Abstract
FGFRL1 is the fifth member of the fibroblast growth factor receptor (FGFR) family. Similar to the other members, it harbors three Ig loops in its extracellular domain, but in contrast to the other receptors, it lacks the intracellular protein tyrosine kinase domain that would be required for signaling by transphosphorylation. FGFRL1 is mainly found in the musculoskeletal system, where it appears to inhibit cell proliferation but to induce cell adhesion and differentiation. Mice with a targeted disruption of the FGFRL1 gene die during birth due to a malformed diaphragm muscle, which is not strong enough to inflate the lungs after birth. Expression of FGFRL1 is highly upregulated during the differentiation of myoblasts to multinucleated myotubes, suggesting an important role for FGFRL1 in cell-cell fusion. Recently we showed that FGFRL1 does indeed induce fusion of cultured cells into large syncytia. A reporter gene assay demonstrated that the third Ig domain and the transmembrane domain of FGFRL1 are both necessary and sufficient to fuse CHO cells into syncytia comprising several hundred nuclei. At the contact site, the fusing cells reveal a peculiar net-like structure with pores of about 1 µm diameter. It is possible that these structures represent membrane areas with fusion pores that set in motion the cell-cell fusion process. FGFRL1 is the first mammalian protein that is capable of triggering cell-cell fusion in vitro.
Key words: fibroblast growth factor (FGF), fibroblast growth factor receptor (FGFR), FGFRL1, cell-cell fusion, fusion pores, muscle formation, CD9
Cell-cell fusion is a tightly controlled process, by which adjacent cells combine into a single, syncytial cell containing several nuclei and a mixed cytoplasm.1–4 During the embryonic development of mammals, cell fusion plays an important role in the initial fertilization process (fusion of sperm and egg) and later in the formation of bones (fusion of macrophages to osteoclasts), skeletal muscles (fusion of myoblasts to myotubes) and the placenta (fusion of trophoblasts to the trophoblast layer). While the fusion of viruses with the host plasma membrane and the fusion of intracellular vesicles have been studied in great detail, the fusion of two entire cells is poorly understood. Nevertheless, it has been demonstrated that this process begins with the tight adhesion of two adjacent cells, followed by the formation of fusion pores that enlarge until cytoplasmic mixing is achieved. During membrane fusion, so called fusogens, i.e., proteins that mingle the two plasma membranes, appear to play a decisive role.2 Only very few fusogens have been described to date and often it is not clear whether these fusogens are really involved in the fusion process or just in the tight adhesion of the two cells. In C. elegans the two transmembrane proteins EFF-1 and AFF-1 play a role in epithelial cell fusion,2 in Drosophila melanogaster the proteins Duf, Rst, Sns and Hbs have some function in myoblast fusion,4 and in mammals the proteins Izumo and CD9 are involved in the fusion process of the gametes.2,5 Interestingly, Duf, Rst, Sns, Hbs and Izumo are all members of the Ig domain superfamily, suggesting that Ig domains are involved in the fusion process.
Recently we have demonstrated that FGFRL1 (fibroblast growth factor receptor-like 1) can induce the fusion of two mammalian cells.6 FGFRL1 represents the fifth member of the fibroblast growth factor receptor (FGFR) family.7 Similar to other potential fusogens, it comprises three extracellular Ig loops, a single transmembrane domain and a relatively short intracellular domain.8,9 In contrast to the classical FGFRs,10 the intracellular domain of FGFRL1 does not exhibit any tyrosine kinase activity but only a short C-terminal tail with a peculiar histidine-rich motif.11 To trigger the fusion process, only the third Ig domain and the transmembrane domain of FGFRL1 are required as demonstrated by a highly reproducible cell fusion assay.6 This assay involves two different cell lines. On the one hand, FGFRL1-transfected HEK-TetOn cells, which can be induced to express FGFRL1 by the addition of doxycycline. These cells constitutively express the gene for the Tet-transactivator protein in the cytoplasm. On the other hand, the fusion assay involves CHO cells, which have been transfected with a GFP construct that is under the control of the Tet-transactivator protein. After mixing of the two cell populations, expression of GFP will indicate whether cell fusion has occurred since GFP can only be transcribed when the Tet-transactivator protein has diffused from the HEK-TetOn cells to the CHO cells. Figure 1A shows a typical fusion experiment performed with this assay. Besides many cells with single nuclei (blue), a large syncytial aggregate is observed, which expresses GFP in its cytoplasm (green) and which comprises many nuclei (light blue). At the periphery of the syncytium, there are remains of HEK-TetOn cells that may just have fused and that contain FGFRL1 on the membrane (red).
Figure 1.
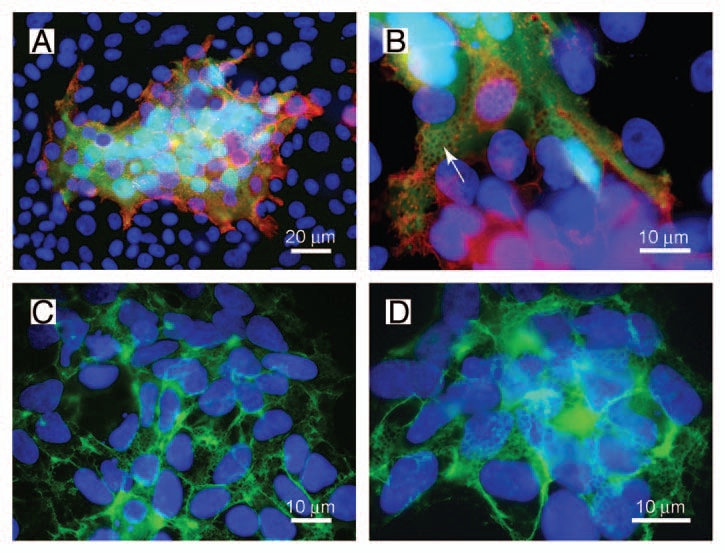
A net-like structure is observed during fusion of HEK-TetOn cells with CHO cells. (A and B) HEK-TetOn cells that express FGFRL1 on their cell surface were seeded together with CHO cells that had been transfected with a GFP construct, which is under the control of the Tet transactivator protein. Photographs of two syncytial cells were taken at different magnifications. FGFRL1 at the surface of the syncytia was stained with a monoclonal antibody, followed by a Cy3 labeled secondary antibody (red). GFP (green) is expressed after the Tet transactivator protein has diffused from the HEK-TetOn cells to the CHO cells. At higher magnification, a net-like structure with pores (arrow) becomes visible. (C and D) The net-like structure is also observed when the HEK-TetOn cells were cultivated without the CHO cells. In this case, FGFRL1 was stained with the monoclonal antibody, followed by a Cy2-labeled secondary antibody (green). When the HEK-TetOn cells were seeded at higher density, the net-like structures are found to be distributed around the entire cells.
When the syncytial aggregates are inspected under higher magnification, a peculiar net-like structure becomes visible (Fig. 1B). The same structure is also observed when the HEK-TetOn cells are cultivated in the absence of the CHO cells (Fig. 1C and D). In this case, the FGFRL1 receptor has been stained with our monoclonal antibody, followed by a Cy2-labeled (green) secondary antibody. The net-like structures are preferentially found at regions where two cells contact each other (Fig. 2 and arrow), but not at sites where the cells touch only the culture dish. When the HEK-TetOn cells are seeded more densely into the culture plate, the nets can be detected around the entire cells (Fig. 1C and D). Typically, the “pores” of the net-like structure have a diameter of 1 µm. Nets with pores have been observed with the full-length FGFRL1 construct as well as with the FGFRL1ΔC construct that is missing most of the intracellular domain. Taken together, our results suggest that FGFRL1 expression induces alterations at the plasma membrane, which microscopically appear as net-like structures when the cells are stained with antibodies for FGFRL1. In the fusing HEK and CHO cells, these alterations are especially prominent at the sites of cell-cell contact where the fusion process occurs.
Figure 2.
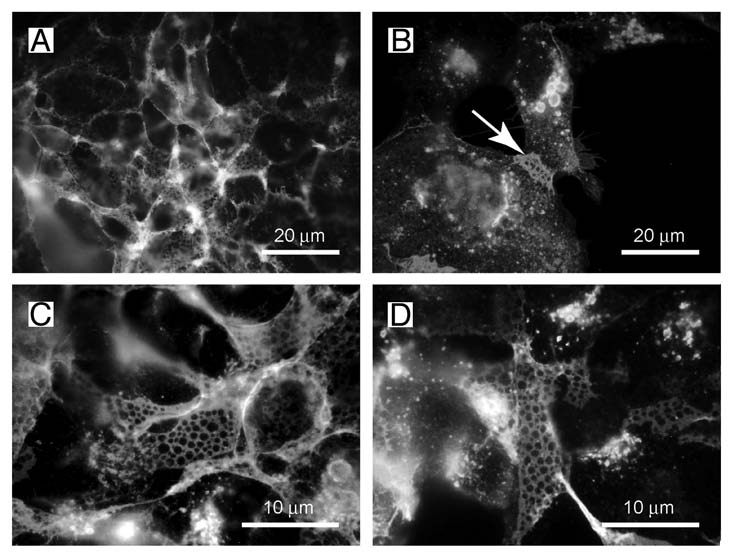
The net-like structures on the surface of HEK-TetOn cells show pores with a diameter of 1 ?m. (A and B) The net-like structures are preferentially observed at membrane regions where two cells touch each other (arrow). (C and D) The diameter of the pores is approximately 1 µm as determined at higher magnification.
We now propose that the net-like structures represent membrane areas with alterations in the lipid bilayer that may predispose two opposing cell membranes to fuse. Our structures are highly reminiscent of the microvilli zippers that have recently been described by Singethan et al. in 2008.12 These authors reported that the candidate fusogen CD9 forms a similar net-like structure on epithelial (Vero, HeLa) and endothelial (HUVEC) cells when stained with a monoclonal antibody against CD9. The authors further analyzed the net like structure by scanning electron microscopy and discovered that the pores represented the light microscopic equivalent of microvilli-like protrusions that emerged from the opposing cells. These microvilli had a length of several µm and appeared to form a zipper between opposing cells.12 CD9 is expressed on many cell types including the mammalian oocyte, where it is involved in the fusion of the sperm and egg.13–15 Consequently, oocytes from CD9-deficient mice are unable to fuse with sperm. By electron microscopy, CD9 has been localized to the microvillar membrane where it appears to affect the radius of membrane curvature at the microvillar tip.16 In CD9 knockout mice, microvillar morphology is impaired and the curvature is altered.
Based on the striking similarities between the net-like structures observed with the CD9 antibodies and the structures presented in Figures 1 and 2, we suggest that FGFRL1 might also be involved in the formation of microvilli zippers between two adjacent cells. The extreme curvature of the FGFRL1 induced microvilli could destabilize the membrane to an extent that is sufficient to cause the fusion of susceptible cells such as CHO cells. The exact mechanism, by which FGFRL1 may induce the formation of the pore-like structure and the protrusion of the microvilli is not yet known. However, it is possible that the Ig3 domains of the FGFRL1 molecules assemble to large aggregates that cause an unusual curvature of the plasma membrane as found for CD9.16 Such a local membrane curvature may stimulate membrane fusion as suggested in the literature.17,18
Thus, FGFRL1 represents a novel membrane protein that fulfills most of the criteria, which should be met in order to define it as a true fusogen2: (1) FGFRL1 is expressed at the time and the location of cell-cell fusion; (2) expression of FGFRL1 is sufficient to fuse two cells that under normal conditions do not fuse; and (3) FGFRL1 induces fusion of heterologous cells in culture. One criterion that is not fulfilled in our fusion system concerns the fact that FGFRL1 is not essential for cell fusion. Cells derived from Fgfrl1 deficient mice19 can still fuse in the absence of FGFRL1.6 We must therefore assume that the fusion machinery of mammals is redundant. Other proteins might exist that also promote cell fusion and that substitute for FGFRL1 in our knock-out animals.
Acknowledgements
This study was supported by grants from the Swiss National Science Foundation (3100A-127046) and from the Swiss Foundation for Research on Muscular Diseases.
References
- 1.Chen EH, Grote E, Mohler W, Vignery A. Cell-cell fusion. FEBS Lett. 2007;581:2181–2193. doi: 10.1016/j.febslet.2007.03.033. [DOI] [PubMed] [Google Scholar]
- 2.Oren-Suissa M, Podbilewicz B. Cell fusion during development. Trends Cell Biol. 2007;17:537–546. doi: 10.1016/j.tcb.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 3.Martens S, McMahon HT. Mechanisms of membrane fusion: disparate players and common principles. Nat Rev Mol Cell Biol. 2008;9:543–556. doi: 10.1038/nrm2417. [DOI] [PubMed] [Google Scholar]
- 4.Abmayr SM, Zhuang S, Geisbrecht ER. Myoblast fusion in Drosophila. Methods Mol Biol. 2008;475:75–97. doi: 10.1007/978-1-59745-250-2_5. [DOI] [PubMed] [Google Scholar]
- 5.Inoue N, Ikawa M, Isotani A, Okabe M. The immunoglobulin superfamily protein Izumo is required for sperm to fuse with eggs. Nature. 2005;434:234–238. doi: 10.1038/nature03362. [DOI] [PubMed] [Google Scholar]
- 6.Steinberg F, Gerber S, Rieckmann T, Trueb B. Rapid fusion and syncytium formation of heterologous cells upon expression of the FGFRL1 receptor. J Biol Chem. 2010;285:37704–37715. doi: 10.1074/jbc.M110.140517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trueb B. Biology of FGFRL1, the fifth fibroblast growth factor receptor. Cell Mol Life Sci. 2011;68:951–964. doi: 10.1007/s00018-010-0576-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiedemann M, Trueb B. Characterization of a novel protein (FGFRL1) from human cartilage related to FGF receptors. Genomics. 2000;69:275–279. doi: 10.1006/geno.2000.6332. [DOI] [PubMed] [Google Scholar]
- 9.Sleeman M, Fraser J, McDonald M, Yuan S, White D, Grandison P, et al. Identification of a new fibroblast growth factor receptor, FGFR5. Gene. 2001;271:171–182. doi: 10.1016/s0378-1119(01)00518-2. [DOI] [PubMed] [Google Scholar]
- 10.Beenken A, Mohammadi M. The FGF family: biology, pathophysiology and therapy. Nat Rev Drug Discov. 2009;8:235–253. doi: 10.1038/nrd2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhuang L, Karotki AV, Bruecker P, Trueb B. Comparison of the receptor FGFRL1 from sea urchins and humans illustrates evolution of a zinc binding motif in the intracellular domain. BMC Biochemistry. 2009;10:33. doi: 10.1186/1471-2091-10-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singethan K, Muller N, Schubert S, Luttge D, Krementsov DN, Khurana SR, et al. CD9 clustering and formation of microvilli zippers between contacting cells regulates virus-induced cell fusion. Traffic. 2008;9:924–935. doi: 10.1111/j.1600-0854.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le Naour F, Rubinstein E, Jasmin C, Prenant M, Boucheix C. Severely reduced female fertility in CD9-deficient mice. Science. 2000;287:319–321. doi: 10.1126/science.287.5451.319. [DOI] [PubMed] [Google Scholar]
- 14.Miyado K, Yamada G, Yamada S, Hasuwa H, Nakamura Y, Ryu F, et al. Requirement of CD9 on the egg plasma membrane for fertilization. Science. 2000;287:321–324. doi: 10.1126/science.287.5451.321. [DOI] [PubMed] [Google Scholar]
- 15.Kaji K, Oda S, Shikano T, Ohnuki T, Uematsu Y, Sakagami J, et al. The gamete fusion process is defective in eggs of Cd9-deficient mice. Nat Genet. 2000;24:279–282. doi: 10.1038/73502. [DOI] [PubMed] [Google Scholar]
- 16.Runge KE, Evans JE, He ZY, Gupta S, McDonald KL, Stahlberg H, et al. Oocyte CD9 is enriched on the microvillar membrane and required for normal microvillar shape and distribution. Dev Biol. 2007;304:317–325. doi: 10.1016/j.ydbio.2006.12.041. [DOI] [PubMed] [Google Scholar]
- 17.McMahon HT, Kozlov MM, Martens S. Membrane curvature in synaptic vesicle fusion and beyond. Cell. 2010;140:601–605. doi: 10.1016/j.cell.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 18.Graham TR, Kozlov MM. Interplay of proteins and lipids in generating membrane curvature. Curr Opin Cell Biol. 2010;22:430–436. doi: 10.1016/j.ceb.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baertschi S, Zhuang L, Trueb B. Mice with a targeted disruption of the Fgfrl1 gene die at birth due to alterations in the diaphragm. FEBS J. 2007;274:6241–6253. doi: 10.1111/j.1742-4658.2007.06143.x. [DOI] [PubMed] [Google Scholar]


