Abstract
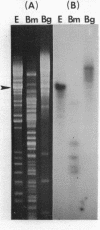
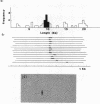
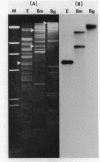
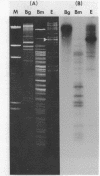
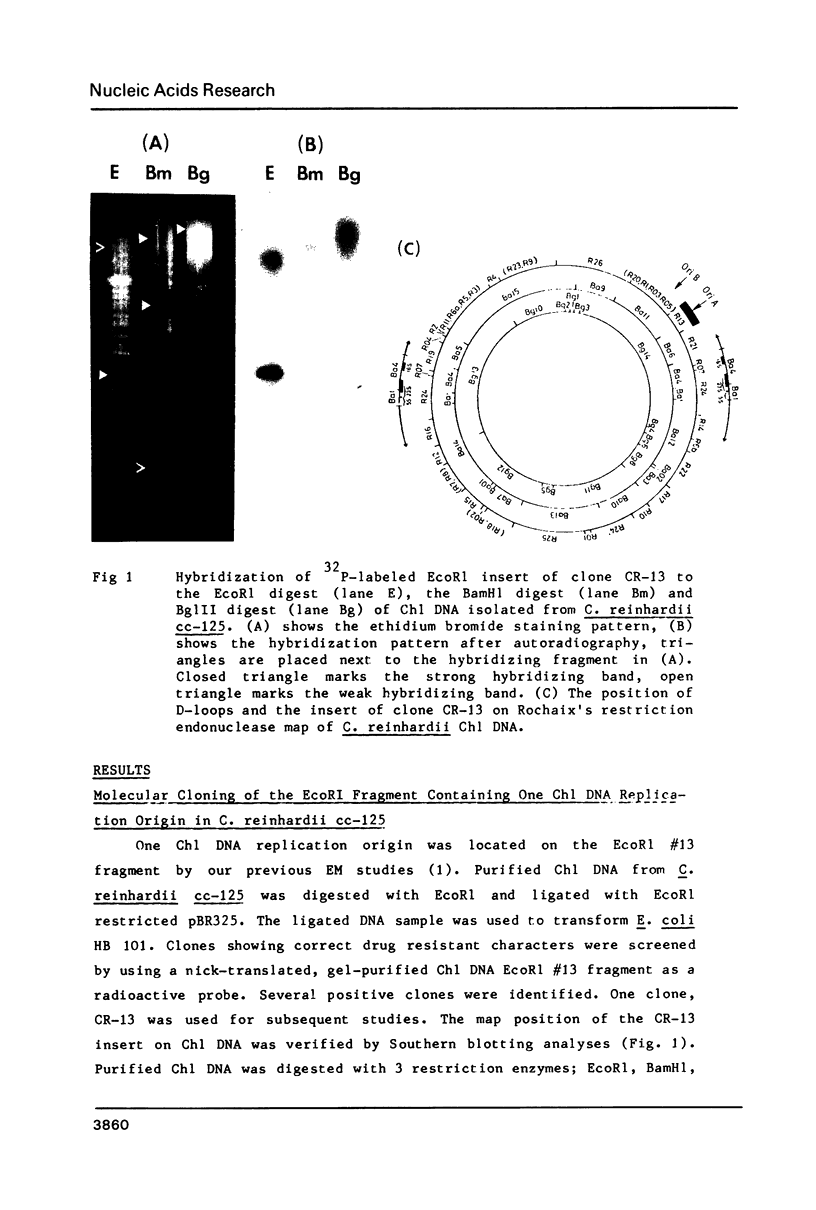
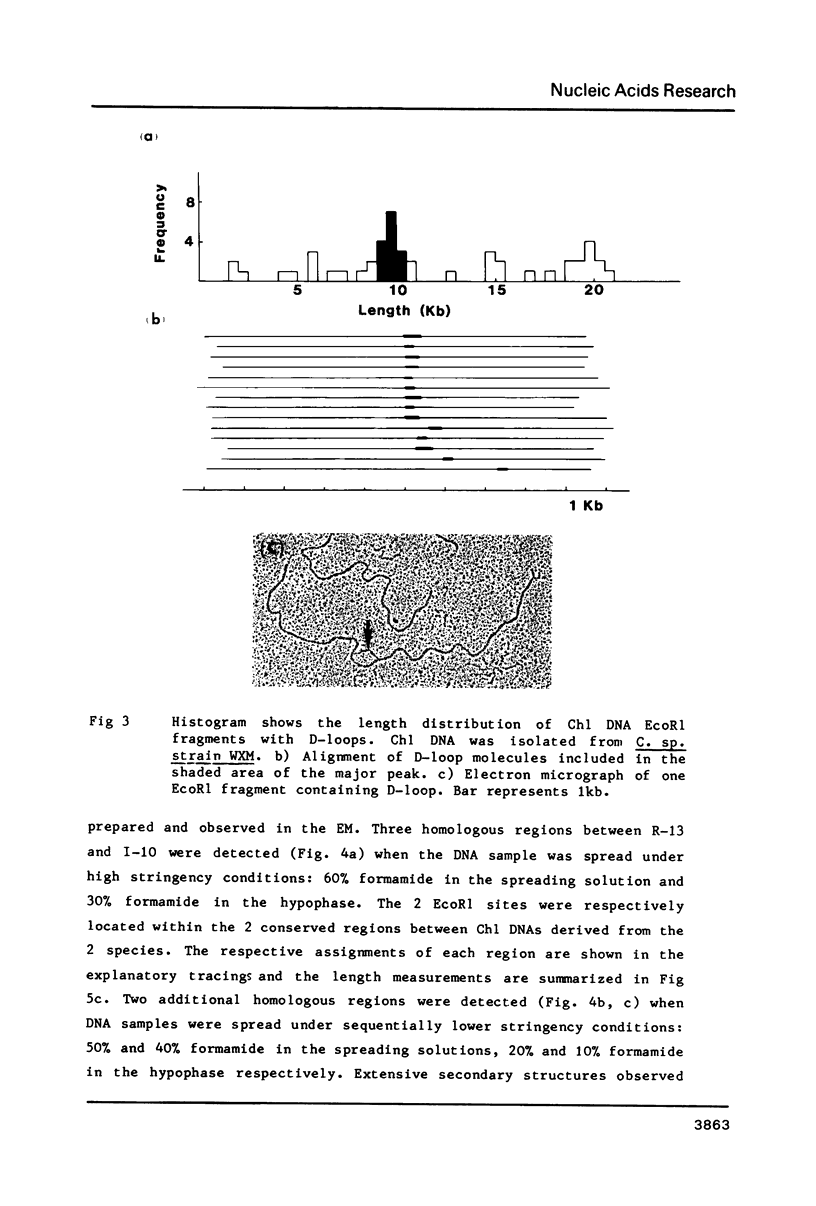
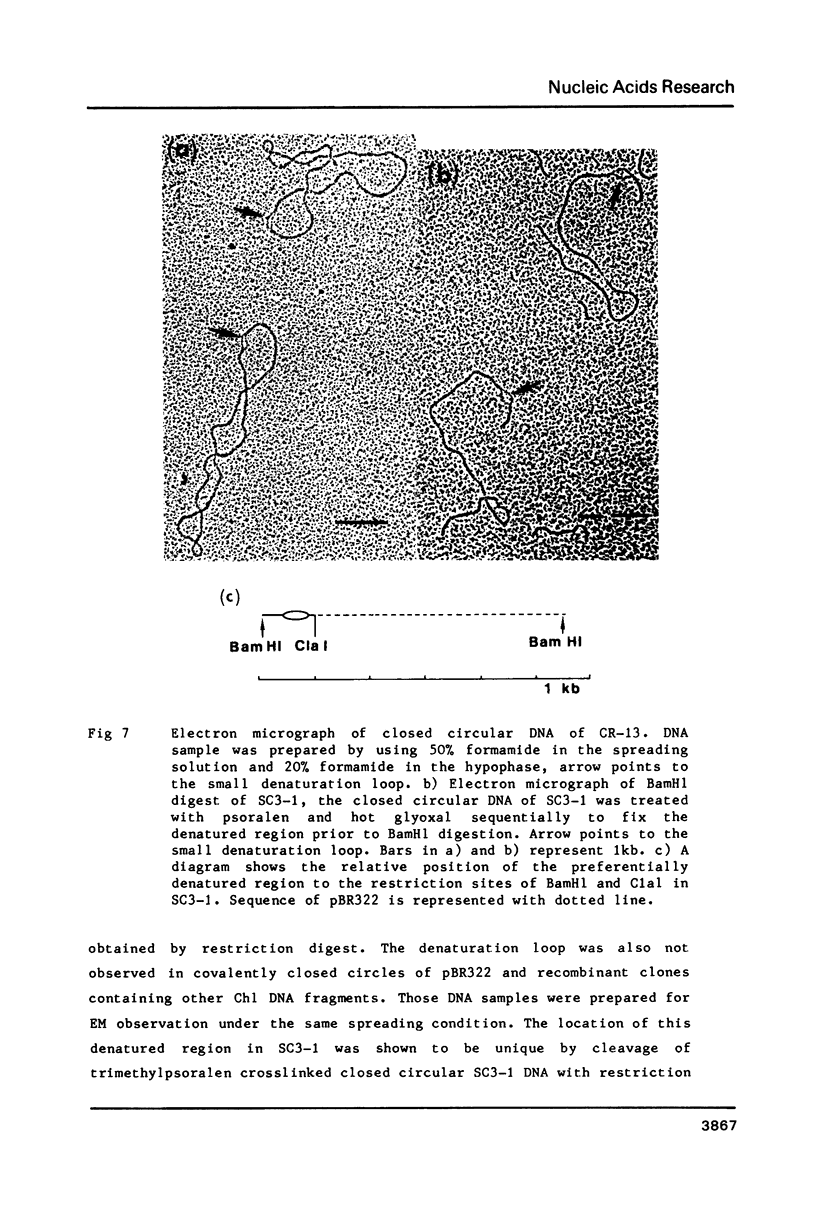
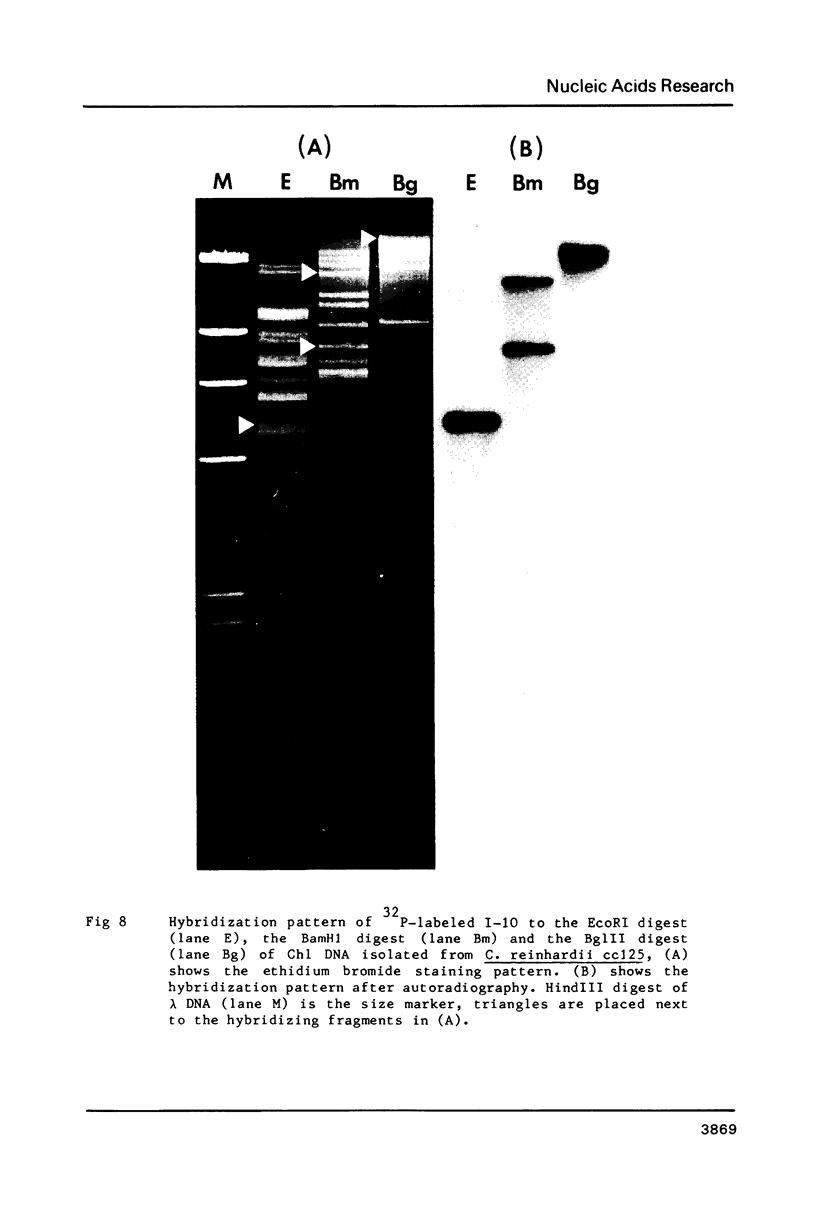
The EcoR1 restriction fragments containing D-loops which marked the replication origin of chloroplast DNA were identified in two different species of Chlamydomonas. Each fragment was cloned in the E. coli plasmid pBR325. The cloned fragments were compared by restriction endonuclease analyses and by heteroduplex analyses in the electron microscope. The relative position of the D-loop regions and the homologous regions between the 2 fragments was determined. The D-loops were located within one short homologous region of 0. 42kb in length between the 2 cloned restriction fragments. The homologous region was subcloned in pBR322. Closed circular plasmid DNAs containing the short homologous region showed preferred denaturation in the D-loop region under physiological salt concentration which suggested that D-loop region was AT rich. Sequence divergence was detected at both ends of the D-loop region. Southern blot analyses indicated the presence of species-specific repetitive sequences within the divergent regions.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chiang K. S., Sueoka N. Replication of chloroplast DNA in Chlamydomonas reinhardi during vegetative cell cycle: its mode and regulation. Proc Natl Acad Sci U S A. 1967 May;57(5):1506–1513. doi: 10.1073/pnas.57.5.1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton D. A. Replication of animal mitochondrial DNA. Cell. 1982 Apr;28(4):693–705. doi: 10.1016/0092-8674(82)90049-6. [DOI] [PubMed] [Google Scholar]
- Davis R. W., Hyman R. W. A study in evolution: the DNA base sequence homology between coliphages T7 and T3. J Mol Biol. 1971 Dec 14;62(2):287–301. doi: 10.1016/0022-2836(71)90428-1. [DOI] [PubMed] [Google Scholar]
- Hershey N. D., Davidson N. Two drosophila melanogaster tRNAGly genes are contained in a direct duplication at chromosomal locus 56F. Nucleic Acids Res. 1980 Nov 11;8(21):4899–4910. doi: 10.1093/nar/8.21.4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys A. J., Flavell R. A. The rabbit beta-globin gene contains a large large insert in the coding sequence. Cell. 1977 Dec;12(4):1097–1108. doi: 10.1016/0092-8674(77)90172-6. [DOI] [PubMed] [Google Scholar]
- Khan S. A., Adler G. K., Novick R. P. Functional origin of replication of pT181 plasmid DNA is contained within a 168-base-pair segment. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4580–4584. doi: 10.1073/pnas.79.15.4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koller B., Delius H. Origin of replication in chloroplast DNA of Euglena gracilis located close to the region of variable size. EMBO J. 1982;1(8):995–998. doi: 10.1002/j.1460-2075.1982.tb01283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodner R. D., Tewari K. K. Chloroplast DNA from higher plants replicates by both the Cairns and the rolling circle mechanism. Nature. 1975 Aug 28;256(5520):708–711. doi: 10.1038/256708a0. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., Kleid D. G. Nucleotide sequence of the rightward operator of phage lambda. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1184–1188. doi: 10.1073/pnas.72.3.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka A., Sugimoto K., Takanami M., Hirota Y. Replication origin of the Escherichia coli K-12 chromosome: the size and structure of the minimum DNA segment carrying the information for autonomous replication. Mol Gen Genet. 1980 Apr;178(1):9–20. doi: 10.1007/BF00267207. [DOI] [PubMed] [Google Scholar]
- Saito H., Tabor S., Tamanoi F., Richardson C. C. Nucleotide sequence of the primary origin of bacteriophage T7 DNA replication: relationship to adjacent genes and regulatory elements. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3917–3921. doi: 10.1073/pnas.77.7.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C. K., Hearst J. E. Psoralen-crosslinked secondary structure map of single-stranded virus DNA. Proc Natl Acad Sci U S A. 1976 Aug;73(8):2649–2653. doi: 10.1073/pnas.73.8.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sueoka N. MITOTIC REPLICATION OF DEOXYRIBONUCLEIC ACID IN CHLAMYDOMONAS REINHARDI. Proc Natl Acad Sci U S A. 1960 Jan;46(1):83–91. doi: 10.1073/pnas.46.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upholt W. B., Dawid I. B. Mapping of mitochondrial DNA of individual sheep and goats: rapid evolution in the D loop region. Cell. 1977 Jul;11(3):571–583. doi: 10.1016/0092-8674(77)90075-7. [DOI] [PubMed] [Google Scholar]
- Walberg M. W., Clayton D. A. Sequence and properties of the human KB cell and mouse L cell D-loop regions of mitochondrial DNA. Nucleic Acids Res. 1981 Oct 24;9(20):5411–5421. doi: 10.1093/nar/9.20.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]