Abstract
Circadian clocks allow organisms to adjust multiple physiological and developmental processes in anticipation of daily and seasonal changes in the environment. At the molecular level these clocks consist of interlocked feedback loops, involving transcriptional activation and repression, but also post-translational modifications. In a recently published work we provided evidence that PRMT5, a protein arginine methyl transferase, is part of a novel loop within the circadian clock of the plant Arabidopsis thaliana by regulating alternative splicing of key clock mRNAs. We also found evidence indicating that PRMT5 has a role in the regulation of alternative splicing and the circadian network in Drosophila melanogaster, although the clock connection in the latter is more elusive and seems to be at the output level. We conclude that alternative precursor messenger RNA (premRNA) splicing is part of the circadian program and could be a main actor in the fine-tuning of biological clocks. Here, we embrace the alternative splicing process as part of the circadian program and discuss the possibility that this mechanism is of fundamental relevance for the fine-tuning of biological clocks.
Key words: Arabidopsis thaliana, circadian clock, PRMT5, arginine methyltransferase, alternative splicing, transcriptional regulation, epigenetic, Drosophila melanogaster
Most organisms have the ability to anticipate their physiological and developmental processes to daily and seasonal environmental changes. This anticipatory behavior to the recurrent conditions has been shown to enhance fitness of organisms1 and relies on an internal circadian clock. This clock is synchronized by different external signals, such as light/dark or high/low temperature cycles.2 In plants and animals, it consists of interlocked negative feedback loops involving transcriptional activation and repression, but also post-translational modifications.3 In Arabidopsis thaliana, the expression of 30% of the genes is regulated by the clock, and up to 90% oscillates at the transcript level under cycling environmental conditions.4 This observation reflects the importance of the circadian clock in the control of basic cellular and organismal processes.
Different steps of gene expression are key processes that could fine-tune the clock in different organisms. One of these steps is pre-mRNA splicing, a mechanism catalyzed by the spliceosome complex, by which introns are removed and exons are joined together to form mature mRNA. In many cases, the splicing process can create alternative mRNAs from the same precursor by varying the selection of the included/excluded regions. This mechanism, known as alternative splicing, is an important source of protein diversity in multicellular eukaryotes.5 Since splicing variants often have different functions and regulatory features, the regulation of alternative splicing represents a powerful tool to control gene expression. In humans, around 95% of the transcripts with at least two exons undergo alternative splicing6 and this mechanism has also been associated with the etiology of several diseases.7 In spite of the great extent of knowledge in humans and other organisms, alternative splicing in plants is just arising as an important process regulating key cellular functions and the transcriptome,8 and very few physiologically relevant events have been described in references 9 and 10.
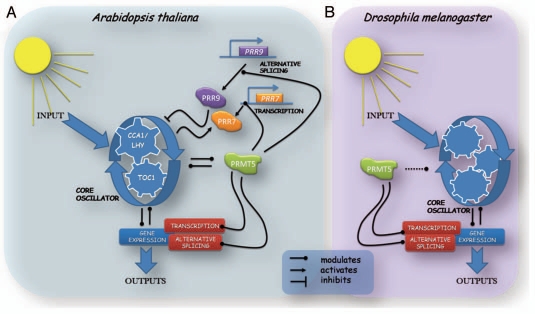
In a recent study we reported a new clock mutant in the plant model Arabidopsis thaliana. This long-period mutant has a defective PRMT5 gene,11 which encodes an enzyme whose sequence is conserved from yeast to humans, and whose biological activity is to catalyze the symmetric dimethylation of arginines. Histones and non-histone proteins have been shown to be targets for this post-translational modification.12 Furthermore, this mark seems to be essential for physiological and morphological processes in Arabidopsis thaliana, since the lack of it induces flowering defects and morphological changes in the leaves.13–15 In addition to previously described roles, we found this protein to be necessary for proper functioning of the circadian clock and to be a main component of a newly described regulatory loop within the central oscillator (Fig. 1A), since it regulates the timing of expression of the core clock genes CCA1, LHY and TOC1, and has itself a circadian pattern of expression, which has been also shown by another recent publication.16 Looking for the mechanism underlying these phenotypes, we found that prmt5 mutants have altered expression of two Pseudo Response Regulator (PRR) family members, PRR7 and PRR9, which are components of a feedback loop close to the core oscillator.17 Surprisingly, these clock components are regulated in a different manner: PRR7 expression is regulated through transcription, while PRR9 expression is regulated by transcription and alternative splicing (Fig. 1A). Furthermore, global analysis using tiling arrays as well as a high resolution RT-PCR panel18 revealed a broad participation of PRMT5 in alternative splicing. This process can occur in different ways: exons can be included or skipped or can be extended (modulating 5′ and 3′ splice site selection), and introns can be retained or eliminated. Our results show an over-representation of events with alterations in 5′ splice site (5′ss) selection. Moreover, the relative amounts of different splicing variants of some genes follow a circadian pattern that is severely disrupted in prmt5 mutants, showing that the circadian clock modulates alternative splicing through PRMT5. Recent evidence supports the idea that other components of the central oscillator suffer alternative splicing of their own pre-mRNA,8 reinforcing the idea of a new feedback loop within the circadian clock (Fig. 1A). Thus, coupling alternative splicing to the circadian clock could contribute to fine tuning circadian rhythms in several cellular processes including the core oscillator itself.
Figure 1.
Schematic representations of circadian clock mechanisms for Arabidopsis thaliana and Drosophila melanogaster. The role of PRMT5 is shown in the model, although the connection with the core oscillator is different for Arabidopsis thaliana (A) and Drosophila melanogaster (B). In both organisms, gene expression can be modulated by alternative splicing or transcription levels, and both of them are controlled by the circadian core and in turn control the core oscillator, which leads to understanding alternative splicing and transcription as regulators of the core oscillator themselves.
Since PRMT5 is conserved from yeast to humans, we next decided to evaluate its role in Drosophila melanogaster another model organism that had characterized mutants for the PRMT5 ortholog. We found that PRMT5 plays a key role in alternative splicing, similar to the one described for Arabidopsis thaliana.11 Even more, we also found an over-representation of events with alterations in 5′ss selection. However, the role of PRMT5 in the control of the circadian clock seems to be more elusive. At the behavioral level, mutant flies lacking PRMT5 activity are arrhythmic. Expression patterns of core oscillator genes are slightly affected, but they are not arrhythmic. Thus, in flies, PRMT5 operates mainly by modulating the connection between the core oscillator and its outputs, rather than on the core oscillator itself. Furthermore, in contrast to what was observed in Arabidopsis, in flies, PRMT5 gene expression is not under the control of the circadian clock, reinforcing the idea of a more elusive connection between PRMT5 and the circadian clock in flies than in plants (Fig. 1B).
Finally, on one hand, PRMT5 modulates the expression of several genes in Arabidopsis and Drosophila, which is expectable due to the fact that it is a global transcriptional repressor.19 On the other hand, independently of the differential roles with respect to the clock, PRMT5 seems to have conserved roles on the intimate alternative splicing mechanism in both plants and flies, by enhancing the recognition and usage of weak 5′ss during the splicing reaction. Alternative splicing is a co-transcriptional process that can be regulated by histone modifications20,21 and also by the assembly of the spliceosome over the pre-mRNA. The latter depends on the post-translational modification state of the proteins involved in the spliceosome machinery. Our evidence supports the hypothesis that PRMT5 methylation of Sm proteins (components of the spliceosome), rather than an epigenetic regulation, is essential for proper recognition of weaker 5′ss. This is supported by recently published findings, where several Arabidopsis thaliana Sm proteins have been identified as targets of PRMT5 methylation.22 Those data are consistent with structural analysis demonstrating the relevance of Sm proteins in the 5′ss recongnition.23
The dual and conserved roles of PRMT5 in different layers of gene expression in distantly related organisms makes it a perfect candidate to be selected or coopted to fine tune the circadian clocks and/or their outputs, even more than once throughout evolution.
References
- 1.Dodd AN, Salathia N, Hall A, Kevei E, Toth R, Nagy F, et al. Plant circadian clocks increase photosynthesis, growth, survival and competitive advantage. Science. 2005;309:630–633. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- 2.Harmer SL. The circadian system in higher plants. Annu Rev Plant Biol. 2009;60:357–377. doi: 10.1146/annurev.arplant.043008.092054. [DOI] [PubMed] [Google Scholar]
- 3.McClung CR, Gutiérrez RA. Network news: prime time for systems biology of the plant circadian clock. Curr Opin Genet Dev. 2010;20:588–598. doi: 10.1016/j.gde.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michael TP, Mockler TC, Breton G, McEntee C, Byer A, Trout JD, et al. Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet. 2008;4:14. doi: 10.1371/journal.pgen.0040014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 7.Faustino NA, Cooper TA. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- 8.Filichkin SA, Priest HD, Givan SA, Shen R, Bryant DW, Fox SE, et al. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 2010;20:45–58. doi: 10.1101/gr.093302.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang N, Portis A., Jr Mechanism of light regulation of Rubisco: A specific role for the larger Rubisco activase isoform involving reductive activation by thioredoxin-f. Proc Natl Acad Sci USA. 1999;96:9438–9443. doi: 10.1073/pnas.96.16.9438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung HS, Cooke TF, DePew CL, Patel LC, Ogawa N, Kobayashi Y, et al. Alternative splicing expands the repertoire of dominant JAZ repressors of jasmonate signaling. Plant J. 63:613–622. doi: 10.1111/j.1365-313X.2010.04265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanchez SE, Petrillo E, Beckwith EJ, Zhang X, Rugnone ML, Hernando CE, et al. A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature. 2010;468:112–116. doi: 10.1038/nature09470. [DOI] [PubMed] [Google Scholar]
- 12.Bedford M, Richard S. Arginine methylation: An emerging regulator of protein function. Mol Cell. 2005;18:263–272. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 13.Pei Y, Niu L, Lu F, Liu C, Zhai J, Kong X, et al. Mutations in the type II protein arginine methyltransferase AtPRMT5 result in pleiotropic developmental defects in Arabidopsis. Plant Physiol. 2007;44:1913–1923. doi: 10.1104/pp.107.099531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, Zhang Y, Ma Q, Zhang Z, Zue Y, Bao S, et al. SKB1-mediated symmetric dimethylation of histone H4R3 controls flowering time in Arabidopsis. EMBO J. 2007;26:1934–1941. doi: 10.1038/sj.emboj.7601647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmitz R, Sung S, Amasino R. Histone arginine methylation is required for vernalization-induced epigenetic silencing of FLC in winter-annual Arabidopsis thaliana. Proc Natl Acad Sci USA. 2008;105:411–416. doi: 10.1073/pnas.0710423104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hong S, Song HR, Lutz K, Kerstetter RA, Michael TP, McClung CR. Type II protein arginine methyltransferase 5 (PRMT5) is required for circadian period determination in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2010;107:21211–21216. doi: 10.1073/pnas.1011987107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farré EM, Harmer SL, Harmon FG, Yanovsky MJ, Kay SA. Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr Biol. 2005;15:47–54. doi: 10.1016/j.cub.2004.12.067. [DOI] [PubMed] [Google Scholar]
- 18.Simpson CG, Fuller J, Maronova M, Kalyna M, Davidson D, McNicol J, et al. Monitoring changes in alternative precursor messenger RNA splicing in multiple gene transcripts. Plant J. 2008;53:1035–1048. doi: 10.1111/j.1365-313X.2007.03392.x. [DOI] [PubMed] [Google Scholar]
- 19.Xu X, Hoang S, Mayo M, Bekiranov S. Application of machine learning methods to histone methylation ChIP-Seq data reveals H4R3me2 globally represses gene expression. BMC Bioinformatics. 11:396. doi: 10.1186/1471-2105-11-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alló M, Buggiano V, Fededa JP, Petrillo E, Schor I, de la Mata M, et al. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat Struct Mol Biol. 2009;16:717–724. doi: 10.1038/nsmb.1620. [DOI] [PubMed] [Google Scholar]
- 21.Schor IE, Rascovan N, Pelisch F, Alló M, Kornblihtt AR. Neuronal cell depolarization induces intragenic chromatin modifications affecting NCAM alternative splicing. Proc Natl Acad Sci USA. 2009;106:4325–4330. doi: 10.1073/pnas.0810666106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng X, Gu L, Liu C, Lu T, Lu F, Lu Z, et al. Arginine methylation mediated by the Arabidopsis homolog of PRMT5 is essential for proper premRNA splicing. Proc Natl Acad Sci USA. 2010;107:19114–19119. doi: 10.1073/pnas.1009669107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pomeranz Krummel DA, Oubridge C, Leung AKW, Li J, Nagai K. Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature. 2009;458:475–480. doi: 10.1038/nature07851. [DOI] [PMC free article] [PubMed] [Google Scholar]