planktonic cells from the same reactors used to harvest the biofilms) where the nutrient content of the medium during pre-growth and induction was varied by diluting the M9-YE growth medium to various concentrations ranging from 10-100%, or by using tryptic soy broth. In all of these experiments (not shown) a unimodal induction pattern similar to that depicted in Figure 3A was observed, suggesting that biofilm growth was a more important determinant of the bimodal response than nutrient content or growth rate.
A structural component of the biofilm that could cause the biofilm cells to undergo different response patterns from planktonic cells is the biofilm matrix. Most simply, the matrix could inhibit pheromone induction of some cells by interfering with signal diffusion. The matrix could also serve to concentrate the pheromone in certain areas to stimulate cell induction in the immediate vicinity. To test for these possibilities, coupons containing biofilm cells were vortexed to release them from the matrix and suspended in a 50% concentration of minimal liquid medium prior to pheromone induction. The overall induction pattern of dispersed biofilm cells was the same as that of attached biofilm cells (Figure 3C) demonstrating that the effects of the biofilm matrix on cCF10 diffusion is not a major factor in the difference responses to pheromone observed between biofilm and planktonic cells.
We also tested the induction profile of a GFP fusion construct derived from pCF10 where transcription was driven by the same promoter, but where the gene encoding pheromone receptor/conjugation repressor protein, PrgX, was deleted. In this case, GFP expression was constitutive, unimodal, and unresponsive to pheromone induction (Figure S3A). Adding prgX in trans rescued the bimodal response (Figure S3B). From this we conclude that the bimodal distribution in GFP expression observed with the pheromone-inducible construct arises from biofilm effects on the native regulatory machinery and not due to random inhibition of GFP expression in a subpopulation of the biofilm cells.
The results described above suggest that at limiting concentrations of pheromone typically produced by recipient cells, the overall frequency of plasmid transfer might be lower in biofilms than in planktonic cultures. We examined this possibility by comparing transfer frequencies in the planktonic and biofilm subpopulations of CDC reactors containing mixed cultures of donors and recipients, and found that the overall efficiency of transfer was significantly lower in the biofilm phase (Table 1).
Table 1.
Conjugation rates in biofilms and liquid culture
| Cell counts** | ||||
|---|---|---|---|---|
| Culture | Donors | Recipients | Transconjugants | Transconjugant:Donor |
| Biofilm | 2.5×109 | 1.3×109 | 5.6×104 | 1:2.2×105 |
| Planktonic | 1.0×109 | 5.9×108 | 1.0×107 | 1:102 |
Numbers are given in CFU/membrane for biofilms and CFU/mL for planktonic cells. Biofilm membranes have surface area of ~190cm2 as described in methods.
Mathematical modeling suggests that pCF10 copy number changes resulting from biofilm growth could alter the pheromone response
To explore possible causes of the differing response to pheromone induction, we employed a mathematical model for the pCF10 genetic network (Figure 1A). The prgQ-prgX gene pair controls a genetic switch for the onset of conjugation in the pCF10 based system. Recent work has demonstrated that transcriptional interference and antisense RNA regulation due to the overlapping configuration of prgQ and prgX operons, confers on the pCF10 system a bistable switch-like behavior in response to induction with pheromone (Chatterjee et al., 2011). Transcriptional interference (TI) occurs due to collision between elongating RNAP within the overlapping DNA between promoters PQ and PX and causes generation of shorter truncated RNA within the overlapping locus. This results in mutual suppression of transcription from both promoters, with expression from the aggressive promoter prevailing over the weak one (Shearwin et al., 2005). In addition to TI, the complementary RNAs originating from the convergent promoters have been shown to negatively regulate one another due to base pairing interactions (Shokeen et al., 2010). The resultant regulatory circuit for this system is summarized in Figure 4A. The reciprocal regulation of the two operons via both TI and antisense regulation results in a double-negative feedback loop (Figure 4A) contributing to the bistable response of this system.
Figure 4. Modeling the effect of pCF10 plasmid copy number (N) on the cCF10 pheromone response.
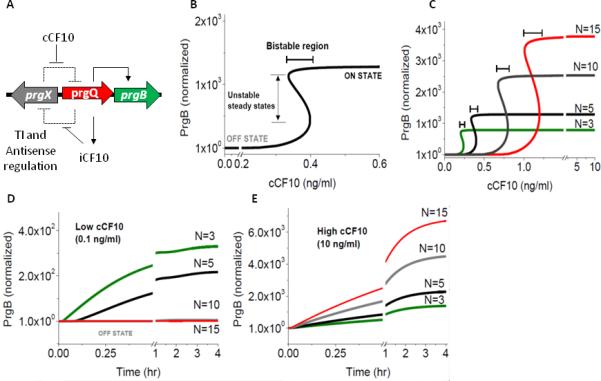
The data illustrated in this figure were produced by simulations based on the mathematical model described in the text and in the supplementary material. A. Regulatory circuit of pCF10-based conjugation. The prgQ operon encodes the pCF10 conjugation machinery, including an aggregation protein Asc10 encoded by prgB. The prgQ orf encodes the inihibitor peptide iCF10, which can bind to PrgX, increasing repression of the prgQ promoter, whereas binding of the chromosomally-encoded pheromone cCF10 to PrgX has the opposite effect. The promoters for these operons are located within the same region, but on the opposite strands of pCF10, and produce transcripts that are complementary for 220 nt at the 5’ ends. This arrangement leads to reciprocal negative regulation by antisense interactions and transcription interference, resulting in the double negative feedback loop illustrated by the dashed lines. B. Mathematical modeling predicts that the steady state response of PrgB levels to induction with cCF10 shows bistable switch behavior. The bistable regions described in the text are indicated. C. Bistable switch response of pCF10 to induction with cCF10 for different plasmid copy numbers. The bistable region increases with plasmid copy number along with an increased threshold cCF10 concentration required to turn “on” the genetic switch. PrgB levels shown in b-c are normalized to initial “off” steady state at 0 ng/mL of cCF10. D-E. Dynamic response of donor cells with different plasmid copy numbers to low (0.1 ng/mL, D) and high (10ng/mL, E) induction concentrations of cCF10. Cells with high plasmid copy number respond slowest to induction and at a higher concentration of cCF10 whereas low copy number cells respond faster and at a lower concentration of cCF10. PrgB levels shown in D-E are normalized to initial state at time t=0 corresponding to steady state levels at 0 ng/mL of cCF10.
Based on the gene regulatory network shown in Figures 1A and 4A, a set of nine ordinary differential equations (ODEs) based on mass-action kinetics for RNA species QS, QL, QAR, X and XAR, PrgB protein and iCF10 and cCF10 were generated as described (Equations S1-S9 in Supplemental Notes). The notation of all species and the parameters used for the simulation are summarized Table S3. The ODEs describing transcripts QS, QL, QAR, X and XAR in Equations S1-S5 are a balance of transcription rate, rate of removal due to antisense interactions, degradation and dilution due to growth. Protein PrgB shown in Equation S6, is balance of rate of PrgB production, degradation and dilution terms. The dynamics of extracellular iCF10 (i), intracellular iCF10 (I) and intracellular cCF10 (C) is shown in Equations S7-S9. The transport of signaling molecules cCF10 and iCF10 across the cell membrane is modeled as a first order reaction (Equation S8-S9). Generation of extracellular iCF10 is modeled as first order with respect to both QS and QL RNA. Dilution due to growth is not considered for extracellular iCF10, however, extracellular degradation is considered in Equation S7.
ODEs S1-S9 were solved for steady state and dynamic solution for fixed extracellular concentration of cCF10 (Figure 4B-E). A characteristic S-shaped bistable response of PrgB to extracellular cCF10 is shown in Figure 4B. The bistable curve is comprised of three sections: the lower and upper parts of the curve correspond, respectively, to “off” (low level of PrgB) and “on” states (high level of PrgB) of the donor cell. The middle section is characterized by multiple steady states, two stable states corresponding to “on” and “off” and an unstable steady state which is not observed experimentally. The separation between “off” and “on” state is characterized by the width of the bistable region, i.e. the region with multiple steady states (Figure 4B). This predicted bistable behavior is intrinsic to the gene regulatory network and would be present both in planktonic cells as well as biofilm cells.
Mathematical modeling suggested that an increase in the copy number of pCF10 could greatly alter induction responses to cCF10 (Figure 4C and Figure S4A). The effect of increased plasmid copy number manifests itself through increased number of operator sites (XBS 1 and 2) available for iCF10 bound PrgX tetramers and cCF10 bound PrgX dimers to bind (Equation S10) and an increase in the number of copies of QS and QL transcripts per donor cell, which consequently results in increased production of inhibitor iCF10. Donor cells with fewer copies of pCF10 are predicted to require lower amounts of cCF10 to turn “on” but respond with a lower PrgB expression level than cells with a higher copy number (Figure 4C). Interestingly, increasing plasmid copy number widens the bistable region, suggesting that the “on” and “off” populations are better separated. Similarly, the model predicts that cells with high plasmid copy number respond slowest to induction and require a higher concentration of cCF10 whereas low copy number cells respond faster and at a lower concentration of cCF10 (Figure 4D-E). A broader plasmid copy number distribution is predicted to give rise to a bimodal population response to low levels of inducer at longer exposure times, such that cells with high plasmid copy number continue to exist in the “off” state even after long exposure time to inducer, whereas cells with lower copy number switch to “on” state (Figure 4D-E and Figure S4B-C), consistent with experimental data shown in Figure S2. Our model predicts that a higher pCF10 copy number and copy number heterogeneity would enhance a bimodal response to induction with pheromone.
pCF10 copy number and heterogeneity is increased in biofilm cells
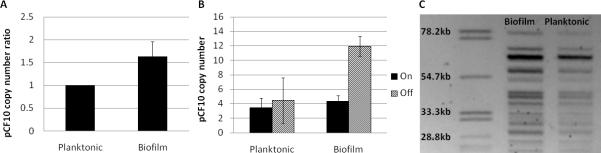
To examine this hypothesis experimentally, we performed qPCR on genomic DNA obtained from pCF10-containing planktonic and biofilm cells to compare the pCF10 copy number of the respective populations (Figure 4C). A statistically significant increase in pCF10 copy number of 1.5-2 times that of planktonic cells was observed using qPCR (Figure 5A). A complementary approach using pulse-field gel electrophoresis showed an increase in pCF10 copy number of approximately1.23 times as determined by band density analysis (Figure 5C). The slightly lower estimate from PFGE is understandable based on the band saturation of the pCF10 band in the biofilm cells.
Figure 5. The copy number of pCF10 as well as the population heterogeneity of copy number is increased in biofilm cells compared to planktonic cells.
A. Analysis of pCF10 copy number in uninduced biofilm and planktonic cells by qPCR. The average copy number of biofilm cells is 1.81±0.49 times great than that of planktonic cells. n=5 independent experiments. B. Comparison of pCF10 copy number in biofilm and planktonic cells exposed to cCF10. Biofilm and planktonic cultures were induced with cCF10 for 60 minutes and sorted into “on” and “off” populations. The sorted subpopulations were then analyzed for pCF10 copy number by qPCR. Induced biofilm cells not expressing GFP had a pCF10 copy number 2.77±0.12 times higher than induced cells expressing GFP. Sorted planktonic populations were not statistically different. All planktonic cells contained 3-5 copies of pCF10/chromosome whereas biofilm cells contain up to 15 copies of pCF10/chromosome with statistically significant heterogeneity between the “on” versus “off” subpopulations. n=4 independent experiments ± and error bars represent standard deviation ** P value <0.02 ***P value < 0.003 P values were calculated using a two-tailed t-test assuming equal variance. C. Pulse field gel electrophoresis (PFGE) of biofilm and planktonic cells shows that biofilm cells have a higher pCF10 DNA/chromosomal DNA ratio than planktonic cells. Arrow indicates pCF10 band. Using three different chromosomal bands for reference, the ratio of Biofilm pCF10/Planktonic pCF10 = 1.23±0.01
To further examine the relationship between biofilm growth plasmid copy number and pheromone response, biofilm cells were induced using conditions that generated two approximately equal subpopulations (e.g. Figure 3B). These cells were then sorted based on GFP expression and qPCR was used to examine pCF10 copy number in the “on” and “off” subpopulations. Even though the entire population of biofilm cells was exposed to the same concentration of inducer, the copy number of the pCF10 plasmid differed significantly between cells that expressed GFP and those that did not. Cells not expressing GFP had a statistically higher copy number of pCF10 than cells that expressed GFP during the same induction course (Figure 5B, biofilm). Planktonic cells were also induced with the same level of cCF10 and cells from the single induced peak were sorted. Cells labeled “on” represent cells from 25% of the peak with the highest GFP expression levels and those labeled “off” are cells from the 25% of the peak with the lowest expression of GFP. When cells from these populations were examined for copy number heterogeneity, no statistical difference was observed (Figure 5B, planktonic). This demonstrates that biofilm cells have, on average, higher copy numbers of the conjugative plasmid pCF10 than their planktonic counterparts and also possess a greater heterogeneity in copy number compared to planktonic populations consistent with our mathematical model (Figure 4C-E). A substantial difference in plasmid copy number was also observed between induced biofilm and planktonic cells. Planktonic cells contained pCF10 at 3-5 copies per chromosome (Figure 5B, planktonic) while induced biofilm cells possessed plasmid copy numbers as high as 8-15 copies per chromosome (Figure 5B, biofilm).
Discussion
Control of expression of conjugation functions in the pCF10 system is complex and involves competing antagonistic activities of two secreted signaling peptides and multiple intracellular regulatory circuits acting at the level of transcription initiation, as well post-transcriptionally (Figure 1A and (Dunny & Johnson, 2011)). There is experimental evidence demonstrating that the two-peptide signaling system increases versatility such that a response can be activated either by the presence of potential recipient cells in close proximity or by growth in the mammalian bloodstream (Chandler et al., 2005b), where expression of the pheromone-inducible prgB (Aggregation Substance) gene increases virulence (Chuang et al., 2009, Hirt et al., 2002). However, until recently the relative importance of the multiple (potentially redundant) intracellular mechanisms of regulation of expression of the pCF10 prgQ conjugation operon was not clear. We recently carried out modeling studies and quantitative analysis of transcript levels from the prgQ and prgX operons in response to cCF10 in planktonic cultures, using genetic constructs to analyze the specific contributions of the individual regulatory mechanisms to the system (Chatterjee et al., 2011). These studies suggested that the pheromone response system could function as a sensitive bistable genetic switch, and that disruption of any of the individual regulatory circuits of the system abolished switch behavior. A key finding consistent with the switch model was the observation of a distinct “jump” in the production of QL RNA following exposure to pheromone concentrations in the range of 1-5 ng/ml. Limitations of this study include the fact that the measurements of transcript levels by qRT-PCR represented the averages for the population neglecting possible heterogeneity in the response, and that only one growth condition was examined.
In the present study, we used a pheromone-inducible GFP reporter construct to allow for expression analysis on a single cell level, and we also examined the effects of biofilm growth on the population dynamics of the response. At first glance, the results reported here for planktonic cells appear to be inconsistent with our previous studies. However, considering the surprising effects of biofilm growth on increasing both the average plasmid copy number and copy number heterogeneity (Figure 5), our cumulative results are consistent with and validate the predictions of mathematical models described here (Figure 4) and in previous experiments (Chatterjee et al., 2011).
While fluorescent reporter proteins facilitate single cell expression analysis, they do have limitations for our system, including the fact that the fermentative metabolism of enterococci may result in a reduced cytoplasmic environment suboptimal for proper GFP folding. Also, the gfp allele we used encodes a stable protein (to date none of the unstable versions tried produce adequate signal levels). These factors probably reduced the effective signal to noise ratio in our experiments. We suspect that the apparent lack of bimodal response in planktonic cultures might reflect the limitations of the detection system. It is likely that the low copy number of pCF10 in planktonic cells created a lower threshold for switch behavior to occur and also caused the cells to respond at a lower level blurring the distinction between “on” and “off” (Figure 4C). In any case, there is a clear difference in the biology of the pheromone response in the two types of cells, with biofilm growth requiring an increased level of signal, but resulting in a more vigorous response to pheromone. This suggested that the average frequency of conjugative plasmid transfer in biofilms could be lower than that of planktonic cells, which was observed experimentally (Table 1).
An unexpected result of this study was the effect of biofilm growth on pCF10 copy number (Figure 5). Our mathematical model suggested that changes in plasmid copy number could account for the observed effects of biofilm growth on the pheromone response (Figure 4). While the average copy number of the entire population of biofilm cells was slightly increased, biofilm cells also exhibited a remarkable increase in the heterogeneity of copy number values, which was revealed by analysis of sorted “on” and “off” populations exposed to threshold inducing concentrations of pheromone. As noted above, cells with more copies of pCF10 are more adequately poised to respond to cCF10 pheromone in an appropriate and highly controlled manner than adjacent cells with lower plasmid copy number. By requiring higher pheromone concentrations to turn on conjugation, these cells avoid the expenditure of large amounts of energy involved in producing conjugative machinery. Biofilms are often seen as a type of multicellular community and allowing for conjugation to occur only in a subset of potential donor cells may be beneficial to the community as a whole.
While the bacteria in biofilms are sometimes described as non-growing, or slowly growing, we observed the bimodal pheromone response in cells grown for as little as 4 hours on surfaces, when adherent cell densities were quite low and populations were increasing rapidly (data not shown). This observation coupled with the apparent unimodal response of planktonic cells grown in a variety of nutrient conditions suggests that aspects of biofilm development not directly related to changes in generation time may affect plasmid copy number and lead to the observed changes in the pheromone response. It will be of interest to examine effects of biofilm growth on the copy number of different plasmid replicons. Finally, the physiological basis for the heterogeneity of copy number and pheromone response within the biofilm population is of great interest, and it should be feasible to carry out transcriptomic analysis of sorted responsive and non-responsive biofilm subpopulations, as has been reported for biofilm cells of Streptococcus mutans exposed to a competence-inducing peptide pheromone (Lemme et al., 2011).
The reduced efficiency of pCF10 transfer in biofilms runs contrary to the popular notion of biofilms being the optimal niche for conjugation (Hausner & Wuertz, 1999). The conjugation experiments reported here employed biofilms grown in vitro with inducing pheromone produced by recipient cells; in the context of a bloodstream infection, we previously showed induction of the transfer system via a plasma component (Chandler et al., 2005b) This mechanism could enhance plasmid transfer during experimental endocarditis, as described by Hirt et al. (2002). The present results also reflect the anatomy of enterococci and differences in the cell attachment mechanisms employed by the conjugative transfer machines of gram positive versus gram negative bacteria. E. faecalis has no known mechanisms for motility in liquids or on solid surfaces. When planktonic cells colonize a surface and initiate biofilm growth, or attach and become part of a pre-existing biofilm, they likely remain in the same location until they either die or re-enter the planktonic phase by detaching from the biofilm. In the pCF10 system, mating pair formation is mediated by the surface adhesin Asc10 (encoded by prgB), which can stably bind the surfaces of cells that randomly collide; there are no extended sex pili that could attach cells that do not come into direct wall-to-wall contact. We believe that the evolution of the pheromone response system has been driven to allow induction of conjugation in biofilms only when donor cells happen to be in extremely close proximity to recipients (perhaps in direct contact), so that the energetically expensive conjugation machinery is only produced when it can be effectively utilized. In planktonic cultures of sufficient population density, random diffusion increases the probability of collision between donors and recipients, and induced donors can form stable mating pairs extremely efficiently under these conditions. In this model, the selective pressures for effective spread of the plasmid are balanced by those operating to minimize the metabolic burden of synthesizing the plasmid-encoded proteins. This hypothesis may be tested experimentally by examining conjugation and the pheromone responses of individual donor cells growing in mixed biofilm communities with recipients. We are currently developing the tools to carry out such experiments.
Experimental Procedures
Bacterial strains and medium
All bacterial strains and plasmids are listed in Table S1. MM9YEG medium (Dunny & Clewell, 1975), a semi-defined M9-based medium supplemented with 0.3% yeast extract, 1% casamino acids, 20mM glucose, 1mM MgSO4 and 0.1mM CaCl2 was used for all experiments unless otherwise states. When required, the following antibiotics were used to supplement the medium in overnight cultures: 10μg/mL tetracycline (Tc), 200 μg/mL rifampicin (Rif), 20 μg/mL chloramphenicol (Cl), 50 μg/mL erythromycin (Erm), and 1000 μg/mL spectinomycin (Spec) and streptomycin (Strep). X-Gal was added at a concentration of 250 μg/mL.
Strain construction of OG1RF+pCF10-GFP
The gfpmut3b gene was inserted into the pCF10 plasmid using a double crossover method previously described (Kristich et al., 2007). Briefly, approximately 800 base pairs of the downstream region of prgB, the ribosomal binding site of prgB, the gfp gene, and approximately 800 base pairs of the upstream region of prgC were joined, in that order, via a two-step PCR method (Table S2). This construct was then inserted into the NotI site of the multiple cloning region of the pCJK47 plasmid creating pCJK47-LCC1. The plasmid was electroporated into E. coli strain EC1000 for propagation and then into E. faecalis strain CK111-pCF10-101. This strain was then conjugated with OG1RF recipient cells to ensure delivery of pCJK47-LCC1 into OG1RF cells. Transconjugants were selected based on antibiotic resistance and were propagated without selection until colonies with the double crossover phenotype arose. These colonies were grown overnight, diluted 1:20 in fresh MM9YEG medium and induced using 10 ng/mL cCF10 pheromone. Correct insertion of GFP gave rise to colonies that fluoresced green following a 60 minute induction as measured by a fluorometer (Turner Biosystems/Promega, Sunnyvale, CA).
Conjugation experiments
Creation of certain strains required conjugative mating between strains of E. faecalis. Matings were done on Brain Heart Infusion (BHI) agar plates (solid mating). Donor and recipient strains were grown to late exponential phase (approximately 2×109 CFU/mL) in BHI and then diluted 1:10 into fresh BHI medium. After 90 minutes of growth at 37°C, the populations were mixed at a donor:recipient ratio of 1:9. 800 μL of this mixture was spun down and plated onto BHI agar plates without antibiotic selection. Plates were incubated at 37° C for 16 hours. Cells were removed from the plate by addition of 1 mL of PBS supplemented with 2mM EDTA followed by scraping. EDTA is used in these and other experiments to break clumps of cells that arise after mating pair formation in to single cells. Serial dilutions were plated onto donor, recipient, and tranconjugant-selective agar plates and incubated for 24 hours at 37°C. For measurement of mating transfer efficiencies of biofilm and planktonic cells, overnight cultures of donors and recipients were diluted 1:10 into fresh BHI medium and grown for 90 minutes at 37°C. The cells were then mixed at a donor:recipient ratio of 1:9 and 1 mL of the mixture was added to each well of a 6 well dish. Autoclaved circular aclar membranes (Honeywell Inc., Morristown, NJ) of 11mm in diameter (~95mm2 surface area/side) were added to each well, providing a surface for biofilm formation. The 6 well dish was placed on a shaker at 37°C for 4 hours. Cells remaining in the liquid phase were left in the plate. The aclar membranes were then removed, rinsed twice in sterile water, and place into a new 6 well plate with 1 mL of fresh sterile BHI medium. Both 6 well dishes were then placed back on the shaker and remained at 37°C for 20 hours. Following 24 hours of mating, the liquid cells were collected from the original 6 well dish. Aclar membranes were removed from the remaining plate and rinsed twice in sterile water. The membranes were then placed in 2 mL microcentrifuge tubes with 1 mL of PBS+2mM EDTA and vortexed for 3-5 minutes at 4°C to remove biofilm cells. Biofilm and planktonic cells were then spun down and washed twice in 1 mL PBS+ 2mM EDTA. Serial dilutions were plated onto donor, recipient, and tranconjugant-selective agar plates and incubated for 24 hours at 37°C.
Biofilm growth and induction
Biofilms were grown using the CDC Biofilm Reactor (CBR) (Bio Surface Technologies Corp., Bozeman, MT). Experiments were done at 37° C except vortexing which was done at 4° C. MM9YEG medium was used as the growth medium for overnight cultures as well as for the batch culture medium. 10% MM9YEG was used in the carboy for the continuous flow portion of growth. The CBR protocol followed the instruction manual guidelines with few minor changes. Briefly, after autoclaving, the reactor and carboy setup were placed in a 37° C room with the reactor on a stir plate rotating at a speed of approximately 125 rotations per minute. 2 mL of overnight culture at a cell density of approximately 2×109 cfu/mL was added to the reactor aseptically. The reactor was run as a batch culture for 4 hours to allow bacterial attachment to the coupons. Tubing was run through a peristaltic pump allowing addition of medium to the reactor at a rate of 8 mL/min . At this point, the coupons were either sampled or subjected to a continuous flow of medium for 20 hours. To retrieve the coupons, one polypropylene rod was removed from the reactor and the three coupons were unscrewed and placed in one well of a 6 well dish. 5 mL of sterile water was then added to each well and the plate was placed onto a rotating platform for three minutes to remove planktonic cells. Coupons were moved to a 24 well plate. Pheromone cCF10 was diluted to various concentrations in 50% MM9YEG. 1 mL of the pheromone mixture was added to each well to completely submerge the coupon and the plate was placed on a rocking platform at 37° C for 0-120 minutes (60 minutes unless otherwise stated). Following induction, coupons were removed to 15 mL conical tubes containing 3 mL of PBS supplemented with 2mM EDTA and vortexed for 3 minutes at 4° C to remove attached biofilm cells and break up cell clumps. Preliminary data showed that vortexing for 3 minutes at 4° C gave the highest cell yield and lowest variability compared to sonication and vortexing at room temperature. Quantification of cells was done by making dilutions into PBS supplemented with 2mM EDTA and plating dilutions onto BHI plates with applicable antibiotics and incubating overnight at 37°C.
Flow cytometry and Fluorescent Activated Cell Sorting (FACS)
Cell suspensions were analyzed via flow cytometry using a FACSCalibur flow cytometer (BD Biosciences, Rockville, MD). Cells were analyzed by size and granularity and dead cells and debris were gated out of the population. Cells did not require filtering and the vast majority of cells fell into the same region on the forward and side scatter plot indicating that the majority of cell clumps were broken up by EDTA treatment (Figure 3D). Expression of GFP was measured for 100,000-200,000 live cells using the 488 nm laser excitation line. Data from flow cytometry experiments are shown as histograms and contour plots. Data was analyzed using FlowJo (Tree Star, Ashland, OR). FACS was performed using a FACSAria II and cells were gated, analyzed, and sorted using FACSDiva software (BD Biosciences).
Genomic DNA preparation and quantification
Genomic DNA was obtained from 24 hour biofilms, liquid from the biofilm reactor, sorted cells, and a reference strain (100-5) containing one copy of prgX in the chromosome. Samples were taken from ≥ 3 independent experiments for each data point shown. If frozen prior to analysis, the samples were immediately pelleted and cell pellets were frozen no more than 16 hours before genomic DNA preparation. DNA preparations were completed using the Qiagen DNeasy Blood and Tissue Kit (Qiagen Inc, Valencia, CA) following the procedure for extraction of genomic DNA from Gram-positive cells. Nucleic acid quantification was done using the Quant-iT PicoGreen dsDNA kit (Invitrogen, Carlsbad, CA). Samples were mixed in black 96 well plates and read on a fluorometer (Ex: 490nm, Em: 510-570nm, Turner Biosystems).
Quantitative Real Time PCR (qRT-PCR)
To determine the copy number of pCF10 in planktonic and biofilm populations, genomic DNA was obtain from stationary phase 24 hour planktonic cultures, 24 hour biofilms on polycarbonate coupons, sorted cells, and a reference strain, 100-5 containing one copy of prgX in the chromosome. Primers used for the reference gene gyrB were previously published (Bourgogne et al., 2007). The primers for prgX are listed in Table S2. qRT-PCR was done using the SYBR green Supermix and an iCycler iQ5 (Bio-Rad) instrument. Each well contained 25μL of total reaction with 200nM primer concentration. Primer efficiencies and Ct measurements were carried out by iQ5 Optical System Software Version 2.0 (2006, Bio-Rad). . All experiments met the following criteria for statistical analysis; efficiencies ≥80%, calibration curve r2 values of ≥0.98, and Ct values for no template control samples ≥35 cycles. Data was analyzed using the Pfaffl method for relative quantification (Pfaffl, 2001). Technical replicates showed no intra-assay variation and were not used in the final statistical analysis.
Statistical analysis
qPCR data are based on n≥3 biological replicates. In each experiment, ≥2 dilutions were used for each sample to compare to ≥4 dilutions of the control 100-5 DNA. All samples were run in triplicate and any wells which had standard deviation of ≥0.4 from the other two samples were not used. If removal of one well did not decrease the standard deviation below 0.4, the dilution was not used. Statistical significance was determined by a paired two-sample t test (Excel 2010, Microsoft, Redmond, WA). P values were considered statistically significant if they were ≤0.01. Exact P values are shown in figure legends.
Microscopy
Coupons with GFP-expressing E. faecalis cells were counterstained with a red fluorescent Alexa Fluor 594: Wheat Germ Agglutinin conjugate (Invitrogen) labeling the cell envelope and mounted in VECTASHIELD (Vector Laboratories, Burlingame, CA) immediately prior to image acquisition. Images were acquired with a Cascade 1K EMCCD camera (Photometrics, Tucson, AZ) as widefield z-stacks with a 60× 1.4 NA objective (Nikon Instruments, Melville, NY). Z-stacks were taken at 0.2 μm intervals, and were deconvolved using Huygens Pro (Scientific Volume Imaging, The Netherlands). Processed images were aligned and projected as a best-focus composites using MetaMorph (Molecular Devices, Sunnyvale, CA).
Pulse Field Gel Electrophoresis
Pulse Field Gel Electrophoresis (PFGE) was done as previously described with minor alterations (Natarajan et al., 1999). Biofilm and planktonic cells from an overnight CBR experiment were grown and harvested as above. Cells were then washed twice and resuspended in 1 mL PBS with 2mM EDTA and chilled on ice. The cells were diluted to final OD600 values of 0.86 for biofilm cells and 0.43 for planktonic cells using sterile TE Buffer (10 mM Tris-Cl, pH 8.0, 1 mM EDTA). Plate counts showed that the CFU/mL values at these empirically derived ODs were similar. The gel standard used in this experiment was Salmonella enterica serotype Braenderup H9812 (ATCC BAA-664) (provided by the Minnesota Department of Health); XhoI was the primary restriction enzyme. The gel was run on a CHEF-DR III System (Bio-Rad) at an initial switch time of 2.0 seconds and a final switch time of 10.0 seconds for 13 hours followed by an initial switch time of 20.0 seconds and final switch time of 25.0 seconds for 6 hours. A Gel Doc XR System with Quantity One software (Bio-Rad) was used to capture and convert the gel images.
Mathematical Modeling
A set of ordinary differential equations based on pCF10 genetic network were developed (Figure 1A). Steady state and dynamic simulations were performed in MATLAB (version 2008a MathWorks, Natick, MA). Steady state solutions of non-linear algebraic equations were obtained using MATLAB solve function. ODEs were integrated using function ode23s in MATLAB to obtain dynamic behavior.
Supplementary Material
Acknowledgments
We thank the Dr. David Boxrud and Selina Jawahir at the Minnesota Department of Health for running Pulse field gel electrophoresis. L.C. is the recipient of the American Academy of University Women American Fellowship (2010-2011) and the Biotechnology Training Grant (2007-2009) T32GM008347, A.B. received additional support from the NIH Medical Scientist Training Grant 2T32GM0082445, A.C. is the recipient of the University of Minnesota Doctoral Dissertation Fellowship. This work was supported by grants from the National Institutes of Health GM081888, GM049530, AI058134.
References
- Bae T, Clerc-Bardin S, Dunny G. Analysis of expression of prgX, a key negative regulator of the transfer of the Enterococcus faecalis pheromone-inducible plasmid pCF10. Journal of Molecular Biology. 2000;297:861–875. doi: 10.1006/jmbi.2000.3628. [DOI] [PubMed] [Google Scholar]
- Bae T, Kozlowicz T, Dunny G. Two targets in pCF10 DNA for PrgX bidning: their role in production of Qa and prgX mRNA and in regulation of pheromone-inducible conjugation. Journal of Moleclular Biology. 2002a;315:995–1007. doi: 10.1006/jmbi.2001.5294. [DOI] [PubMed] [Google Scholar]
- Bae T, Kozlowicz T, Dunny G. Two targets in pCF10 DNA for PrgX bidning: their role in production of Qa and prgX mRNA and in regulation of pheromone-inducible conjugation. Journal of Moleclular Biology. 2002b;315:995–1007. doi: 10.1006/jmbi.2001.5294. [DOI] [PubMed] [Google Scholar]
- Bae T, Kozlowicz B, Dunny G. Characterization of cis-active prgQ mutants: evidence for two distinct repression mechanisms by Anti-Q RNA and PrgX protein in pheromone-inducible enterococcal plasmid pCF10. Molecular Microbiology. 2004;51:271–281. doi: 10.1046/j.1365-2958.2003.03832.x. [DOI] [PubMed] [Google Scholar]
- Bensing B, Manias D, Dunny G. Pheromone cCF10 and plasmid pCF10-encoded regulatory molecules act post-transcriptionally to activate expression of downstream conjugation functions. Molecular Mircobiology. 1997;24:285–294. doi: 10.1046/j.1365-2958.1997.3301710.x. [DOI] [PubMed] [Google Scholar]
- Boles B, Horswill A. agr-Mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathogens. 2008:4. doi: 10.1371/journal.ppat.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgogne A, Singh K, Fox K, Pflughoeft K, Murray B, Garsin D. EbpR is important for biofilm formation by activating expression of the endocarditis and biofilm-associated pilus operon (ebpABC) of Enterococcus faecalis OG1RF. Journal of Bacteriology. 2007;19:4. doi: 10.1128/JB.00594-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler J, Flynn A, Bryan E, Dunny G. Specific control of endogenous cCF10 pheromone by conserved domain of the pCF10-encoded regulatory protein PrgY in Enterococcus faecalis. Journal of Bacteriology. 2005a;187:13. doi: 10.1128/JB.187.14.4830-4843.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler J, Hirt H, Dunny G. A paracrine peptide sex pheromone also acts as an autocrine signal to induce plasmid transfer and virulence factor expression in vivo. Proc Natl Acad Sci. 2005b;102:15617–15622. doi: 10.1073/pnas.0505545102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A, Johnson CM, Shu C-C, Kaznessis YN, Ramkrishna D, Dunny GM, Hu W-S. Convergent transcription confers a bistability switch in Enterococcus faecalis conjugation. Proceedings of the National Academy of Sciences. 2011;108:9721–9726. doi: 10.1073/pnas.1101569108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang O, Schlievert P, Wells C, Manias D, Tripp T, Dunny G. Multiple functional domains of Enterococcus faecalis aggregation substance Asc10 contribute to endocarditis virulence. Infection and Immunity. 2009;77:539–548. doi: 10.1128/IAI.01034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang-Smith O, Wells C, Henry-Stanley M, Dunny G. Acceleration of Enterococcus faecalis biofilm formation by aggregation substance expression in an ex vivo model of cardiac valve colonization. PLoS One. 2010;5:e15798. doi: 10.1371/journal.pone.0015798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J, Dunny G. Transcriptional analysis of a region of Enterococcus faecalis plasmid pCF10 involved in positive regulation of conjugative transfer functions. Journal of Bacteriology. 1995;177:2118–2124. doi: 10.1128/jb.177.8.2118-2124.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvitkovitch D. Genetic competence and transformation in oral streptococci. Critical Reviews in Oral Biology and Medicine. 2001;12:217–243. doi: 10.1177/10454411010120030201. [DOI] [PubMed] [Google Scholar]
- Dunny G, Clewell D. Transmissible toxin (hemolysin) plasmid in Streptococcus faecalis and its mobilization of a noninfectious drug resistance plasmid. Journal of Bacteriology. 1975;124:784–790. doi: 10.1128/jb.124.2.784-790.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunny G, Brown B, Clewell D. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proceedings of the National Academy of Sciences. 1978;75:3479–3483. doi: 10.1073/pnas.75.7.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunny G, Funk C, Adsit J. Direct stimulation and transfer of antibiotic resistance by sex pheromones in Streptococcus faecalis. Plasmid. 1981;6:270–278. doi: 10.1016/0147-619x(81)90035-4. [DOI] [PubMed] [Google Scholar]
- Dunny G, Johnson C. Regulatory circuits controlling enterococcal conjugation: lessons for functional genomics. Current Opinions in Microbiology. 2011 doi: 10.1016/j.mib.2011.01.008. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi Y, Itoh T, J T. Antisense RNA. Annual Reviews in Biochemistry. 1991;60:631–652. doi: 10.1146/annurev.bi.60.070191.003215. [DOI] [PubMed] [Google Scholar]
- Fixen K, Chandler J, Le T, Kozlowicz B, Manias D, Dunny G. Analysis of amino acid sequence specificity determinants of enterococcal cCF10 sex pheromone in interactions with the pheromone-sensing machinery. Journal of Bacteriology. 2007;189:1399–1406. doi: 10.1128/JB.01226-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghigo J. Natural conjugative plasmids induce bacterial biofilm development. Nature. 2001;412:442–445. doi: 10.1038/35086581. [DOI] [PubMed] [Google Scholar]
- Gold O, Jordan H, van Houte J. The prevalence of enterococci in the human mouth and their pathogenicity in animal models. Archives of Oral Biology. 1975;20:473–477. doi: 10.1016/0003-9969(75)90236-8. [DOI] [PubMed] [Google Scholar]
- Hausner M, Wuertz S. High rates of conjugation in bacterial biofilms as determined by quantitative in situ analysis. Applied and Environmental Microbiology. 1999;65:3710–3713. doi: 10.1128/aem.65.8.3710-3713.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt H, Manias D, Bryan E, Klein J, Marklund J, Staddon J, Paustian M, Kapur V, Dunny G. Characterization of the Pheromone Response of the Enterococcus faecalis Conjugative Plasmid pCF10: Complete Sequence and Comparative Analysis of the Transcriptional and Phenotypic Responses of pCF10-Containing Cells to Pheromone Induction. Journal of Bacteriology. 2005;187:1044–1054. doi: 10.1128/JB.187.3.1044-1054.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt H, Schlievert P, Dunny G. In vivo induction of virulence and antibiotic resistance transfer in Enterococcus faecalis mediated by the sex pheromone-sensing system of pCF10. Infection and Immunity. 2002;70:716–723. doi: 10.1128/iai.70.2.716-723.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C, Manias D, Haemig H, Shokeen S, Weaver K, Henkin T, Dunny G. Direct evidence for control of the pheromone-inducible prgQ operon of Enterococcus faecalis plasmid pCF10 by a countertranscript-driven attenuation mechanism. Journal of Bacteriology. 2010;192:1634–1642. doi: 10.1128/JB.01525-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlowicz B, Bae T, Dunny G. Enterococcus faecalis pheromone-responsive protein PrgX: genetic separation of positive autoregulatory functions from those involved in negative regulation of conjugative plasmid transfer. Molecular Microbiology. 2004;54:13. doi: 10.1111/j.1365-2958.2004.04286.x. [DOI] [PubMed] [Google Scholar]
- Kozlowicz B. Microbiology. University of Minnesota; Minneapolis, MN: 2005. The molecular mechanism and peptide signaling response of PrgX used to control pheromone-induced conjugative transfer of pCF10. [Google Scholar]
- Kristich C, Chandler J, Dunny G. Development of a host-genotype-independent counterselectable marker and a high-frequency conjugative delivery system and their use in genetic anaylsis of Enterococcus faecalis. Plasmid. 2007;57:131–144. doi: 10.1016/j.plasmid.2006.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leenhouts K, Buist G, Bolhuis A, tend Berge A, Kiel J, Mierau I, Dabrowska M, Venema G, Kok J. A general system for generating unlabelled gene replacements in bacterial chromosomes. Mol Gen Genet. 1996;353:217–224. doi: 10.1007/s004380050315. [DOI] [PubMed] [Google Scholar]
- Lemme A, Grobe L, Reck M, Tomasch J, Wagner-Dobler I. Subpopulation specific transcriptome analysis of CSP induced Streptococcus mutans. Journal of Bacteriology. 2011 doi: 10.1128/JB.01363-10. epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Lau P, Lee J, Ellen R, Cvitkovitch D. Natural genetic transformation of Streptococcus mutans growing in biofilms. Journal of Bacteriology. 2001;183:897–908. doi: 10.1128/JB.183.3.897-908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujan S, Guogas L, Ragonese H, Matson S, Redinbo M. Disrupting antibiotic resistance propagation by inhibiting the conjugative DNA relaxase. Proceedings of National Academy of Sciences. 2007;104:12282–12287. doi: 10.1073/pnas.0702760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehra S, Charaniya S, Takano E, Hu W. A bistable gene switch for antibiotic biosynthesis: the butyrolactone regulon in Streptomyces coelicolor. PLoS One. 2008;3:e2724. doi: 10.1371/journal.pone.0002724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molin S, Tolker-Nielsen T. Gene transfer occurs with enhanced efficiency in biofilms and induced enhanced stabilisation of the biofilm structure. Current Opinion in Biotechnology. 2003;14:255–261. doi: 10.1016/s0958-1669(03)00036-3. [DOI] [PubMed] [Google Scholar]
- Mori M, Sakagami Y, Isogai A, Kitada C, Fujino M, Adsit J, Dunny G, Suzuki A. Structre of cCF10nsfer of the Streptococcus faecalis tetracycline resistant plasmid, pCF10. Journal of Biological Chemistry. 1988;263:14574–14578. [PubMed] [Google Scholar]
- Nakayama J, Ruhfel R, Dunny G, Isogai A, Suzuki A. The prgQ gene of Enterococcus faecalis tetracycline resistance plasmid pCF10 encodes a peptide inhibitor, iCF10. Journal of Bacteriology. 1994;176:4. doi: 10.1128/jb.176.23.7405-7408.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan A, Boxrud D, Dunny G, Srienc F. Flow cytometric analysis of growth of two Streptococcus gordonii derivatives. Journal of Microbiological Methods. 1999;34:223–233. [Google Scholar]
- Nguyen K, Piastro K, Gray T, Derbyshire K. Mycobacterial biofilms facilitate horizontal DNA transfer between strains of Mycobacterium smegmatis. Journal of Bacteriology. 2010;192:5134–5142. doi: 10.1128/JB.00650-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux A, Payne S, Gilmore M. Microbial telesensing: probing the environment for friends, foes, and food. Cell Host Microbe. 2009;6:115–124. doi: 10.1016/j.chom.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearwin KE, Callen BP, Egan JB. Transcriptional interference - a crash course. Trends in Genetics. 2005;21:339–345. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shokeen S, Johnson CM, Greenfield TJ, Manias DA, Dunny GM, Weaver KE. Structural analysis of the Anti-Q-Qs interaction: RNA-mediated regulation of E. faecalis plasmid pCF10 conjugation. Plasmid. 2010;64:26–35. doi: 10.1016/j.plasmid.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiropoulos V, Kaznessis Y. Synthetic tetracycline-inducible regulatory networks: computer-aided design of dynamic phenotypes. BMC Systems Biology. 2007;1:7. doi: 10.1186/1752-0509-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staddon J, Bryan E, Manias D, Chen Y, Dunny G. Genetic characterization of the conjugative DNA processing system of enterococcal plasmid pCF10. Plasmid. 2006;56:102–111. doi: 10.1016/j.plasmid.2006.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.