Abstract
In cancerous cells, physiologically tight regulation of protein synthesis is lost, contributing to uncontrolled growth and proliferation. We describe a novel experimental cancer therapy approach based on genetically recombinant poliovirus that targets an intriguing aberration of translation control in malignancy. This strategy is based on the confluence of several factors enabling specific and efficacious cancer cell targeting. Poliovirus naturally targets the vast majority of ectodermal/neuroectodermal cancers expressing its cellular receptor. Evidence from glioblastoma patients suggests that the poliovirus receptor is ectopically upregulated on tumor cells and may be associated with stem cell-like cancer cell populations and proliferating tumor vasculature. We exploit poliovirus’ reliance on an unorthodox mechanism of protein synthesis initiation to selectively drive viral translation, propagation and cytotoxicity in glioblastoma. PVSRIPO, a prototype nonpathogenic poliovirus recombinant, is scheduled to enter clinical investigation against glioblastoma.
Keywords: eIF4E, eIF4G, Erk, glioblastoma, IRES, Mnk1, Necl-5, PKC-α, poliovirus, translation
Originally, the concept of ‘oncolytic’ viruses, or viruses used to target cancerous cells for destruction, stems from anecdotal reports of spontaneous cancer remissions coincidental with natural infection [1] or the use of live attenuated vaccines [2]. Improved understanding of the molecular basis for viral host cell tropism, cytotoxicity and cell type-specificity has opened up possibilities for targeted design of antineoplastic strategies based on viruses. Contemporary efforts to harness viruses for cancer therapy are based on viruses with inherently low human pathogenic potential (e.g., the orphan reovirus [3]), veterinary pathogens with unknown human pathogenicity (e.g., vesicular stomatitis virus [4]) or human pathogens genetically engineered to selectively kill cancerous cells without concomitant cytotoxicity in normal cells (e.g., herpes simplex virus-1 [5]). Achieving success with oncolytic viruses in the clinic has been more challenging than the encouraging results achieved in many tissue culture or animal studies would suggest. The reason for this is that viruses depend on intricate relationships with their hosts that determine tropism for the intended tumor target(s) and the efficiency of target cell killing. The favorable conditions encountered by many viruses (or genetically engineered variants thereof) in select transformed cell lines most often are poor representations of the clinical situation. Moreover, issues such as delivery, pre-existing immunity or innate immune responses complicate therapeutic uses of viruses. In the following paragraphs, we describe an experimental cancer therapy approach based on poliovirus. The focus of this article is on defining the molecular determinants for viral tumor tropism and target cell killing to enable rational design of clinical studies. The presented approach rests on specific targeting of the poliovirus receptor, nectin-like molecule-5 (Necl-5), and manipulation of an unorthodox translation initiation mechanism exemplified by poliovirus. Therefore, and because there are few mechanistic parallels with other proposed oncolytic virus species, our article will focus on oncolytic poliovirus recombinants.
Mechanism(s) of oncolytic virus efficacy
Oncolytic viruses may exert direct effects on tumors, stemming from virus-mediated host cell killing, and/or secondary effects owing to host responses to infected and/or destroyed tumor. Recent, encouraging clinical results and many experiments in animal models indicate that both are important components contributing to efficacy in the clinic. For example, an oncolytic herpes simplex virus-1 engineered to simultaneously infect and kill tumor cells and stimulate the immune system (through expression of granulocyte–macrophage colony-stimulating factor [6]) demonstrated remarkable antineoplastic efficacy after intratumoral administration in several clinical trials [7,8].
The approach described here is targeted to exert the two-pronged effects of direct tumor cell killing and engaging the host immune system. Infection of susceptible transformed cells with poliovirus produces rapid cell death and lysis. While many events early in the viral life-cycle may contribute to this, a particularly important event in producing host cell death is expression of the viral 2A protease (2Apro). Executing poliovirus’ strategy to intercept host cell gene expression while stimulating viral translation, 2Apro engages in rapid cleavage of key host cell components involved in mRNA export [9] and translation [10]. Demonstrating its potent cytolytic properties, 2Apro alone is sufficient to trigger cell death [11].
Poliovirus’ brief and destructive interactions with host cells elicit potent host responses against infected/lysed tumor. Owing to its austere genetic repertoire and limited lifespan inside infected hosts, poliovirus does not dispose of sophisticated strategies to impede host defense mechanisms. Accordingly, efficient host-mediated, antineoplastic responses were observed with oncolytic polioviruses tested in a syngeneic murine neuroblastoma model [12]. The mouse tumor, Neuro2A, was transduced with the human poliovirus receptor gene to enable viral targeting [12]. Destruction of Neuro2A cells by oncolytic poliovirus produced a robust antitumor response [12], which was T-cell mediated [13]. In addition, prior oncolytic virus treatment mediated long-term immunity to tumor rechallenge [12]. To allow testing of poliovirus oncolysis in the clinic, a vast effort was undertaken to: first, generate nonpathogenic poliovirus recombinants with tumor-selective replication phenotypes and demonstrate their safety in nonhuman primates; second, to define the molecular mechanisms for tumor cell targeting; and third, to elucidate the molecular mechanisms responsible for selective tumor cytotoxicity.
Poliovirus tropism for cancerous cells
It is reasonable to assume that to succeed, oncolytic viruses must recognize host cell surface signatures mediating viral attachment and entry in the intended (tumor) target tissue. Lacking insight into the molecular mechanisms of viral host cell entry or missing correlative evidence from the intended tumor target impedes proper design of clinical trials. Identification of host cell determinants of viral tropism and documentation of their expression/distribution in the intended target can help to properly identify suitable target neoplasias or profile patients in clinical studies. Retargeting viruses towards confirmed tumor cell surface markers through genetic engineering may help to overcome restricted tropism of some proposed oncolytic agents [14–16]. The strategy described in this article is motivated by poliovirus’ natural tropism for an intriguing cell surface molecule broadly associated with neoplasia.
The poliovirus receptor is a tumor antigen
The poliovirus receptor (synonymous with CD155), now classified as Necl-5 [17], was originally identified via virus-neutralizing antibodies raised against fractionated HeLa cell membranes [18]. Based on all available empirical evidence, Necl-5 alone is necessary and sufficient to confer susceptibility of mammalian cells to wild-type (WT) poliovirus [19]. Poliovirus’ host range is restricted to humans and old-world primates, owing to the viruses reliance on species-specific Necl-5. This restriction can be overcome by supplying exogenous human Necl-5. For example, transgenic mice expressing the human Necl-5 gene, upon poliovirus infection, develop a syndrome consistent with paralytic poliomyelitis in humans [20,21]. In susceptible primates, Necl-5 expression patterns overlap with the susceptibility to poliovirus. For example, cells at the primary site of virus infection in the GI tract, which are currently undefined cell populations in the epithelial lining and associated lymphatic structures express Necl-5 [22]. Similarly, selective replication of poliovirus in spinal cord motor neurons in Necl-5-transgenic mice is explained by restrictive Necl-5 expression in this cellular compartment [23].
The nectin/nectin-like family of genes comprises a group of cell adhesion molecules characterized by three extracellular immunoglobulin-like domains, a transmembrane region and a cytoplasmic domain. Nectin/Necl molecules have been implicated in various cell adhesion functions in a variety of key physiological processes; for example, development or regenerative tissue responses (reviewed in [17]). Apart from Necl-5’s role as a receptor for poliovirus, members of the nectin/Necl family also serve as cell attachment factors for α-herpesviruses [24]. Although a precise biological function has not been assigned, Necl-5 may be expressed in the embryonic CNS [25] and, like other nectin/Necl molecules, may be involved in the development of CNS structure [17]. In contrast to restrictive expression in the adult organism, Necl-5 is broadly associated with malignancy. Ecoptic nectin/Necl molecule expression may contribute to basic biological properties of cancer cells (e.g., invasion and metastasis, altered contact inhibition or unhinged proliferation control [reviewed in [17]]). Ectopic expression of Necl-5 (or its rodent homolog TagE4) has been reported for glial [26–29], colorectal [30], breast [31], lung [32] or hepatocellular [33] carcinomas in patients. With the exception of select lymphoma cell lines [34], Necl-5 expression is universal in cell lines established from ectodermal/neuroectodermal tumors.
Several cis-acting regulatory elements within the Necl-5 promoter have been identified and the corresponding trans-acting factors were implicated in regulation of the Necl-5 gene [35–37]. While the involvement of certain morpho genic factors, such as sonic hedgehog [36], is suggestive of a role in ectopic upregulation in malignancy, the basic mechanisms contributing to widespread expression of Necl-5 in cancers are not understood. Studies in rat livers indicate that expression of Necl-5 (or its rat TagE4 homolog) may be upregulated following acute or chronic injury and the ensuing regenerative response [33]. As in analogous human studies, tightly regulated rat Necl-5 expression in normal hepatocytes was substantially elevated upon induced transformation of hepatocellular carcinomas [33].
Necl-5 is prominently associated with glioblastoma
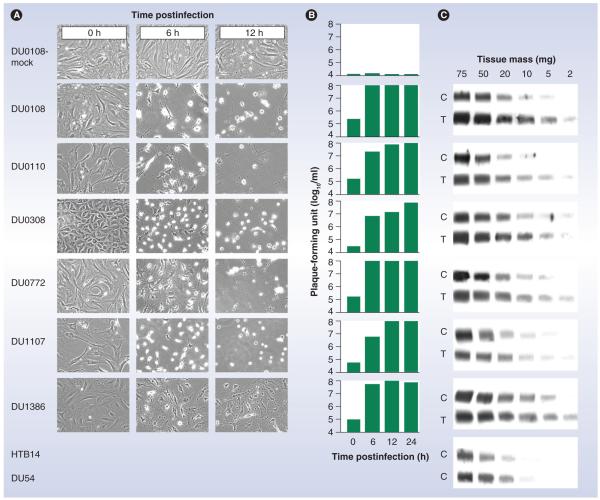
There is ample evidence for a particularly intricate association of Necl-5 with primary CNS neoplasia, for example, glioblastoma (GBM). Functional studies have implicated Necl-5 in GBM cell invasion and intracerebral dispersion [28,29] and immunohistochemical studies have located the molecule in tumor cells at the invasive front of tumors [32]. Fluorescence-activated cell sorting and immunohistochemical studies of GBM patient tumors confirmed universal and abundant expression of Necl-5, detected by immunoblot of lysates obtained from such tumors [38]. Interestingly, these studies also revealed strong expression in CD133+ stem cell-like GBM cell populations in patients and in proliferating tumor vasculature [38]. The presence of Necl-5 in such prominent tumor components may be particularly important for efficacious targeting of GBM. We reported analyses from a series of patient tumors, which demonstrated abundant Necl-5 expression in GBM tissues and corresponding receptor levels in primary explant cultures derived thereof (Figure 1) [27]. All primary-explant GBM cultures supported efficient poliovirus propagation, which produced killing of all cells in all cultures by 12 h postinfection, implying expression of Necl-5 in every cell (the assays were conducted using a nonpathogenic poliovirus recombinant, PVSRIPO [see following sections]) (Figure 1).
Figure 1. Expression of nectin-like molecule-5 in glioblastoma.
(A) A panel of six primary-explant glioblastoma (GBM) cultures were infected with PVSRIPO and images of the infected cultures were acquired at the indicated time points of hours postinfection. At a total of 12 h postinfection, all cells were lysed in all samples. The cytopathic effects of PVSRIPO in laboratory glioma cell lines (e.g., HTB14 or DU54) have been reported previously (Figure 5D) [26,27]. (B) Quantification of PVSRIPO levels in infected cultures indicates proficient propagation of the virus in infected primary-explant GBM cells. (C) Immunoblot analyses of nectin-like molecule-5 in homogenates obtained from primary explant cells (C) or in homogenates from GBM tissues directly (T). HTB14 and DU54 are established laboratory glioma cell lines. Abundant nectin-like molecule-5 expression in primary-explant GBM cultures equals the observed levels in GBM patient tissues or in established laboratory cell lines.
C: Cell; T: Tissue.
Reproduced from [27] by permission of Oxford University Press.
Apart from simple, mechanistic considerations, such as Necl-5 expression in GBMs, these tumors are obvious and attractive targets for therapeutic intervention with oncolytic polioviruses. Owing to the lack of effective therapies, the outlook for GBM patients is particularly bleak. Poliovirus, a natural neuropathogen with inherent neuroinvasive properties, may be particularly apt at inducing antitumor responses against a type of neoplasm that is relatively resistant to other, more conventional forms of treatment.
Mechanisms of tumor cell specificity
There is solid empirical evidence suggesting that poliovirus is capable of targeting, infecting and killing cancer cells derived from ectodermal/neuroectodermal tumors in vitro and in animal tumor models [26]. Unfortunately, the Necl-5 tumor antigen mediating this property is also present on select normal cells (e.g., spinal cord motor neurons). Therefore, to enable clinical use of targeting Necl-5 with poliovirus, a strategy to selectively ablate viral cytotoxicity for normal CNS cells is needed.
Poliovirus success depends on early translation of viral genomes
Owing to the fact that poliovirus is a positive-strand RNA virus, the viral life-cycle is exceedingly simple. Poliovirus does not engage in intricate parasitic relationships with its host cells, but rapidly overruns and kills them to achieve maximal propagation efficiency. Immediately after the virus genome is uncoated, it is available for translation of viral proteins. This step may be the critical, rate-limiting event in the virus’ life-cycle, because it constitutes the main strategy of poliovirus to pre-empt defensive responses of the infected host cell. The first nonstructural polypeptide generated by poliovirus is the viral 2Apro [39]. This enzyme is cotranslationally released from the nascent viral polyprotein by autoproteolytic cleavage. 2Apro broadly intercepts host responses to infection by cleaving the central scaffold of the protein synthesis apparatus, eukaryotic initiation factor (eIF)4G [10] and the nuclear pore complex [9]. These events occur with remarkable efficiency; for example, in infected transformed cells, eIF4G cleavage is complete by 2–3 h postinfection. The effects of 2Apro proteolysis (including cleavage of eIF4G and nuclear pore components, but possibly involving other, as-yet unidentified events [40]) lead to a shut down of host cell gene expression, thus limiting antiviral responses requiring protein biosynthesis. Meanwhile, none of this affects viral functions, because poliovirus employs an unorthodox translation mechanism that persists in the absence of intact eIF4G (see later) and the viral replication cycle does not involve the nuclear compartment. In fact, 2Apro itself enhances translation of the viral genome directly [41]. Therefore, in essence, at 2–3 h postinfection, host cells cease to maintain normal cellular function and are subverted for production of viral progeny.
Cap-independent translation of poliovirus
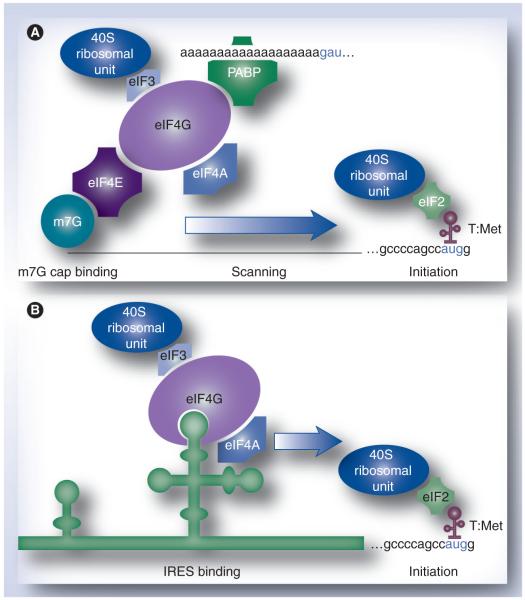
The core principle to subvert host cells employed by poliovirus is alternative translation initiation at the incoming viral genome. In eukaryotes, conventional translation occurs upon recruitment of ribosomal subunits via the canonical 5′ 7-methyl-guanidine (m7G) cap on mRNAs. This involves association of the cap-binding protein, eIF4E [42], with the cap structure and recruitment of its binding partner, the ‘ribosome adaptor’ eIF4G, which supplies the bridge to 40S ribosomal subunits (Figure 2A). Poliovirus is incapable of translating in this manner because the viral RNA genome lacks a 5′ cap structure [43]. Instead, poliovirus uses a cis-acting genetic element in its 5′ untranslated region, the internal ribosomal entry site (IRES), to recruit ribosomal subunits independent of a 5′ cap, or eIF4E (Figure 2B) [44,45]. For some viral IRESs, including poliovirus, this process depends on recruiting eIF4G to the IRES directly (Figure 2B) [46]. Poliovirus translation via the IRES functions equally well with intact eIF4G or a C-terminal eIF4G fragment (Ct), generated by 2Apro cleavage. Ct harbors the RNA-binding domains of eIF4G and contains the binding motif for eIF3 (and thus retains the ability to recruit 40S ribo somal subunits). Therefore, Ct is sufficient to satisfy the minimalist requirements for IRES-mediated translation of viral proteins. However, host cap-dependent translation cannot proceed because eIF4G cleavage removes the bridge linking the m7G-cap/eIF4E to 40S ribosomal subunits (Figure 2A).
Figure 2. Mechanisms of translation initiation.
(A) Conventional, cap-dependent translation occurs upon binding of eIF4E to the canonical m7G cap on eukaryotic mRNAs. This enables recruitment of the preinitiation complex, including 40S ribosomal subunits, to mRNAs. Assembly of the preinitiation complex at the 5′ cap precedes scanning of the 5′ untranslated region and initiation proper at the initiation codon. (B) Cap-independent translation at the poliovirus genome occurs upon direct recruitment of eIF4G to the IRES element in the viral 5′ untranslated region.
eIF: Eukaryotic initiation factor; IRES: Internal ribosomal entry site; m7G: 7-methyl-guanidine; PABP: Poly(A) binding protein; T:Met: Initiator (Met) tRNA.
Generating polioviruses with conditional replication in malignant cells
Prior to contemplating the strategy described here, we discovered that the IRES is a critical pathogenesis determinant. Exchanging the poliovirus IRES with its counterpart from human rhinovirus type 2 (HRV2), which generated the polio/rhinovirus chimera, RIPO, efficiently eliminated the ability of poliovirus to translate, propagate in and kill spinal cord motor neurons [47,48]. This was documented in a variety of neuron-like cell lines (e.g., neuroblastoma cells [47] and HEK293 cells [49]), in Necl-5-transgenic mice [47] or in cynomolgus macaques [48]. Meanwhile, viral replication in transformed cells, such as HeLa or GBM cells, is little changed compared with WT virus [26]. This indicates that the heterologous HRV2 IRES in RIPO mediates defective viral replication specifically in the CNS.
To maximize attenuation of RIPO, we generated PVSRIPO. PVSRIPO is the serotype 1, live attenuated SABIN poliovirus vaccine containing the HRV2 IRES. This virus combines neuroattenuation mediated by the hetero logous HRV2 IRES, with attenuating mutations mapping to the capsid and the coding region for the RNA-dependent RNA polymerase (reviewed in [50]). Two recent Investigational New Drug application-directed toxicology, biodistribution and shedding studies involving intracerebral administration of PVSRIPO in 42 cynomolgus macaques did not reveal morbidity or mortality.
Poliovirus CNS competence may be determined by the ability of the IRES to recruit eIF4G
What is the mechanistic basis for defective CNS replication of PVSRIPO? Genetic recombination experiments provided a possible answer to this key question. PVSRIPO shares defective CNS competence with the three live attenuated (SABIN) poliovirus vaccines. Each of these contains a single point mutation in their respective IRES elements, which is located in a distinctive stem-loop domain (SLD), SLD V (Figure 3A) [50]. The SABIN IRES mutations are critical determinants for CNS competence, as they categorically convert to the WT sequence in SABIN vaccine revertants that regained neurovirulence in patients with vaccine-associated paralytic poliomyelitis (reviewed in [50]). It is therefore compelling to speculate that SLD V is also involved in CNS incompetence, mediated by the HRV2 IRES.
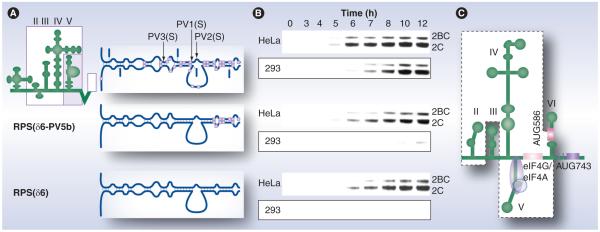
Figure 3. Stem-loop domain V in the human rhinovirus type 2 internal ribosomal entry site harbors neuronal incompetence.
(A) PVSRIPO constructs featuring diverse stem-loop domain (SLD) V with variable sequence content derived from human rhinovirus type 2 or wild-type PV type 1 (MAHONEY; gray boxes). (B) Viral translation in universally permissive HeLa cells or in PSVRIPO nonpermissive HEK293 cells (boxed). The loss of internal ribosomal entry site competence in HEK293 cells coincides with a discrete region of SLD V comprising the portion containing the attenuating point mutations in the SABIN vaccines. Viral translation in HeLa cells occurs at similar levels with all constructs, independent of the primary sequence of SLD V. (C) The eIF4G ‘landing pad’ on SLD V of the poliovirus internal ribosomal entry site is schematically indicated [46].
2BC: Poliovirus protein 2BC; 2C: Poliovirus protein 2C; eIF: Eukaryotic initiation factor; PV: Poliovirus.
Data taken from [49].
We generated PVSRIPO variants with various parts of SLD V derived from WT polio virus (Figure 3A) [49]. The capacity to translate in HEK293 cells, a neuroblastoid cell line that recapitulates the non-neurovirulent phenotype of PVSRIPO [49], maps to a portion of SLD V (Figure 3B). This region overlaps with the location of the SABIN-attenuating IRES mutations (Figure 3A) [49]. For experiments shown in Figure 3, PVSRIPO constructs with SLD VI deletions were used to focus on SLD V specifically [49]. We reported previously that the tip of IRES SLD VI may participate in determining IRES competence in neuron-like cells [48]. Intriguingly, elegant footprinting studies demonstrated that those regions in SLD V/VI identified in our genetic screens [48,49] are involved in recruiting eIF4G to the poliovirus IRES (Figure 3C) [46]. Therefore, CNS incompetence of PVSRIPO may reflect an inability to efficiently recruit eIF4G to the heterologous HRV2 IRES in normal neuronal cells.
This hypothesis would explain the relative genetic stability of PVSRIPO compated with the SABIN vaccines. As mentioned previously, the SABIN vaccines readily revert to neurovirulence, which involves conversion of attenuating point mutations in IRES SLD V to the WT sequence [51]. If the SABIN vaccine’s SLD V mutations alter the virus’ ability to recruit eIF4G to its IRES (as suggested by the available empirical evidence [46]), this defect is easily corrected with substitution of a single nucleotide, in particular when considering the notorious infidelity of positive-strand RNA virus RNA-dependent polymerases. By contrast, the heterologous HRV2 IRES represents a functionally intact regulatory unit, which may inherently exhibit reduced eIF4G affinity. HRV2, a minor-group rhinovirus, targets respiratory epithelial cells expressing the low-density lipoprotein receptor [52]. HRV infections do not produce histologically evident lesions in the human respiratory tract, suggesting a viral replication strategy that does not lead to overt host cell killing in vivo [53]. Part of this strategy may entail less efficient viral cap-independent translation, which would in turn favor ongoing host cell protein synthesis and host cell survival. These hypotheses are difficult to verify empirically owing to the absence of a practical animal model for enterovirus respiratory tract infections, but work in progress on such a murine model supports our assumptions [54,55]. If reduced eIF4G affinity is integral to HRV2 IRES structure, it may not be easily overcome by genetic adaptation, thus benefiting the genetic stability of PVSRIPO. This was borne out in serial in vivo passage studies of PVSRIPO in animal GBM models, which did not produce adaptation events leading to changing genotype or phenotype [56].
Our data suggest that in cancerous cells, such as GBM efficient viral cap-independent translation occurs no matter what the specific sequence context is in IRES SLD V. Therefore, all SLD V recombinants in our study exhibited similar propagation potential in HeLa cells (Figure 3B) [49]. This suggests that in cancerous cells, physiological controls that limit eIF4G recruitment to IRES-bearing RNAs are absent.
Therefore, the molecular basis for PVSRIPO cytotoxicity in cancer may be unhinged regulation of eIF4G recruitment to the heterologous HRV2 IRES. Two (not mutually exclusive) hypotheses may explain our findings: First, limited access of eIF4G to the IRES in normal neurons due to binding of IRES trans-acting factors to PVSRIPO RNA. PVSRIPO RNA could be masked by RNA-binding proteins that associate with viral RNA specifically in CNS cells and preclude eIF4G binding in such cells. Our research suggests that PVSRIPO RNA associates with proteins of the nuclear factor associated with dsRNA (NFAR) complex (dsRNA binding protein 76 [DRBP76] and its binding partner, nuclear factor 45) specifically in neuroblastoid HEK293 cells [57], impairing translation via the hetero logous HRV2 IRES [58]. A possible role for NFAR proteins in host defenses involving suppression of ribosome recruitment to viral RNAs has recently been described [59]. We reported an intriguing phenotypic distinction of DRBP76 in normal CNS neurons (the natural target for polioviruses) versus GBM cells [60]. DRBP76 is exceedingly abundant in the cytoplasm of normal CNS neurons, which is the site of PVSRIPO replication. Meanwhile, in GBM cells, DRBP76 is electrophoretically distinct from the normal neuronal protein (most likely representing a distinct isoform with variable post-translational modifications) and located exclusively in the nucleus [60]. Therefore, if abundant cytoplasmic NFAR proteins bind to PVSRIPO RNA in normal neurons and prevent ribosome recruitment to the HRV2 IRES, this obstacle is removed in GBM, where these proteins are found exclusively in the nucleus [60].
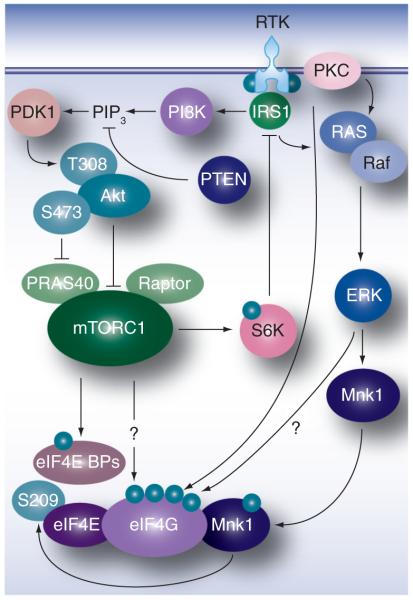
Second, enhanced eIF4G affinity for IRESs owing to post-translational modification of eIF4G in GBM may explain our findings. Translation initiation factor function may be altered in cancerous cells in a manner to indiscriminately favor cap-independent translation. This hypothesis is supported by the fact that in cancer cells, viral translation and propagation driven by the WT poliovirus IRES or the HRV2 IRES are equally efficient [26]. Major mitogenic signaling pathways that are universally deregulated in malignancy converge on eIF4G and eIF4E (Figure 4). Since eIF4G functions as the adaptor, recruiting PVSRIPO RNA to the host cell translation apparatus, and eIF4E targets eIF4G function to m7G-capped host mRNAs, these proteins are likely to influence viral IRES efficiency in infected host cells.
Figure 4. Mitogenic signaling pathways via PI3K–mTOR and Ras–Erk1/2 converge on translation machinery.
BP: Binding protein; eIF: Eukaryotic initiation factor; RTK: Receptor tyrosine kinase.
Mitogenic signal transduction controls PVSRIPO translation & propagation
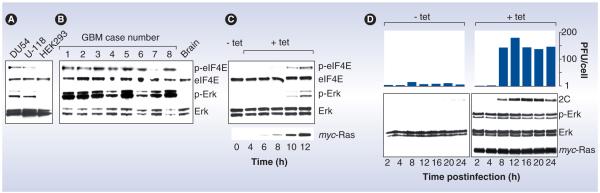
Protein kinase inhibition experiments established that signaling pathways involved in control over translation modulate PVSRIPO translation, replication and cell killing. Blocking the activity of the downstream eIF4E kinase and Erk1/2 substrate, MAPK signal-integrating kinase 1 (Mnk1) – using the Mnk1 inhibitor CGP57380 – depresses PVSRIPO translation, proliferation and cell killing in GBM cells (Figure 5B–D) [61]. Interestingly, in PVSRIPO nonpermissive HEK293 cells, Ras–Erk signaling is inherently low (Figure 6A) [61]. In fact, the relative levels of p-Erk1/2 and p-eIF4E in HEK293 versus GBM cells resembles those in normal human brain versus patient GBM tissues (Figure 6A & 6B) [61]. Activating Ras–Erk signaling in HEK293 cells by introducing a tetracycline-inducible form of oncogenic Ras induced phosphorylation of Erk1/2 and eIF4E to levels comparable with GBM cells or patients’ tumors (Figure 6C). This was accompanied by significantly enhanced PVSRIPO replication (Figure 6D) [61]. These studies implicated Ras–Erk signals in PVSRIPO translation, propagation and tumor cell killing. We fine-tuned our approach to investigate the precise molecular events responsible for mediating PVSRIPO cell killing. Introducing constitutively active Mnk1 into HEK293 cells, which are naturally resistant to PVSRIPO cytotoxicity [49,62], mediates enhanced viral cytotoxicity. Conversely, a dominant-negative Mnk1 variant had the opposite effect [61]. Mnk1 binding to eIF4G [63], which is strongly responsive to Ras–Erk activation [64], results in phosphorylation of Ser209 in eIF4E [65]. It therefore appears that downstream Ras–Erk signals converging on Mnk1, its binding partner eIF4G and its substrate eIF4E control susceptibility to PVSRIPO in cancer cells. This may involve differential regulation of eIF4G’s ability to bind to the IRES in PVSRIPO (see following section).
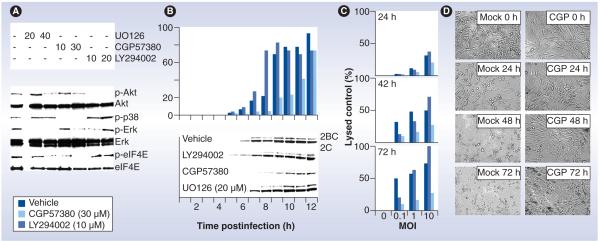
Figure 5. Protein kinase inhibitors modulate PVSRIPO oncolysis in glioblastoma cells.
(A) Immunoblots of kinase substrates in the PI3K (Akt) and Ras (p38, Erk and eIF4E) pathways 2 h after treatment with vehicle, the Mek1 inhibitor UO126, the Mnk1 inhibitor CGP57380 or the PI3K inhibitor LY294002 (concentrations in µM are shown at the top). (B) Kinetics of viral growth (top) and translation (bottom) in mock- or inhibitor-treated U-118 cells infected with PVSRIPO. Progeny was quantified by plaque assay and viral translation was measured by immunoblot of the viral nonstructural proteins 2BC/2C. Viral propagation in mock- and UO126-treated cells was similar (data not shown). (C) PVSRIPO cytotoxicity in mock- or inhibitor-treated U-118 cells at the indicated intervals. (D) Photomicrographs of PVSRIPO-infected (MOI = 1) and vehicle (mock)- or CGP57380 (CGP; 30 μM)-treated U-118 cells at the indicated intervals.
2BC: Poliovirus protein 2BC; 2C: Poliovirus protein 2C; eIF: Eukaryotic initiation factor; MOI: Multiplicity of infection.
Reproduced with permission from [61].
Figure 6. Oncogenic H-Ras rescues PVSRIPO growth in nonpermissive HEK293 cells.
(A) Erk1/2 signaling and inherent phosphorylation of eIF4E in HEK293 cells is reduced compared with DU54 and U-118 GBM cells. (B) Universally active Erk1/2 and eIF4E phosphorylation in GBM patients and absent signal in the normal primate brain. (C) tet-inducible expression of oncogenic Ras in HEK293 cells produces Erk1/2 and eIF4E phosphorylation, and a signaling signature similar to U-118 GBM cells or GBM patient tissues. (D) PVSRIPO growth (top) and translation (bottom) in mock- or tet-induced cells. Immunoblots confirm myc-Ras expression and Erk1/2 signaling. Poliovirus 1(S) progeny recovered from infected (multiplicity of infection = 10) mock- or tet-induced cells are shown at the indicated intervals.
2C: Poliovirus protein 2C; eIF: Eukaryotic initiation factor; GBM: Glioblastoma; PFU: Plaque-forming unit; tet: Tetracycline.
Reproduced with permission from [61].
Inhibition of the PI3K pathway (using the PI3K inhibitor LY294002) slightly, but reproducibly, enhanced PVSRIPO translation, replication and cell killing in GBM (Figure 5B & 5C) [61]. In addition, treating GBM xenografts with PVSRIPO in combination with PI3K inhibitors led to accelerated loss of viable tumor [61]. While mTORC1 inhibition modestly enhances PVSRIPO translation, replication and tumor cell killing in vitro and in vivo, Ras–Erk signaling pathways have a decisive influence over PVSRIPO oncolysis (Figure 5) [61]. This may indicate that Erk1/2 signaling to the protein synthesis apparatus exerts dominant effects on the regulation of cap-independent translation in cancer cells.
Mechanistic basis for PVSRIPO dependence on mitogenic signal transduction
Several proposed oncolytic viruses share a bias towards certain signaling pathways. For example, inhibition of mTORC1 stimulates vesicular stomatitis virus-mediated tumor cell killing by impeding mTORC1-dependent cytokine responses [66]. Since mitogenic signaling pathways are pleiotropic and host cell cytotoxicity of oncolytic viruses is influenced by multiple factors, unraveling the mechanistic basis for the effect of signal transduction on viral cancer cell killing can be daunting. In this regard, poliovirus’ singular dependence on a specific translation initiation event (binding of eIF4G to the IRES) at a defined moment during the viral life-cycle (early after uncoating of the viral RNA) presents a uniquely simple scenario. As outlined previously, the virus’ strategy to combat host cell defenses and unleash viral translation and propagation depend on immediate cap-independent translation of viral nonstructural proteins. Thus, the events controlling ribosome recruitment (via eIF4G) at the incoming viral genome may determine the outcome of the infection. It is obvious that once 2Apro is expressed, host eIF4G is cleaved and the host cell has been subverted, the activity of specific signal transduction pathways no longer matter to the outcome of infection.
The factors determining eIF4G binding to viral IRESs are not immediately clear. Although eIF4G is a confirmed RNA-binding protein [67], it lacks a classic RNA recognition motif or other defined structures known to mediate RNA binding. Similarly, IRESs are defined by their function, not their structure. IRESs known to initiate translation via direct recruitment of eIF4G (e.g., the IRESs of poliovirus [46], the c-myc oncogene mRNA [68,69] or the VEGF mRNA [69]) have no apparent structural similarities. Therefore, there is no defined RNA structure or motif that can be examined to determine a capacity for eIF4G binding.
According to phosphoproteomic screens, eIF4G has approximately 17 mitogen-responsive phosphorylation sites [70]. It is thus conceivable that signal transduction to eIF4G (e.g., via Ras–Erk) leads to phosphorylation events that alter its RNA-binding properties. Despite strong evidence for phosphorylation of eIF4G upon activation of mitogenic signal transduction pathways [71] and a prominent role for eIF4G in protein synthesis regulation, no specific kinases or signal transduction pathways converging on eIF4G have been identified. In addition, the biological effects of signaling to eIF4G remain a mystery. Signal transduction to eIF4G and its effect on translation regulation (e.g., via the poliovirus IRES) is an active area of investigation in our laboratory. While it is too early to communicate definitive insight into this difficult regulatory system, we propose a number of hypotheses.
First, a cluster of mitogen-response phosphorylation sites in eIF4G maps to the ‘interdomain linker’, a flexible loop connecting HEAT domains 1 and 2 of eIF4G [72]. This region also contains the RNA-binding determinants of eIF4G [67]. It is thus conceivable that phosphorylation of residues in the eIF4G interdomain linker influence cap-independent translation by regulating eIF4G’s ability to bind to IRESs in target mRNAs.
Second, our research suggests that Ras–Erk activation controls the association of eIF4G with Mnk1 [73]. Binding of Mnk1 to eIF4G is essential for phosphorylation of the Mnk1 substrate, eIF4E [63]. It is currently not understood whether Mnk1 binding to eIF4G alone modulates eIF4G function (e.g., by altering the RNA-binding capacity).
Third, the availability of eIF4G for IRES-mediated translation may be codetermined by the eIF4E binding proteins (BPs) (Figure 4) [74]. Activity of the eIF4E-BPs is controlled by mTORC1. In the nonphosphorylated state, the eIF4E-BPs bind to eIF4E and prevent its association with eIF4G, leading to repression of cap-dependent translation [75]. Activation of mTORC1, the eIF4E-BP kinase, leads to eIF4E-BP hyperphosphorylation, dissociation from eIF4E and stimulation of cap-dependent translation [76]. It is well established that inhibitors of the PI3K–mTORC1 signaling pathway, in addition to repression of cap-dependent translation, induce alternative, cap-independent protein synthesis. For example, the classic mTORC1 inhibitor rapamycin stimulates WT poliovirus replication [77]. The mechanism responsible for this effect may be that rapamycin-induced eIF4E-BP dephosphorylation enhances the proportion of ‘free’ (not committed in m7G-cap binding complexes) eIF4G. This may result in enhanced IRES binding of eIF4G [74].
Clinical trials of PVSRIPO
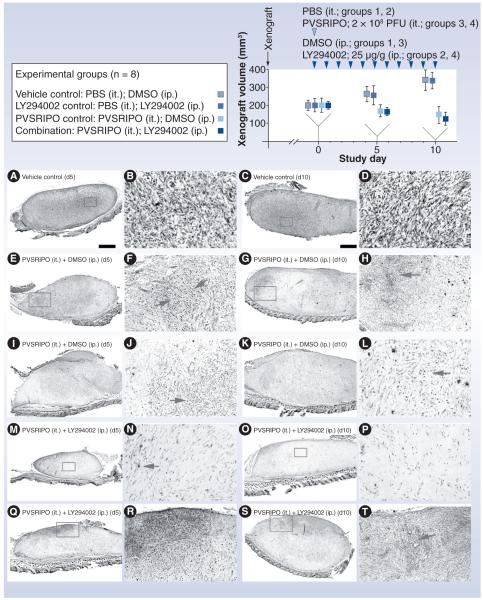
Treatment of GBM xenografts with intratumoral inoculation of PVSRIPO (the intended clinical route) produces rampant tumor cell death, potent host-mediated inflammatory reactions against infected tumor and rapid tumor decline (Figure 7) [26,56,61]. These experiments document the two major components of oncolytic viral activity: direct viral tumor cell killing and resulting host inflammatory responses against the infected tumor.
Figure 7. Testing PVSRIPO and PI3K inhibitor synergy in vivo.
Experimental groups and the study regimen are indicated at the top. U-118 xenografts were measured at study days 0 (when PVSRIPO/vehicle and LY294002/vehicle treatment was initiated), 5 and 10. Four animals from each group were euthanized at study days 5 and 10 for histology and virus recovery. The bottom panels show histology from xenografts recovered at day 5 (left columns) and 10 (right columns). Low-magnification images in the left columns are accompanied by higher-magnification images from the same section (inserts) in the column to their right. (A–D) Tumor histology of a representative xenograft from group 1 shows the characteristic dense arrangement of tumor cells. (E–L) Histology of two representative tumors from group 3 at study days 5 and 10 as indicated. Note extensive tumor cell loss and ‘empty’ appearance of the former xenograft in all cases. Arrows point to areas of intense tumor cell killing and active tissue rearrangement. (M–T) Histology of four representative tumors from group 4 at study days 5 and 10 as indicated. (M & N) Complete tumor regression at study day 5. (N & P) The area of the former tumor was invaded by cells with fibroblast morphology surrounded by dense extracellular matrix. Isolated viable tumor cells (arrow [N]) may remain. (Q & R) Active tumor, which was still present in three out of four animals of group 4.
DMSO: Dimethyl sulfoxide; ip.: Intraperitoneal; it.: Intratumoral; PBS: Phosphate-buffered saline; PFU: Plaque-forming unit.
Reproduced with permission from [61].
Since Necl-5 expression is widespread in many cancers and PVSRIPO exhibits similar cytotoxic properties in the vast majority of cancer cells, the agent may be broadly applicable against many types of neoplasia. We favor a more discriminating approach, at least at this stage. Poliovirus is a naturally neuroinvasive virus, supporting targeted use against a local disseminating lesion in the brain. There are compelling arguments to assume that susceptibility of GBM cells to PVSRIPO oncolysis may be universal. Necl-5 expression in GBM patients includes priority compartments such as tumor vasculature or ‘stem-cell like’ cell populations. Lastly, targeting GBM offers a unique opportunity to test PVSRIPO without interference of serum neutralizing antibodies (tests of CSF from GBM patients revealed the absence of poliovirus-neutralizing antibodies; patients with a history of paralytic poliomyelitis [implying prior poliovirus replication in the CNS] are excluded from the planned trials). Tests in the syngeneic Neuro2A tumor model (see previously) indicate that poliovirus oncolysis not only is not impeded by pre-existing immunity, but that therapeutic efficacy may actually benefit from it [12].
Multiple biomarkers are available to study susceptibility to PVSRIPO in tumors from individual patients. Based on all available empirical evidence, correlative markers of efficacy, such as Necl-5 expression, active Erk1/2 or tumor-specific NFAR, isoform/subcellular distribution are universal features of GBM. Therefore, no patient profiling is currently planned for Phase I trials.
The advanced insight into the molecular mechanisms determining PVSRIPO translation, propagation and tumor cell killing offer intriguing possibilities for synergistic combination with protein kinase inhibitors. For example, PI3K–mTORC1 inhibitors stimulate viral, cap-independent translation and favor virus propagation by blocking hyperphosphorylation of the eIF4E-BPs [77].
Future perspective
Complex biologicals, such as live viruses, unfold intricate relationships with their host cells, either natural target cells in the normal human organism or ‘artificially’ targeted tumor cells. Such complexity is attractive from a therapeutic standpoint, as viruses have the potential to exert multiple effects on target tumors, including direct effects such as tumor cell killing, combined with indirect effects such as inflammatory reactions to dead tumor cells. The ‘mechanistic complexity’ of biologicals, such as oncolytic viruses, requires correlative mechanistic studies that better connect empirical observations in vitro, in tissue culture or in experimental animal systems with the clinical situation in patients. We believe that to encourage further developments and for the refinement of strategies to target cancer with viruses, these should be based on rigorous empirical investigations of factors determining cancer cell tropism and cell killing/specificity. Since many viruses alter host cell function in ways that emulate malignancy, such studies might not only reveal mechanistic insight regarding specific oncolytic viruses, but may improve our understanding of fundamental regulatory processes in transformed cells.
Executive summary.
▪ PVSRIPO is a nonpathogenic, recombinant poliovirus with a conditional replication phenotype in malignant cells.
▪ The poliovirus receptor nectin-like molecule (Necl-5) is a broadly expressed tumor antigen that is associated with ectodermal/neuroectodermal tumors.
▪ Necl-5 is universally expressed in glioblastoma, including CD133+ ‘stem-cell like’ tumor cells and proliferating tumor vasculature.
▪ PVSRIPO selectively translates and propagates in malignant cells with abnormally permissive conditions for viral, cap-independent translation.
▪ Targeting of Necl-5-expressing tumor cells with PVSRIPO elicits efficient cell killing and secondary, host-mediated inflammatory responses directed against the infected tumor.
▪ PVSRIPO was successfully tested in comprehensive, Investigational New Drug application-directed primate neurovirulence assays by the clinically intended intracerebral route.
▪ PVSRIPO has Investigational New Drug application approval for Phase I clinical trials in glioblastoma patients.
Footnotes
Financial & competing interests disclosure The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties. No writing assistance was utilized in the production of this manuscript.
Bibliography
- 1.Bluming AZ, Ziegler JL. Regression of Burkitt’s lymphoma in association with measles infection. Lancet. 1971;2(7715):105–106. doi: 10.1016/s0140-6736(71)92086-1. [DOI] [PubMed] [Google Scholar]
- 2.De Pace N. Sulla scomparsa di un enorme cancro vegetante del callo dell’utero senze cura chirurgica. Ginecologia. 1912;9:82–88. [Google Scholar]
- 3.Coffey MC, Strong JE, Forsyth PA, Lee PW. Reovirus therapy of tumors with activated Ras pathway. Science. 1998;282(5392):1332–1334. doi: 10.1126/science.282.5392.1332. [DOI] [PubMed] [Google Scholar]
- 4.Stojdl DF, Lichty B, Knowles S, et al. Exploiting tumor-specific defects in the interferon pathway with a previously unknown oncolytic virus. Nat. Med. 2000;6(7):821–825. doi: 10.1038/77558. [DOI] [PubMed] [Google Scholar]
- 5.Martuza RL, Malick A, Markert JM, Ruffner KL, Coen DM. Experimental therapy of human glioma by means of a genetically engineered virus mutant. Science. 1991;252(5007):854–856. doi: 10.1126/science.1851332. [DOI] [PubMed] [Google Scholar]
- 6.Liu BL, Robinson M, Han ZQ, et al. ICP34.5 deleted herpes simplex virus with enhanced oncolytic, immune stimulating, and anti-tumour properties. Gene Ther. 2003;10(4):292–303. doi: 10.1038/sj.gt.3301885. [DOI] [PubMed] [Google Scholar]
- 7.Senzer NN, Kaufman HL, Amatruda T, et al. Phase II clinical trial of a granulocyte-macrophage colony-stimulating factor-encoding, second-generation oncolytic herpesvirus in patients with unresectable metastatic melanoma. J. Clin. Oncol. 2009;27(34):5763–5771. doi: 10.1200/JCO.2009.24.3675. [DOI] [PubMed] [Google Scholar]
- 8.Kaufman HL, Kim DW, DeRaffele G, Mitcham J, Coffin RS, Kim-Schulze S. Local and distant immunity induced by intralesional vaccination with an oncolytic herpes virus encoding GM-CSF in patients with stage IIIc and IV melanoma. Ann. Surg. Oncol. 2010;17(3):718–730. doi: 10.1245/s10434-009-0809-6. [DOI] [PubMed] [Google Scholar]
- 9.Gustin KE, Sarnow P. Effects of poliovirus infection on nucleo-cytoplasmic trafficking and nuclear pore complex composition. EMBO J. 2001;20(1–2):240–249. doi: 10.1093/emboj/20.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Etchison D, Milburn SC, Edery I, Sonenberg N, Hershey JW. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J. Biol. Chem. 1982;257(24):14806–14810. [PubMed] [Google Scholar]
- 11.Goldstaub D, Gradi A, Bercovitch Z, et al. Poliovirus 2A protease induces apoptotic cell death. Mol. Cell. Biol. 2000;20(4):1271–1277. doi: 10.1128/mcb.20.4.1271-1277.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toyoda H, Yin J, Mueller S, Wimmer E, Cello J. Oncolytic treatment and cure of neuroblastoma by a novel attenuated poliovirus in a novel poliovirus-susceptible animal model. Cancer Res. 2007;67(6):2857–2864. doi: 10.1158/0008-5472.CAN-06-3713. [DOI] [PubMed] [Google Scholar]
- 13.Toyoda H, Wimmer E, Cello J. Oncolytic poliovirus therapy and immunization with poliovirus-infected cell lysate induces potent antitumor immunity against neuroblastoma in vivo. Int. J. Oncol. 2011;38(1):81–87. [PubMed] [Google Scholar]
- 14.Menotti L, Cerretani A, Campadelli-Fiume G. A herpes simplex virus recombinant that exhibits a single-chain antibody to HER2/neu enters cells through the mammary tumor receptor, independently of the gD receptors. J. Virol. 2006;80(11):5531–5539. doi: 10.1128/JVI.02725-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mathis JM, Stoff-Khalili MA, Curiel DT. Oncolytic adenoviruses – selective retargeting to tumor cells. Oncogene. 2005;24(52):7775–7791. doi: 10.1038/sj.onc.1209044. [DOI] [PubMed] [Google Scholar]
- 16.Allen C, Vongpunsawad S, Nakamura T, et al. Retargeted oncolytic measles strains entering via the EGFRvIII receptor maintain significant antitumor activity against gliomas with increased tumor specificity. Cancer Res. 2006;66(24):11840–11850. doi: 10.1158/0008-5472.CAN-06-1200. [DOI] [PubMed] [Google Scholar]
- 17.Takai Y, Miyoshi J, Ikeda W, Ogita H. Nectins and nectin-like molecules: roles in contact inhibition of cell movement and proliferation. Nat. Rev. Mol. Cell. Biol. 2008;9(8):603–615. doi: 10.1038/nrm2457. [DOI] [PubMed] [Google Scholar]
- 18.Nobis P, Zibirre R, Meyer G, Kühne J, Warnecke G, Koch G. Production of a monoclonal antibody against an epitope on HeLa cells that is the functional poliovirus binding site. J. Gen. Virol. 1985;66(Pt 12):2563–2569. doi: 10.1099/0022-1317-66-12-2563. [DOI] [PubMed] [Google Scholar]
- 19.Mendelsohn CL, Wimmer E, Racaniello VR. Cellular receptor for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell. 1989;56(5):855–865. doi: 10.1016/0092-8674(89)90690-9. [DOI] [PubMed] [Google Scholar]
- 20.Ren RB, Costantini F, Gorgacz EJ, Lee JJ, Racaniello VR. Transgenic mice expressing a human poliovirus receptor: a new model for poliomyelitis. Cell. 1990;63(2):353–362. doi: 10.1016/0092-8674(90)90168-e. [DOI] [PubMed] [Google Scholar]
- 21.Koike S, Taya C, Kurata T, et al. Transgenic mice susceptible to poliovirus. Proc. Natl Acad. Sci. USA. 1991;88(3):951–955. doi: 10.1073/pnas.88.3.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iwasaki A, Welker R, Mueller S, Linehan M, Nomoto A, Wimmer E. Immunofluorescence ana lysis of poliovirus receptor expression in Peyer’s patches of humans, primates, and CD155 transgenic mice: implications for poliovirus infection. J. Infect. Dis. 2002;186(5):585–592. doi: 10.1086/342682. [DOI] [PubMed] [Google Scholar]
- 23.Koike S, Aoki J, Nomoto A. Transgenic mouse for the study of poliovirus pathogenicity. In: Wimmer E, editor. Cellular Receptors for Animal Viruses. Cold Spring Harbor Laboratory Press; NY, USA: 1994. pp. 463–480. [Google Scholar]
- 24.Geraghty RJ, Krummenacher C, Cohen GH, Eisenberg RJ, Spear PG. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science. 1998;280(5369):1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 25.Gromeier M, Solecki D, Patel DD, Wimmer E. Expression of the human poliovirus receptor/CD155 gene during development of the central nervous system: implications for the pathogenesis of poliomyelitis. Virology. 2000;273(2):248–257. doi: 10.1006/viro.2000.0418. [DOI] [PubMed] [Google Scholar]
- 26.Gromeier M, Lachmann S, Rosenfeld MR, Gutin PH, Wimmer E. Intergeneric poliovirus recombinants for the treatment of malignant glioma. Proc. Natl Acad. Sci. USA. 2000;97(12):6803–6808. doi: 10.1073/pnas.97.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merrill MK, Bernhardt G, Sampson JH, Wikstrand CJ, Bigner DD, Gromeier M. Poliovirus receptor CD155-targeted oncolysis of glioma. Neuro Oncol. 2004;6(3):208–217. doi: 10.1215/S1152851703000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sloan KE, Stewart JK, Treloar AF, Matthews RT, Jay DG. CD155/PVR enhances glioma cell dispersal by regulating adhesion signaling and focal adhesion dynamics. Cancer Res. 2005;65(23):10930–10937. doi: 10.1158/0008-5472.CAN-05-1890. [DOI] [PubMed] [Google Scholar]
- 29.Sloan KE, Eustace BK, Stewart JK, et al. CD155/PVR plays a key role in cell motility during tumor cell invasion and migration. BMC Cancer. 2004;4:73. doi: 10.1186/1471-2407-4-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masson D, Jarry A, Baury B, et al. Overexpression of the CD155 gene in human colorectal carcinoma. Gut. 2001;49(2):236–240. doi: 10.1136/gut.49.2.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ochiai H, Moore SA, Archer GE, et al. Treatment of intracerebral neoplasia and neoplastic meningitis with regional delivery of oncolytic recombinant poliovirus. Clin. Cancer Res. 2004;10(14):4831–4838. doi: 10.1158/1078-0432.CCR-03-0694. [DOI] [PubMed] [Google Scholar]
- 32.Nakai R, Maniwa Y, Tanaka Y, et al. Overexpression of Necl-5 correlates with unfavorable prognosis in patients with lung adenocarcinoma. Cancer Sci. 2010;101(5):1326–1330. doi: 10.1111/j.1349-7006.2010.01530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Erickson BM, Thompson NL, Hixson DC. Tightly regulated induction of the adhesion molecule Necl-5/CD155 during rat liver regeneration and acute liver injury. Hepatology. 2006;43(2):325–334. doi: 10.1002/hep.21021. [DOI] [PubMed] [Google Scholar]
- 34.Solecki D, Schwarz S, Wimmer E, Lipp M, Bernhardt G. The promoters for human and monkey poliovirus receptors. Requirements for basic and cell type-specific activity. J. Biol. Chem. 1997;272(9):5579–5586. doi: 10.1074/jbc.272.9.5579. [DOI] [PubMed] [Google Scholar]
- 35.Solecki D, Bernhardt G, Lipp M, Wimmer E. Identification of a nuclear respiratory factor-1 binding site within the core promoter of the human polio virus receptor/CD155 gene. J. Biol. Chem. 2000;275(17):12453–12462. doi: 10.1074/jbc.275.17.12453. [DOI] [PubMed] [Google Scholar]
- 36.Solecki DJ, Gromeier M, Mueller S, Bernhardt G, Wimmer E. Expression of the human poliovirus receptor/CD155 gene is activated by sonic hedgehog. J. Biol. Chem. 2002;277(28):25697–25702. doi: 10.1074/jbc.M201378200. [DOI] [PubMed] [Google Scholar]
- 37.Solecki D, Wimmer E, Lipp M, Bernhardt G. Identification and characterization of the cis-acting elements of the human CD155 gene core promoter. J. Biol. Chem. 1999;274(3):1791–1800. doi: 10.1074/jbc.274.3.1791. [DOI] [PubMed] [Google Scholar]
- 38.Castriconi R, Daga A, Dondero A, et al. NK cells recognize and kill human glioblastoma cells with stem cell-like properties. J. Immunol. 2009;182(6):3530–3539. doi: 10.4049/jimmunol.0802845. [DOI] [PubMed] [Google Scholar]
- 39.Toyoda H, Nicklin MJ, Murray MG, et al. A second virus-encoded proteinase involved in proteolytic processing of poliovirus polyprotein. Cell. 1986;45(5):761–770. doi: 10.1016/0092-8674(86)90790-7. [DOI] [PubMed] [Google Scholar]
- 40.Lloyd RE. Translational control by viral proteinases. Virus Res. 2006;119(1):76–88. doi: 10.1016/j.virusres.2005.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dobrikova EY, Grisham RN, Kaiser C, Lin J, Gromeier M. Competitive translation efficiency at the picornavirus type 1 internal ribosome entry site facilitated by viral cis and trans factors. J. Virol. 2006;80(7):3310–3321. doi: 10.1128/JVI.80.7.3310-3321.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sonenberg N, Morgan MA, Merrick WC, Shatkin AJ. A polypeptide in eukaryotic initiation factors that crosslinks specifically to the 5′-terminal cap in mRNA. Proc. Natl Acad. Sci. USA. 1978;75(10):4843–4847. doi: 10.1073/pnas.75.10.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nomoto A, Lee YF, Wimmer E. The 5′ end of poliovirus mRNA is not capped with m7G(5′)ppp(5′)Np. Proc. Natl Acad. Sci. USA. 1976;73(2):375–380. doi: 10.1073/pnas.73.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334(6180):320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 45.Jang SK, Kräusslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 1988;62(8):2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Breyne S, Yu Y, Unbehaun A, Pestova TV, Hellen CU. Direct functional interaction of initiation factor eIF4G with type 1 internal ribosomal entry sites. Proc. Natl Acad. Sci. USA. 2009;106(23):9197–9202. doi: 10.1073/pnas.0900153106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gromeier M, Alexander L, Wimmer E. Internal ribosomal entry site substitution eliminates neurovirulence in intergeneric poliovirus recombinants. Proc. Natl Acad. Sci. USA. 1996;93(6):2370–2375. doi: 10.1073/pnas.93.6.2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gromeier M, Bossert B, Arita M, Nomoto A, Wimmer E. Dual stem loops within the poliovirus internal ribosomal entry site control neurovirulence. J. Virol. 1999;73(2):958–964. doi: 10.1128/jvi.73.2.958-964.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Campbell SA, Lin J, Dobrikova EY, Gromeier M. Genetic determinants of cell type-specific poliovirus propagation in HEK 293 cells. J. Virol. 2005;79(10):6281–6290. doi: 10.1128/JVI.79.10.6281-6290.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wimmer E, Hellen CU, Cao X. Genetics of poliovirus. Annu. Rev. Genet. 1993;27:353–436. doi: 10.1146/annurev.ge.27.120193.002033. [DOI] [PubMed] [Google Scholar]
- 51.Horie H, Miyazawa M, Ota Y. Analysis of the accumulation of mutants in Sabin attenuated polio vaccine viruses passaged in Vero cells. Vaccine. 2001;19(11–12):1456–1459. doi: 10.1016/s0264-410x(00)00350-9. [DOI] [PubMed] [Google Scholar]
- 52.Hofer F, Gruenberger M, Kowalski H, et al. Members of the low density lipoprotein receptor family mediate cell entry of a minor-group common cold virus. Proc. Natl Acad. Sci. USA. 1994;91(5):1839–1842. doi: 10.1073/pnas.91.5.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Couch RB. Rhinoviruses. In: Knipe DM, Howley PM, editors. Fields Virology. Lippincott Williams and Wilkins; PA, USA: 2001. pp. 777–797. [Google Scholar]
- 54.Wang ES, Dobrikova E, Goetz C, Dufresne AT, Gromeier M. Adaptation of an ICAM-1 tropic enterovirus to the mouse respiratory tract. J. Virol. 2011;85(11):5606–5617. doi: 10.1128/JVI.01502-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dufresne AT, Gromeier M. A nonpolio enterovirus with respiratory tropism causes poliomyelitis in intercellular adhesion molecule 1 transgenic mice. Proc. Natl Acad. Sci. USA. 2004;101(37):13636–13641. doi: 10.1073/pnas.0403998101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dobrikova EY, Broadt T, Poiley-Nelson J, et al. Recombinant oncolytic poliovirus eliminates glioma in vivo without genetic adaptation to a pathogenic phenotype. Mol. Ther. 2008;16(11):1865–1872. doi: 10.1038/mt.2008.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Merrill MK, Dobrikova EY, Gromeier M. Cell-type-specific repression of internal ribosome entry site activity by double-stranded RNA-binding protein 76. J. Virol. 2006;80(7):3147–3156. doi: 10.1128/JVI.80.7.3147-3156.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Merrill MK, Gromeier M. The double-stranded RNA binding protein 76:NF45 heterodimer inhibits translation initiation at the Rhinovirus type 2 internal ribosome entry site. J. Virol. 2006;80(14):6936–6942. doi: 10.1128/JVI.00243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harashima A, Guettouche T, Barber GN. Phosphorylation of the NFAR proteins by the dsRNA-dependent protein kinase PKR constitutes a novel mechanism of translational regulation and cellular defense. Genes Dev. 2010;24(23):2640–2653. doi: 10.1101/gad.1965010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Neplioueva V, Dobrikova EY, Mukherjee N, Keene JD, Gromeier M. Tissue type-specific expression of the dsRNA-binding protein 76 and genome-wide elucidation of its target mRNAs. PLoS One. 2010;5(7):E11710. doi: 10.1371/journal.pone.0011710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goetz C, Everson RG, Zhang LC, Gromeier M. MAPK signal-integrating kinase controls cap-independent translation and cell type-specific cytotoxicity of an oncolytic poliovirus. Mol. Ther. 2010;18(11):1937–1946. doi: 10.1038/mt.2010.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang X, Chen E, Jiang H, et al. Evaluation of IRES-mediated, cell-type-specific cytotoxicity of poliovirus using a colorimetric cell proliferation assay. J. Virol. Methods. 2009;155(1):44–54. doi: 10.1016/j.jviromet.2008.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pyronnet S, Imataka H, Gingras AC, Fukunaga R, Hunter T, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) recruits mnk1 to phosphorylate eIF4E. EMBO J. 1999;18(1):270–279. doi: 10.1093/emboj/18.1.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shveygert M, Kaiser C, Bradrick SS, Gromeier M. Regulation of eIF4E phosphorylation by MAPK occurs through modulation of Mnk1–eIF4G interaction. Mol. Cell. Biol. 2010;30(21):5160–5167. doi: 10.1128/MCB.00448-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Waskiewicz AJ, Flynn A, Proud CG, Cooper JA. Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J. 1997;16(8):1909–1920. doi: 10.1093/emboj/16.8.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alain T, Lun X, Martineau Y, et al. Vesicular stomatitis virus oncolysis is potentiated by impairing mTORC1-dependent type I IFN production. Proc. Natl Acad. Sci. USA. 2010;107(4):1576–1581. doi: 10.1073/pnas.0912344107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pestova TV, Shatsky IN, Hellen CU. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol. Cell. Biol. 1996;16(12):6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hundsdoerfer P, Thoma C, Hentze MW. Eukaryotic translation initiation factor 4GI and p97 promote cellular internal ribosome entry sequence-driven translation. Proc. Natl Acad. Sci. USA. 2005;102(38):13421–13426. doi: 10.1073/pnas.0506536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kaiser C, Dobrikova EY, Bradrick SS, Shveygert M, Herbert JT, Gromeier M. Activation of cap-independent translation by variant eukaryotic initiation factor 4G in vivo. RNA. 2008;14(10):2170–2182. doi: 10.1261/rna.1171808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dephoure N, Zhou C, Villén J, et al. A quantitative atlas of mitotic phosphorylation. Proc. Natl Acad. Sci. USA. 2008;105(31):10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Raught B, Gingras A-C, Gygi SP, et al. Serum-stimulated, rapamycin-sensitive phosphorylation sites in the eukaryotic translation initiation factor 4GI. EMBO J. 2000;19(3):434–444. doi: 10.1093/emboj/19.3.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marintchev A, Edmonds KA, Marintcheva B, et al. Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell. 2009;136(3):447–460. doi: 10.1016/j.cell.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dobrikov M, Dobrikova E, Shveygert M, Gromeier M. Phosphorylation of eIF4G1 by PKCa regulates eIF4G1 binding to Mnk1. Mol. Cell. Biol. 2010;31(14):2947–2959. doi: 10.1128/MCB.05589-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Svitkin YV, Herdy B, Costa-Mattioli M, Gingras A-C, Raught B, Sonenberg N. Eukaryotic translation initiation factor 4E availability controls the switch between cap-dependent and internal ribosomal entry site-mediated translation. Mol. Cell. Biol. 2005;25(23):10556–10565. doi: 10.1128/MCB.25.23.10556-10565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Haghighat A, Mader S, Pause A, Sonenberg N. Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J. 1995;14(22):5701–5709. doi: 10.1002/j.1460-2075.1995.tb00257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pause A, Belsham GJ, Gingras AC. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 1994;371(6500):762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 77.Beretta L, Gingras AC, Svitkin YV, Hall MN, Sonenberg N. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. EMBO J. 1996;15(3):658–664. [PMC free article] [PubMed] [Google Scholar]