Abstract
Nucleolin is a multifunctional phosphoprotein ubiquitously distributed in the nucleolus, nucleus and cytoplasm of the cell. Nucleolin has a bipartite nuclear localization signal sequence and is conserved in animals, plants and yeast. Its levels are correlated with the rate of functional activity of the nucleolus in exponentially growing cells. Nucleolin contains intrinsic DNA and RNA helicase, nucleic-acid-dependent ATPase and self-cleaving activities. It binds RNA through its RNA recognition motifs. It regulates various aspects of DNA and RNA metabolism, chromatin structure, rDNA transcription, rRNA maturation, cytokinesis, nucleogenesis, cell proliferation and growth, the folding, maturation and ribosome assembly and nucleocytoplasmic transport of newly synthesized pre-RNAs. In this review we present an overview on nucleolin, its localization, structure and various functions. We also describe the discovery and important studies of nucleolin in plants.
Keywords: cytoplasm, helicase, nucleolus, phosphoprotein, ribosome
Introduction
The nucleolus is the most prominent structure in a cell nucleus and is the site of the biogenesis of ribosomal RNA (rRNA) and 40S and 60S ribosomal subunits (r-subunits). In metabolically active animal and plant somatic cells and in yeast, the nucleolus contains tens to hundreds of active rRNA and r-subunit genes, which account for about one-half of the total cellular RNA production.1 There are several nuclear proteins in the nucleolus. Nucleolin is one of the most abundant non ribosomal proteins, which accounts for approximately 10% of the protein content within the nucleolus.2–4 This protein was first identified in Chinese hamster ovary (CHO) cells and Novikoff hepatoma cells.5 Whilst mammalian nucleolin has a predicted molecular mass of approximately 77 kDa (depending on the species), the apparent molecular mass is between 100 and 110 kDa, and has been attributed to the amino acid composition of the N-terminal domain, which is highly phosphorylated.4 Cell culture studies have shown that nucleolin is stable in proliferating cells, but undergoes self-cleavage in quiescent cells.6
Although the principal function of the nucleolus is thought to be rRNA synthesis and ribosome biogenesis, this sub-nuclear structure has been implicated in many aspects of cell biology that include functions such as gene silencing, senescence and cell cycle regulation.7,8 Its interaction with ribosomal proteins and with specific pre-rRNA sequences and its implication in the first step of pre-rRNA maturation also suggest that nucleolin could be an important ribosome assembly factor.9–12 The stability of nucleolin in vivo has been studied in the developing embryo. During mouse embryogenesis, nucleolin increases at day 12 of gestation and in Xenopus oocytes, the levels of nucleolin mRNA are high compared to protein expression which is low.13,14 Nucleolin production during growth is under control at translational level and there is no significant difference at mRNA levels.4
Localization
The eukaryotic nucleus contains a number of domains or subcompartments that include nucleoli, nuclear Cajal bodies, nuclear speckles, transcription and replication foci, and chromosome territories.15 Proteomic analyses have revealed that nucleoli are composed of over 400 proteins, which reflect the varying functions and state of the nucleolus. In interphase cells, the nucleolus is formed by three basic components: the fibrillar center (FC), the dense fibrillar component (DFC) and the granular component (GC).16 Occasionally, depending on cell type and stage, other structures such as vacuoles can also be observed in plant nucleolus.17 The transcription of pre-rRNA occurs at the border of the FC and DFC; the early processing steps of pre-rRNA occur in the DFC; and the later processing and RNA modification steps, together with the formation of preribosomal particles is observed in the GC.18
In plant cells, nuclear matrix is an organized matrix that roughly retains the size and shape of the nucleolus and shows resistance to RNase, and does not depend on a disulfide bond or heat stabilization, differing from the results reported for animal systems.19,20 The major task of nucleolus is ribosome biogenesis but there are other functions like regulation of tumor suppressor and oncogene activities, signal recognition particle (SRP) assembly, cell cycle regulation, modification of small RNAs, control of aging, modulating telomerase function, nuclear export and stress sensor.21
In mammalian nucleoli the relatively opaque FCs is surrounded by the highly contrasted fine strands of DFCs. In turn, the DFCs are surrounded by GCs, which fill out the peripheral parts of the nucleolus. The FCs harbor copies of rRNA genes that are well established in the initial pre-rRNA of DFCs, whereas late processing events take place in the GCs. Finally, pre-ribosomal particles are translocated to the cytoplasm.22 Nucleolin protein is localized in the DFCs and GCs of nucleoli.23 In interphase and during mitosis, nucleolin is located in the peripheral region including the vicinity of the outer kinetochore of chromosomes and from prometaphase to anaphase it is associated with the spindle poles. Depletion of nucleolin in the cell shows disruption of chromosome congression. This defect is due to improper kinetochore attachments, resulting in reduced tension and syntelic attachments. Depletion of nucleolin also appears to affect spindle assembly.24
Nucleus and nucleolus are inherently dynamic because of entering and leaving proteins to the nucleolus. Proteins with high affinity to the nucleolar components stay for longer time and others leave soon.21 The microscopic estimations of the purity of the final fractions, as well as the specific localization of nucleolin in the nucleolar matrix by immunofluorescence, immunogold labeling and bismuth staining, discards a contamination of the residual fraction with unextracted nucleoli, confirming nucleolin is actually a component of the residual nucleolar structure.19 The nucleolus exhibits an osmotically regulated gatekeeping activity that controls the spatial dynamics and functions of nucleolin.25
There are reports that, in some cell types, nucleolin can be found in the cytoplasm under certain conditions.26 Daniely and colleagues demonstrated that, during heat shock, nucleolin relocalizes to the nucleoplasm, where it sequesters RPA (replication protein A) in nuclear foci away from sites of ongoing DNA synthesis. They also showed that nucleolin mobilization occurs in a p53-dependent manner in response to γ-irradiation and CPT (camptothecin) treatment.27,28
Structure
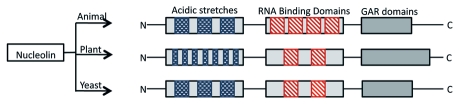
The primary sequence of nucleolin has been determined from several species.14,29 The sequence comparison of nucleolin from different species reveals a high degree of evolutionary conservation: the protein consists of three structural and multifunctional domains: an N-terminal portion containing several acidic stretches; two to four RNA-binding domains called RNA recognition motifs (RRM) in the central region; and a glycine/arginine-rich domain or GAR domain at the C-terminus.12 Animal nucleolin possesses four RRMs, whereas yeast and plants homologs possess two (Fig. 1). The length of the GAR/RGG domain is variable among nucleolins and the sequence and arrangement of the repeats is not well conserved. For example, plant nucleolin-like proteins have a longer GAR domain than mammalian nucleolins.30 The N-terminal acidic and basic region and the C-terminal domain rich in RGG repeats mediate protein-protein interactions with histone H1, U3 snoRNP and ribosomal proteins. The N-terminal part of nucleolin from various eukaryotes contains variable numbers of acidic stretches that are similar to those of nuclear high-mobility group proteins. The number of acidic stretches differs in different species. The plant nucleolin proteins contain considerably more but shorter acidic repeats as compared with others. This N-terminal region interacts with nontranscribed spacer regions in rDNA repeats and histone H1 to influence rDNA transcription.
Figure 1.
Schematic representation of domain diagram of the primary sequence of nucleolin and nucleolin-like proteins in animal, plant and yeast. Acidic stretches are represented in N-terminus region, RRMs are colored in boxes and GAR domains are shown in different length blocks.
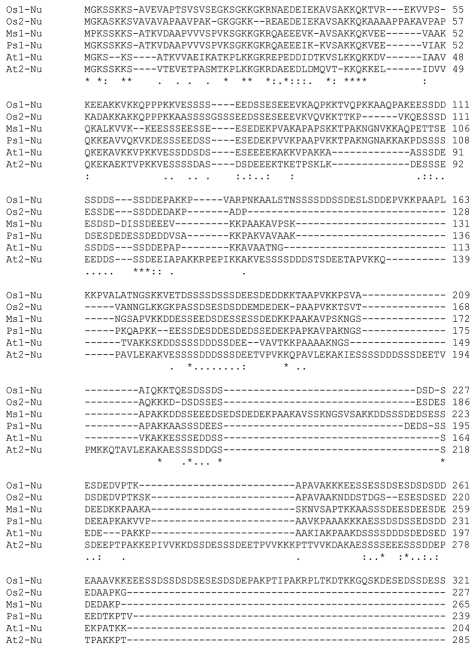
Nucleolin interacts with the stem-loop structure of RNA through its RRM and participates in the modification and processing of pre-rRNA.31 RBD domain of nucleolin interacts with telomerase and alters its subcellular localization.32 The C-terminal GAR domains are implicated in ribosomal assembly and nuclear import of ribosomal proteins.31 This GAR domain most likely functions in protein-protein interactions. The entire protein sequences of nucleolin from some plants were aligned which showed acidic region in N-terminal region (Fig. 2). The alignment study has shown two RRM domains and the sequence of RRM1 is (R/K)G(F/Y)(G/A)(F/Y)VX(F/Y) and RRM2 contains (L/I)(F/Y)(V/I)(G/K)(G/N)L and C-terminal region contains Arg-Gly-Gly (RGG) repeats interspersed with mostly aromatic amino acids (Fig. 2).
Figure 2.
Multiple sequence aligment of plant nucleolin protein. Nucleolin protein sequences from the indicated plant species were aligned using the Clustal W algorithm. Acidic residues stretched at the N-terminus. Towboxes show conserved RRMs and highlights represent RGG repeats at the C-terminus.
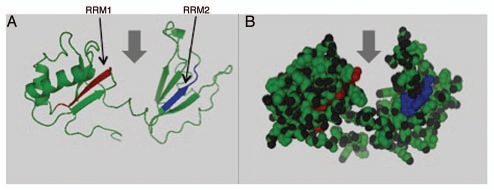
Using NMR spectroscopy Allain and colleagues (2000) determined the structure of the 28 kDa complex of the first two RNA binding domains (RBDs) of nucleolin (RBD12) with an RNA stem-loop that includes the nucleolin recognition element UCCCGA in the loop.10 They showed that the two RBDs bind on opposite sides of the RNA loop, forming a molecular clamp that brings the 5′ and 3′ ends of the recognition sequence close together stabilizing the stem-loop (Fig. 3). Interaction of the nucleolin protein with the RNA suggests that nucleolin may act as an RNA chaperone to prevent improper folding of the nascent pre-rRNA.33
Figure 3.
Predicted secondary structure and two RRM of Nucleolin protein. Secondary structural predictions of nucleolin were made using Phyre server and visualized using PyMOL software. (A) Ribbon diagram of nucleolin protein structure with two RRM shown in red and blue. (B) Three dimensional structure of nucleolin and arrows show the groove that RNA binds to the RRM motifs.
Nucleolin is highly phosphorylated and its phosphorylation is highly regulated during the cell cycle.34,35 Extensive phosphorylation by casein kinase 2 (CK2) occurs at interphase and by CDC2 during mitosis and this regulated phosphorylation of nucleolin probably regulates nucleolin functions during the cell cycle.12,34 It has been demonstrated that phosphorylation of nucleolin at interphase is correlated with active rRNA transcription. Despite a previous study describing the localization of nucleolin phosphorylated by CDC2, no information about its functions during mitosis has been obtained to date. Studies have revealed that nucleolin associates with a number of other proteins. For example, it was shown to be a component of LR1 (lipopolysaccharide-responsive factor), a DNA-binding complex that regulates transcription in activated B cells.36 Nucleolin is also part of a B cell specific DNA recombination complex.37
Function
Although the principal function of the nucleolin is rRNA synthesis and ribosome biogenesis, this protein has been implicated in many aspects of cell biology that include functions such as gene silencing, senescence and cell cycle regulation.7,8 Most of our knowledge on the function of this protein in higher eukaryotic cells comes from in vitro studies using acellular systems, microinjection experiments, or overexpression of a nucleolin transgene.38–40 Nucleolin plays important roles in various steps of ribosomal synthesis, such as the transcription of rDNA repeats, the modification and processing of pre-rRNA, the assembly of pre-ribosomal particles and nuclear-cytoplasmic transport of ribosomal proteins and subunits.12,31,41 The interaction of nucleolin with pre-rRNA is required for the first RNA processing step, which occurs in the 5′ ETS (External Transcribed Sequence).33
Nucleolin is a DNA-dependent ATPase and is capable of auto-degradation and it plays a role in regulating cell growth and DNA replication.42 In addition, the protein undergoes extensive post-translational modification.12,31 Nucleolin has been proposed to be a nuclear matrix-binding protein, to interact with telomerase and to be involved in the regulation of apoptosis, intestinal cell differentiation and remodeling of nucleosomes.32,43–45
Few reports have described the effect of downregulation of nucleolin. These reports revealed that downregulation of the nucleolin by RNA interference (RNAi) in human cells increases expression of p53 protein and inhibits RNA polymerase I transcription.45,46 This inactivation also causes nucleolar disruption, cell cycle arrest and defects in centrosome duplication.42 Nucleolin is present in a complex with Rad51 in nuclear protein extracts prepared from mammalian somatic cells. De and colleagues showed that nucleolin and Rad51 participate in a common pathway of homologous recombinational repair and nucleolin may function to regulate DNA repair activity of Rad51.47 Nucleolin has role in different cell cycle stages such as prometaphase. Nucleolin depleted cells show delay at prometaphase with misaligned or non-aligned chromosomes and defects in chromosome biorientation. Nucleolin is involved in efficient kinetochore microtubule interactions and/or the stabilization of such interactions, but nucleolin is not required for kinetochore assembly. Nucleolin depletion does not affect the localizations of kinetochore proteins and kinetochores can capture microtubules in the absence of nucleolin.24
The role of nucleolin in some viruses has been revealed through various studies. Nucleolin can promote replication in HDV (Hepatitis Delta Virus) and poliovirus. Its expression on the cell surface allows Coxsackie B virus and HIV binding. Nucleolin is known to be modified by phosphorylation, methylation and ADP-ribosylation, which may render its targeting to various compartments to exert different functions.48 Nucleolin associates with a number of other proteins. For example, it was shown to be a component of LR1, a DNA-binding complex that regulates transcription in activated B cells.36 Also, it is part of a B-cell specific DNA recombination complex.37 In addition, it was shown that nucleolin, along with topoisomerase I, controls the holoenzyme cohesion of the SV40 large T antigen helicase at DNA replication forks in vitro.49
It has been shown that the Rad51-binding replication protein A p53 physically associate with nucleolin.28,50 Nucleolin interacts with a death domain which is p53-induced protein death domain (PIDD). Nucleolin itself is relocalized from nucleoli into the nucleoplasm upon cellular stress (heat shock, ionizing radiation and camptothecin) by complex formation with p53.28,51 Grinstein and colleagues reported the interaction of nucleolin with retinoblastoma (Rb) protein, which is cell cycle-dependent and takes place in G1 phase, suggesting that the ability of nucleolin to interact with Rb depends on the phosphorylation status of Rb.52
Nucleolin binds to the human telomerase reverse transcriptase catalytic subunit hTERT.32 RNA binding domain 4 and carboxyl-terminal RGG domain are involved in this interaction and through these domains nucleolin interacts with telomerase RNA subunit hTERT. This interaction between nucleolin and hTERT is critical for the nucleolar localization of hTERT. Nucleolin binds the active telomerase complex through both protein-protein and protein-RNA interactions. Two distinct regions of nucleolin are involved in the interaction with hTERT: a central region containing the RBD1 and a carboxyl-terminal region containing both the RBD4 and RGG domains.32
Nucleolin is a histone chaperone that is able to drastically increase the remodeling efficiency of the chromatin remodelers SWI/SNF (SWItch/Sucrose Non Fermentable) and ACF (ATP-dependent chromatin-assembly factor). Interestingly, nucleolin promotes the remodeling of nucleosomes containing macroH2A, but not H2ABbd histone variant, which are otherwise resistant to remodeling. Furthermore, nucleolin is able to remove H2A-H2B dimers from assembled nucleosomes. Nucleolin is acting as a FACT-like protein (facilitates chromatin transcription) helping the passage of the RNA polymerase II through the nucleosomal particles. This work defines new functions for histone chaperones in chromatin remodeling and regulation of transcription.45
The gatekeeping activity of the nucleolus is the target of a fast-acting signaling mechanism that modulates the biological activities of nucleolin by affecting its subcellular localization. An increase in nucleoplasmic nucleolin levels has been observed in response to some pathological stimuli: a chronic thermal challenge (up to 90-min exposure to 44°C), or after chemically induced genotoxicity.25,27,53,54 Yang and colleagues (2008) demonstrate that physiologically relevant perturbations in the osmotic environment rheostatically regulate a gate keeping function for the nucleolus that controls the spatial dynamics and functions of nucleolin by affecting its subcellular localization (HeLa cells and U2-OS osteosarcoma cells were osmotically challenged) whether the intracellular compartmentalization of nucleolin responds to hyperosmotic stress andswitches its functions following hyperosmotic stress.25 They analyzed endogenous nucleolin in an effort to maintain a physiologically relevant context. In osteosarcoma cells (U2OS) incubated in standard culture media, nucleolin was found to be concentrated in the nucleolus. The absence of nucleolin from the fibrillar core, where it normally facilitates transcription, replication and recombination of rDNA suggests that these early stages of ribosome biogenesis are halted following hyperosmotic stress.25
Nucleolin is involved in processes other than the ribosome synthesis. It has role in the different steps involved in ribosome biogenesis, including RNA polymerase (Pol) I transcription and processing of pre-rRNA, assembly and nucleocytoplasmic transport of ribosome particles.9,11,40 It also acts as a transcriptional repressor and a post-transcriptional regulator to stabilize amyloid precursor protein mRNA. Jiang and colleagues demonstrated an essential role of nucleolin/C23 in the antiapoptotic effects of Hsp70. They provided evidence that nucleolin/C23 is an essential downstream effecter of Hsp70 in the protection of cardiomyocytes against oxidative stress-induced apoptosis.55
Nucleolin possesses DNA helicase activity and interacts with replication protein A, suggesting that it participates in DNA unwinding and replication.56–58 Nucleolin in animals interacts with various transcription factors and nuclear components, and is involved in the regulation of RNA polymerase II-dependent gene expression.59,60 The nucleolin protein binds DNA and RNA. Nucleolin is a DNA-dependent ATPase and is capable of autodegradation.47 Nucleolin possesses a histone chaperone activity that activates chromatin remodeling complexes and facilitates transcription through the nucleosomes.45 This protein binds to single stranded human telomeric DNA and it is highly specific for the G-rich strand.61
Disruption of the AtNUC-L (Arabidopsis nucleolin-like protein) gene provokes changes in growth and plant development such as changes in the ratio of primary pre-rRNA and processed pre-rRNA at the P site. It also causes major effect on nucleolus structure.16 In vitro assays and microinjection experiments in X. laevis oocytes showed a role of nucleolin in transcription of pre-rRNA.39 Nucleolin, along with topoisomerase I, controls the holoenzyme cohesion of the SV40 large T antigen helicase at DNA replication forks in vitro.49 Furthermore, nucleolin binds to topoisomerase and the growth factor midkine to localize them at the nucleolus.48 One of the most remarkable consequences of nucleolin inactivation is the apparent blockage of cells in the G2 phase, and the significant increase in multinuclear cells and cells with micronuclei. By contrast, the number of cells in mitosis was drastically reduced in nucleolin depleted cells.42 It was recently reported that nucleolin was required for chromosome congression and the maintenance of mitotic spindle integrity.24 Nucleolin interacts with telomerase and alters its subcellular localization.32 Nucleolin also switches its functions following hyperosmotic stress.25 In a recent study it has been demonstrated that nucleolin can function as a repressor of c-Myc transcription by binding to its promoter.62 It was further reported that nucleolin's RNA binding domains 3 and 4 and its RGG domain are required to repress c-Myc transcription.62
Nucleolin in Plants
Nucleolin is not conserved, but genes encoding proteins with similar structural organization are found in other eukaryotes. In Saccharomyces cerevisiae the Nsr1 gene encodes a nucleolinlike protein, characterized by only two RRMs. The major milestones in the discovery of nucleolin in plants are summarized in Table 1. Structural organizations of nucleolin like proteins is similar to nucleolin but, like yeast nucleolin, nucleolin-like proteins from alfalfa (Medicago sativa),30 pea (Pisum sativum) and Arabidopsis (Arabidopsis thaliana) possess two RRMs.63–65 Therefore plant nucleolin can be considered as nucleolin like proteins. A. thaliana encodes two nucleolin-like proteins, AtNuc-L1 (Arabidopsis nucleolin like protein) and AtNuc-L2. In normal growth conditions only AtNUC-L1 gene is constitutively expressed. Pontvianne et al. disrupted the AtNUC-L1 gene and found the AtNUC-L2 gene is expressed and rescue, at least partially, AtNuc-L1disruption. They showed the AtNUC-L1 gene affects plant growth and development and demonstrate that AtNUC-L1 gene expression is required to preserve the ultrastructure of the nucleolus and nucleolus organising region (NOR) condensation.16
Table 1.
Historical background of plant nucleolin proteins
| No. | Year | Major discoveries/event | Reference |
| 1 | 1996 | Nucleolin is a component of nucleolar matrix | Minguez & Espinana19 |
| 2 | 1996 | Nucleolin role in cell cycle of alfalfa | Bogre, et al.30 |
| 3 | 1997 | Pea nucleolin expression is induced by redlight | Tong, et al.63 |
| 4 | 1997 | Nop64A, onion nucleolin, shares immunological determinant with mammalian nucleolin | De Carcer, et al.71 |
| 5 | 2001 | Light Differentially Regulates Cell Division and the mRNA Abundance of Pea Nucleolin | Reichler, et al.64 |
| 6 | 2004 | Nucleolin role in rRNA binding and pre-RNA processing in Arabidopsis | Saez-Vasquez, et al.65 |
| 7 | 2005 | pea nucleolin is a DNA helicase | Nasirudin, et al.58 |
| 8 | 2006 | Arabidopsis nucleolin is inducible by sugar | Kojima, et al.68 |
| 9 | 2006 | Role of NopA100, a nucleolin-like protein, during the cell cycle in proliferating plant cells | Gonza lez-Camacho, et al.72 |
| 10 | 2007 | Arabidopsis nucleolin has role in plant development and patterning | Patricka and Nelson70 |
| 11 | 2007 | AtNucL2 will rescue disruption of AtNucL1 | Pontvianne, et al.16 |
AtNUC-L1 has structural similarities to Nsr1 of S. cerevisiae and Gar2 of Schizosaccharomyce s pombe.66,67 Sugar causes a rapid increase in the level of AtNuc-L1 mRNA and AtNuc-L1 protein and this sugar regulation is closely associated with regulation of de novo ribosome synthesis.68 Pontvianne and colleagues showed that disruption of the AtNUC-L gene provokes changes in growth and plant development and AtNUC-L1 disruption has a major effect on nucleolus structure. AtNUC-L1 gene disruption induces changes in the ratio of primary pre-rRNA and processed pre-rRNA at the P site. Absence of this nucleolin like protein in mutated plants induces nucleolar disorganization, nucleolus organizer region decondensation and affects the accumulation levels of pre-rRNA precursors.16 AtNuc-L1 presents in NF D (nuclear factor D) and could act as an assembly factor for this processing complex and direct its binding to pre-RNA. NFD is a multiprotein factor of 600 kDa that dissociates into smaller complexes. Two polypeptides of NFD identified by microsequencing are homologues of nucleolin and fibrillarin. AtNuc-L1 can bind to the 5′ETS RNA implicates only two contiguous RRMs,33 while the interaction with U3 snoRNP and the processing factors implicate the N-terminal acidic domain.11,12 Thus, AtNuc-L1 could fulfill functions similar to those of nucleolin and Nsr1 in pre-rRNA cleavage. It is interesting to note that nucleophosmin interacts directly with nucleolin,69 and that a nucleolin-like protein, together with other nucleolar proteins has been found associated with centrosomal structure.2 But apart from similarities the role of the plant nucleolin-like proteins in pre-rRNA synthesis has not been investigated.65
Patricka and Nelson found Arabidopsis nucleolin parallel1 (parl1) mutant (AtNuc-L1) displays several aberrations in venation pattern of all foliar organs, including the parallel alignment of veins along the proximal/distal leaf axis, the parallel exit of multiple veins from the petiole, and a reduction in higher order venation. These defects in leaves are associated with the mislocalization of activity from the provascular Athb8:GUS and auxinresponsive DR5:GUS reporters. Arabidopsis parl1 mutants also have an accumulation of unprocessed 35S pre-rRNA that is consistent with other results demonstrating PARL1 is able to rescue mutants in the yeast nucleolin gene NSR1. These data suggest that auxin-dependent growth and patterning processes, including vein patterning, are particularly sensitive to perturbations in ribosomal processing.70
Nucleolin has been shown to be light regulated and to have DNA helicase and ATPase activity in pea (P. sativum),58,63,64 to be cell cycle regulated in alfalfa (M. sativa),30 and to be part of a complex responsible for rRNA binding and in vitro pre-rRNA processing in Arabidopsis (A. thaliana). 65 Red light treatment induces pea nucleolin transcription 1 h prior to rRNA synthesis and coincides with increased cell division rates in pea apical internodes as well as increased nuclear number/weight of pea plumules.64 Bogre and colleagues30 suggested nucMs1 alfalfa expression is tightly linked to cell proliferation but does not depend on a particular cell cycle phase. No nucMs1 expression was observed in cells that had exited the cell cycle and were undergoing differentiation or polar growth, indicating that nucMs1 may not be necessary for processes other than cell proliferation. Also, nucMs1 express predominantly in meristematic, where its expression is limited to cells actively engaged in cell division.
Acknowledgements
This work is partially supported by the Department of Biotechnology, Department of Science and Technology and Defence Research and Development Organization grants. Infrastructural support from the Department of Biotechnology, Government of India is gratefully acknowledged.
References
- 1.Raska I, Shaw PJ, Cmarko D. New insights into nucleolar architecture and activity. Int Rev Cytol. 2006;255:177–235. doi: 10.1016/S0074-7696(06)55004-1. [DOI] [PubMed] [Google Scholar]
- 2.Andersen JS, Lam YW, Leung AKL, Ong S, Lyon CE, Lamond AI, et al. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- 3.Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- 4.Bicknell K, Brooks G, Kaiser P, Chen H, Dove BK, Hiscox JA. Nucleolin is regulated both at the level of transcription and translation. Biochem Biophys Res Comm. 2005;332:817–822. doi: 10.1016/j.bbrc.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 5.Bugler B, Caizergues-Ferrer M, Bouche G, Bourbon H, Amalric F. Detection and localization of a class of proteins immunologically related to a 100 KDa nucleolar protein. Eur J Biochem. 1982;128:473–480. doi: 10.1111/j.1432-1033.1982.tb06989.x. [DOI] [PubMed] [Google Scholar]
- 6.Chen CM, Chiang SY, Yeh NH. Increased stability of nucleolin in proliferating cells by inhibition of its self-cleavingactivity. J Biol Chem. 1991;266:7754–7758. [PubMed] [Google Scholar]
- 7.Pederson T. The plurifunctional nucleolus. Nucleic Acids Res. 1998;26:3871–3876. doi: 10.1093/nar/26.17.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olson MOJ, Dundr M, Szebeni A. The nucleolus: an old factory with unexpected capabilities. Trends Cell Biol. 2000;10:189–196. doi: 10.1016/s0962-8924(00)01738-4. [DOI] [PubMed] [Google Scholar]
- 9.Bouvet P, Diaz J, Kindbeiter K, Madjar J, Amalric F. Nucleolin interacts with several ribosomal proteins through its RGG domain. J Biol Chem. 1998;273:19025–19029. doi: 10.1074/jbc.273.30.19025. [DOI] [PubMed] [Google Scholar]
- 10.Allain FH, Bouvet P, Dieckmann T, Feigon J. Molecular basis of sequence-specific recognition of pre-ribosomal RNA by nucleolin. EMBO J. 2000;19:6870–6881. doi: 10.1093/emboj/19.24.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ginisty H, Amalric F, Bouvet P. Nucleolin functions in the first step of ribosomal RNA processing. EMBO J. 1998;17:1476–1486. doi: 10.1093/emboj/17.5.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ginisty H, Sicard H, Roger B, Bouvet P. Structure and functions of nucleolin. J Cell Sci. 1999;112:761–772. doi: 10.1242/jcs.112.6.761. [DOI] [PubMed] [Google Scholar]
- 13.Biggiogera M, Kaufmann SH, Shaper JH, Gas N, Amalric F, Fakan S. Distribution of nucleolar proteins B23 and nucleolin during mouse spermatogenesis. Chromosoma. 1991;100:162–172. doi: 10.1007/BF00337245. [DOI] [PubMed] [Google Scholar]
- 14.Caizergues-Ferrer M, Mariottini P, Curie C, Lapeyre B, Gas N, Amalric F, et al. Nucleolin from Xenopus laevis: cDNA cloning and expression during development. Genes Dev. 1989;3:324–333. doi: 10.1101/gad.3.3.324. [DOI] [PubMed] [Google Scholar]
- 15.Lamond AI, Earnshaw WC. Structure and function in the nucleus. Science. 1998;24:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- 16.Pontvianne F, Matía I, Douet J, Tourmente S, Medina FJ, Echeverria M, et al. Characterization of AtNUC-L1 reveals a central role of nucleolin in nucleolus organization and silencing of AtNUC-L2 gene in Arabidopsis. Mol Biol Cell. 2007;18:369–379. doi: 10.1091/mbc.E06-08-0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.González-Melendi P, Wells B, Beven AF, Shaw PJ. Single ribosomal transcription units are linear, compacted Christmas trees in plant nucleoli. Plant J. 2001;27:223–233. doi: 10.1046/j.1365-313x.2001.01091.x. [DOI] [PubMed] [Google Scholar]
- 18.Thiry M, Lafontaine DL. Birth of a nucleolus: the evolution of nucleolar compartments. Trends Cell Biol. 2005;15:194–199. doi: 10.1016/j.tcb.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 19.Minguez A, Moreno Diaz De La Espina S. In situ localization of nucleolin in the plant nucleolar matrix. Exp Cell Res. 1996;222:171–178. doi: 10.1006/excr.1996.0022. [DOI] [PubMed] [Google Scholar]
- 20.Bourgeois CA, Bouvier D, Seve AP, Hubert J. Evidence for the existence of a nuclear skeleton attached to the pore complex lamina in human fibroblasts. Chromosoma. 1987;95:315–323. doi: 10.1007/BF00293178. [DOI] [PubMed] [Google Scholar]
- 21.Olson MO, Dundr M. The moving parts of the nucleolus. Histochem Cell Biol. 2005;123:203–216. doi: 10.1007/s00418-005-0754-9. [DOI] [PubMed] [Google Scholar]
- 22.Shaw P, Doonan J. The nucleolus: playing by different rules. Cell Cycle. 2005;4:102–105. doi: 10.4161/cc.4.1.1467. [DOI] [PubMed] [Google Scholar]
- 23.Biqqioqera M, Burki K, Kaufmann SH, Shaper JH, Gas N, Amalric F, et al. Nucleolar distribution of proteins B23 and nucleolin in mouse preimplantation embryos as visualized by immunoelectron microscopy. Development. 1990;110:1263–1270. doi: 10.1242/dev.110.4.1263. [DOI] [PubMed] [Google Scholar]
- 24.Ma N, Matsunaga S, Takata H, Ono-Maniwa R, Uchiyama S, Fukui K. Nucleolin functions in nucleolus formation and chromosome congression. J Cell Science. 2007;120:2091–2105. doi: 10.1242/jcs.008771. [DOI] [PubMed] [Google Scholar]
- 25.Yang L, Reece JM, Cho J, Bortner CD, Shears SB. The nucleolus exhibits an osmotically regulated gatekeeping activity that controls the spatial dynamics and functions of nucleolin. J Biol Chem. 2008;283:11823–11831. doi: 10.1074/jbc.M800308200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caffrey JJ, Hidaka K, Matsuda M, Hirata M, Shears SB. The human and rat forms of multiple inositol polyphosphate phosphatase: functional homology with a histidine acid phosphatase upregulated during endochondral ossification. FEBS Lett. 1999;442:99–104. doi: 10.1016/s0014-5793(98)01636-6. [DOI] [PubMed] [Google Scholar]
- 27.Daniely Y, Borowiec JA. Formation of a complex between nucleolin and replication protein A after cell stress prevents initiation of DNA replication. J Cell Biol. 2000;149:799–810. doi: 10.1083/jcb.149.4.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daniely Y, Dimitrova DD, Borowiec JA. Stress-dependent nucleolin mobilization mediated by p53-nucleolin complex formation. Mol Cell Biol. 2002;22:6014–6022. doi: 10.1128/MCB.22.16.6014-6022.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maridor G, Nigg EA. cDNA sequences of chicken nucleolin/C23 and NO38/B23, two major nucleolar proteins. Nucleic Acids Res. 1990;18:1286. doi: 10.1093/nar/18.5.1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bogre L, Jonak C, Mink M, Meskiene I, Traas J, Ha DT, et al. Developmental and cell cycle regulation of alfalfa nucMs1, a plant homolog of the yeast Nsr1 and mammalian nucleolin. Plant Cell. 1996;8:417–428. [PMC free article] [PubMed] [Google Scholar]
- 31.Tuteja R, Tuteja N. Nucleolin: A multifunctional major nucleolar Phosphoprotein. Crit Rev Biochem Mol Biol. 1998;33:407–436. doi: 10.1080/10409239891204260. [DOI] [PubMed] [Google Scholar]
- 32.Khurts S, Masutomi K, Delgermaa L, Arai K, Oishi N, Mizuno H, et al. Nucleolin interacts with telomerase. J Biol Chem. 2004;279:51508–51515. doi: 10.1074/jbc.M407643200. [DOI] [PubMed] [Google Scholar]
- 33.Allain FH, Gilbert DE, Bouvet P, Feigon J. Solution structure of the two N-terminal RNA-binding domains of nucleolin and NMR study of the interaction with its RNA target. J Mol Biol. 2000;303:227–241. doi: 10.1006/jmbi.2000.4118. [DOI] [PubMed] [Google Scholar]
- 34.Peter M, Nakagawa J, Dorée M, Labbé JC, Nigg EA. Identification of major nucleolar proteins as candidate mitotic substrates of cdc2 kinase. Cell. 1990;60:791–801. doi: 10.1016/0092-8674(90)90093-t. [DOI] [PubMed] [Google Scholar]
- 35.Morimoto H, Ozaki A, Okamura H, Yoshida K, Amorim BR, Tanaka H, et al. Differential expression of protein phosphatase type 1 isotypes and nucleolin during cell cycle arrest. Cell Biochem Funct. 2005;25:369–375. doi: 10.1002/cbf.1300. [DOI] [PubMed] [Google Scholar]
- 36.Hanakahi LA, Dempsey LA, Li MJ, Maizels N. Nucleolin is one component of the B cell-specific transcription factor and switch region binding protein, LR1. Proc Natl Acad Sci USA. 1997;94:3605–3610. doi: 10.1073/pnas.94.8.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borggrefe T, Wabl M, Akhmedov AT, Jessberger R. A B-cell-specific DNA recombination complex. J Biol Chem. 1998;273:17025–17035. doi: 10.1074/jbc.273.27.17025. [DOI] [PubMed] [Google Scholar]
- 38.Yanagida M, Shimamoto A, Nishikawa K, Furuichi Y, Isobe T, Takahashi N. Isolation and proteomic characterization of the major proteins of the nucleolin-binding ribonucleoprotein complexes. Proteomics. 2001;1:1390–1404. doi: 10.1002/1615-9861(200111)1:11<1390::AID-PROT1390>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 39.Roger B, Moisand A, Amalric F, Bouvet P. Nucleolin provides a link between RNA polymerase I transcription and pre-ribosome assembly. Chromosoma. 2003;111:399–407. doi: 10.1007/s00412-002-0221-5. [DOI] [PubMed] [Google Scholar]
- 40.Roger B, Moisand A, Amalric F, Bouvet P. Repression of RNA polymerase I transcription by nucleolin is independent of the RNA sequence that is transcribed. J Biol Chem. 2002;277:10209–10219. doi: 10.1074/jbc.M106412200. [DOI] [PubMed] [Google Scholar]
- 41.Srivastava M, Pollard HB. Molecular dissection of nucleolin's role in growth and cell proliferation: new insights. FASEB J. 1999;13:1911–1922. [PubMed] [Google Scholar]
- 42.Ugrinova I, Monier K, Ivaldi C, Thiry M, Storck S, Mongelard F, et al. Inactivation of nucleolin leads to nucleolar disruption, cell cycle arrest and defects in centrosome duplication. BMC Mol Biol. 2007;8:66. doi: 10.1186/1471-2199-8-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mi Y, Thomas SD, Xu X, Casson LK, Miller DM, Bates PJ. Apoptosis in leukemia cells is accompanied by alterations in the levels and localization of nucleolin. J Biol Chem. 2003;278:8572–8579. doi: 10.1074/jbc.M207637200. [DOI] [PubMed] [Google Scholar]
- 44.Turck N, Lefebvre O, Gross I, Gendry P, Keninger M, Simon-Assman P, et al. Effect of laminin-1 on intestinal cell differentiation involves inhibition of nuclear nucleolin. J Cell Physiol. 2006;206:545–555. doi: 10.1002/jcp.20501. [DOI] [PubMed] [Google Scholar]
- 45.Angelov D, Bondarenko VA, Almagro S, Menoni H, Monge' lard F, Hans F, et al. Nucleolin is a histone chaperone with FACT-like activity and assists remodeling of nucleosomes. EMBO J. 2006;25:1669–1679. doi: 10.1038/sj.emboj.7601046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takagi M, Absalon MJ, McLure KG, Kastan MB. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123:49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- 47.De A, Donahue SL, Tabah A, Castro NE, Mraz N, Cruise JL, Campbell C. A novel interaction of nucleolin with Rad51. Biochem Biophys Res Comm. 2006;344:206–213. doi: 10.1016/j.bbrc.2006.03.113. [DOI] [PubMed] [Google Scholar]
- 48.Lo SJ, Lee CC, Lai HJ. The nucleolus: reviewing oldies to have new understandings. Cell Res. 2006;16:530–538. doi: 10.1038/sj.cr.7310070. [DOI] [PubMed] [Google Scholar]
- 49.Seinsoth S, Uhlmann-Schiffler H, Stahl H. Bidirectional DNA unwinding by a ternary complex of T antigen, nucleolin and topoisomerase I. EMBO Rep. 2003;4:263–268. doi: 10.1038/sj.embor.embor770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Golub EI, Gupta RC, Haaf T, Wold MS, Radding CM. Interaction of human Rad51 recombination protein with single-stranded DNA binding protein, RPA. Nucl Acids Res. 1998;26:5388–5393. doi: 10.1093/nar/26.23.5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pick R, Badura S, Bosser S, Zornig M. Upon intracellular processing, the C-terminal death domain-containing fragment of the p53-inducible PIDD/LRDD protein translocates to the nucleoli and interacts with nucleolin. Biochem Biophys Res Comm. 2006;349:1329–1338. doi: 10.1016/j.bbrc.2006.08.176. [DOI] [PubMed] [Google Scholar]
- 52.Grinstein E, Shan Y, Karawajew L, Snijders PJ, Meijer CJ, Royer HD, et al. Cell cycle-controlled interaction of nucleolin with the retinoblastoma protein and cancerous cell transformation. J Biol Chem. 2006;281:22223–22235. doi: 10.1074/jbc.M513335200. [DOI] [PubMed] [Google Scholar]
- 53.Shears SB. Assessing the omnipotence of inositol hexakisphosphate. Cell Signal. 2001;13:151–158. doi: 10.1016/s0898-6568(01)00129-2. [DOI] [PubMed] [Google Scholar]
- 54.Wang Y, Guan J, Wang H, Leeper D, Iliakis G. Regulation of DNA replication after heat shock by replication protein A-nucleolin interactions. J Biol Chem. 2001;276:20579–20588. doi: 10.1074/jbc.M100874200. [DOI] [PubMed] [Google Scholar]
- 55.Jiang B, Zhang B, Liang P, Song J, Deng H, Tu Z, et al. Nucleolin/C23 mediates the antiapoptotic effect of heat shock protein 70 during oxidative stress. FEBS J. 2009;277:642–652. doi: 10.1111/j.1742-4658.2009.07510.x. [DOI] [PubMed] [Google Scholar]
- 56.Tuteja N, Huang NW, Skopac D, Tuteja R, Hrvatic S, Zhang J, et al. Human DNA helicase IV is nucleolin, an RNA helicase modulated by phosphorylation. Gene. 1995;160:143–148. doi: 10.1016/0378-1119(95)00207-m. [DOI] [PubMed] [Google Scholar]
- 57.Kim K, Dimitrova DD, Carta KM, Saxena A, Daras M, Borowiec JA. Novel checkpoint response to genotoxic stress mediated by nucleolin-replication protein A complex formation. Mol Cell Biol. 2005;25:2463–2474. doi: 10.1128/MCB.25.6.2463-2474.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nasirudin KM, Ehtesham NZ, Tuteja R, Sopory SK, Tuteja N. The Gly-Arg-rich C-terminal domain of pea nucleolin is a DNA helicase that catalytically translocates in the 5′ to 3′ direction. Arch Biochem Biophys. 2005;434:306–315. doi: 10.1016/j.abb.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 59.Masumi A, Fukazawa H, Shimazu T, Yoshida M, Ozato K, Komuro K, et al. Nucleolin is involved in interferon regulatory factor-2-dependent transcriptional activation. Oncogene. 2006;25:5113–5124. doi: 10.1038/sj.onc.1209522. [DOI] [PubMed] [Google Scholar]
- 60.Huddleson JP, Ahmad N, Lingrel JB. Upregulation of the KLF2 transcription factor by fluid shear stress requires nucleolin. J Biol Chem. 2006;281:15121–15128. doi: 10.1074/jbc.M513406200. [DOI] [PubMed] [Google Scholar]
- 61.Pollice A, Zibella PM, Bilaud T, Laroche T, Pulitzer JF, Gilson E. In vitro binding of nucleolin to double-stranded telomeric DNA. Biochem Biophys Res Comm. 2000;268:909–915. doi: 10.1006/bbrc.2000.2237. [DOI] [PubMed] [Google Scholar]
- 62.González V, Hurley LH. The C-terminal of nucleolin promotes the formation of the c-MYC G-Quadruplex and inhibits c-MYC promoter activity. Biochemistry. 2010;49:9706–9714. doi: 10.1021/bi100509s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tong CG, Reichler S, Blumenthal S, Balk J, Hsieh HL, Roux SJ. Light regulation of the abundance of mRNA encoding a nucleolin-like protein localized in the nucleoli of pea nuclei. Plant Physiol. 1997;114:643–652. doi: 10.1104/pp.114.2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reichler SA, Balk J, Brown ME, Woodruff K, Clark GB, Roux SJ. Light differentially regulates cell division and the mRNA abundance of Pea nucleolin during de-etiolation. Plant Phys. 2001;125:339–350. doi: 10.1104/pp.125.1.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sa'ez-Vasquez J, Caparros-Ruiz D, Barneche F, Echeverría M. A plant snoRNP complex containing snoRNAs, fibrillarin and nucleolin-like proteins is competent for both rRNA gene binding and Pre-rRNA processing in vitro. Mol Cell Biol. 2004;24:7284–7297. doi: 10.1128/MCB.24.16.7284-7297.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kondo K, Inouye M. Yeast NSR1 protein that has structural similarity to mammalian nucleolin is involved in pre-rRNA processing. J Biol Chem. 1992;267:16252–16258. [PubMed] [Google Scholar]
- 67.Gulli MP, Girard JP, Zabetakis D, Lapeyre B, Melese T, Caizergues-Ferrer M. gar2 is a nucleolar protein from Schizosaccharomyces pombe required for 18S rRNA and 40S ribosomal subunit accumulation. Nucleic Acids Res. 1995;23:1912–1918. doi: 10.1093/nar/23.11.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kojima H, Suzuki T, Kato T. Sugar-inducible expression of the nucleolin-1 gene of Arabidopsis thaliana and its role in ribosome synthesis, growth and development. Plant J. 2007;49:1053–1063. doi: 10.1111/j.1365-313X.2006.03016.x. [DOI] [PubMed] [Google Scholar]
- 69.Liu HT, Yung BY. In vivo interaction of nucleophosmin/B23 and protein C23 during cell cycle progression in HeLa cells. Cancer Lett. 1999;144:45–54. doi: 10.1016/s0304-3835(99)00184-6. [DOI] [PubMed] [Google Scholar]
- 70.Petricka JJ, Nelson TM. Arabidopsis nucleolin affects plant development and patterning. Plant Physiol. 2007;144:173–186. doi: 10.1104/pp.106.093575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.De Carcer G, Cerdido A, Medina FJ. NopA64, a novel nucleolarphosphoprotein from proliferating onion cells, sharing immunological determinants with mammalian nucleolin. Planta. 1997;201:487–495. doi: 10.1007/s004250050093. [DOI] [PubMed] [Google Scholar]
- 72.Gonza'lez-Camacho F, Medina FJ. The nucleolar structure and the activity of NopA100, a nucleolin-like protein, during the cell cycle in proliferating plant cells. Histochem Cell Biol. 2006;125:139–153. doi: 10.1007/s00418-005-0081-1. [DOI] [PubMed] [Google Scholar]