Abstract
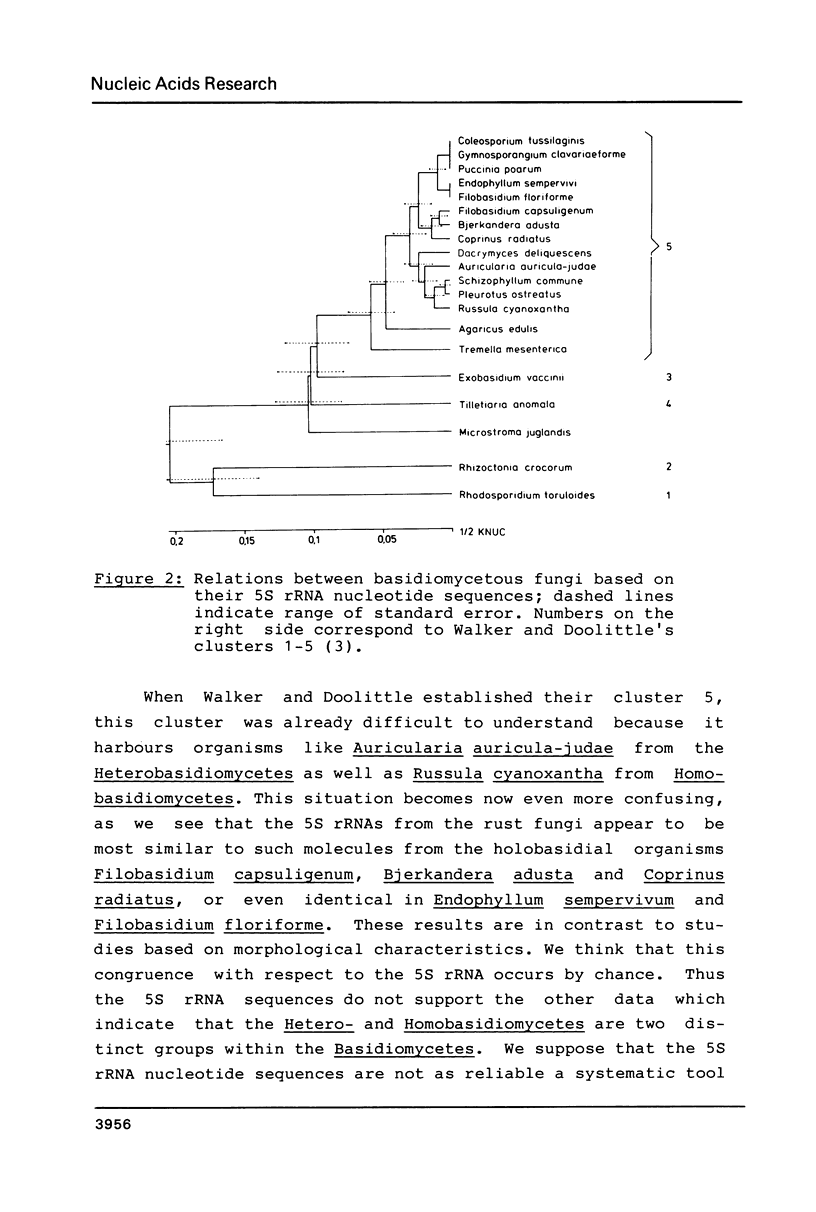
The 5S ribosomal RNA nucleotide sequences of five basidiomycetous fungi, Coleosporium tussilaginis , Gymnosporangium clavariaeforme , Puccinia poarum , Endophyllum sempervivi and Microstroma juglandis were determined. Despite high differentiation in their host spectra the four rust species are highly conserved with respect to their 5S rRna sequences, which fit with the basidiomycete cluster 5 described by Walker and Doolittle (1). The sequences obtained from the first three rust fungi were proven to be identical while the sequence from Endophyllum sempervivi showed two base substitutions compared with the other rust fungi. The Microstroma juglandis 5S rRNA sequence differs from all other basidiomycete 5S rRNA sequences published so far in respect to its secondary structure which shows an atypical 'CCA' loop in helix D, but it reveals typical basidiomycetous signature nucleotides. Therefore Microstroma juglandis represents a cluster of its own within the Basidiomycetes. A dendrogram was constructed based on Kimura's "Neutral Theory of Molecular Evolution".
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- De Wachter R., Chen M. W., Vandenberghe A. Conservation of secondary structure in 5 S ribosomal RNA: a uniform model for eukaryotic, eubacterial, archaebacterial and organelle sequences is energetically favourable. Biochimie. 1982 May;64(5):311–329. doi: 10.1016/s0300-9084(82)80436-7. [DOI] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donis-Keller H. Phy M: an RNase activity specific for U and A residues useful in RNA sequence analysis. Nucleic Acids Res. 1980 Jul 25;8(14):3133–3142. doi: 10.1093/nar/8.14.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donk M. A. The Heterobasidiomycetes: a reconnaissance. II. Some problems connected with the restricted emendation. Proc K Ned Akad Wet C. 1972;75(5):376–390. [PubMed] [Google Scholar]
- Hindley J., Page S. M. Nucleotide sequence of yeast 5 S ribosomal RNA. FEBS Lett. 1972 Oct 1;26(1):157–160. doi: 10.1016/0014-5793(72)80563-5. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G., Klotz L. C., Blanken R. L., Loeblich A. R., 3rd An evaluation of the phylogenetic position of the dinoflagellate Crypthecodinium cohnii based on 5S rRNA characterization. J Mol Evol. 1981;17(6):334–337. doi: 10.1007/BF01734355. [DOI] [PubMed] [Google Scholar]
- Huysmans E., Dams E., Vandenberghe A., De Wachter R. The nucleotide sequences of the 5S rRNAs of four mushrooms and their use in studying the phylogenetic position of basidiomycetes among the eukaryotes. Nucleic Acids Res. 1983 May 11;11(9):2871–2880. doi: 10.1093/nar/11.9.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980 Dec;16(2):111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Komiya H., Miyazaki M., Takemura S. The nucleotide sequence of 5S ribosomal RNA from Schizosaccharomyces pombe. J Biochem. 1981 May;89(5):1663–1666. doi: 10.1093/oxfordjournals.jbchem.a133365. [DOI] [PubMed] [Google Scholar]
- Kumazaki T., Hori H., Osawa S. Phylogeny of protozoa deduced from 5S rRNA sequences. J Mol Evol. 1983;19(6):411–419. doi: 10.1007/BF02102316. [DOI] [PubMed] [Google Scholar]
- Levy C. C., Karpetsky T. P. The purification and properties of chicken liver RNase: An enzyme which is useful in distinguishing between cytidylic and uridylic acid residues. J Biol Chem. 1980 Mar 10;255(5):2153–2159. [PubMed] [Google Scholar]
- Levy C. C., Karpetsky T. P. The purification and properties of chicken liver RNase: An enzyme which is useful in distinguishing between cytidylic and uridylic acid residues. J Biol Chem. 1980 Mar 10;255(5):2153–2159. [PubMed] [Google Scholar]
- Lockard R. E., Alzner-Deweerd B., Heckman J. E., MacGee J., Tabor M. W., RajBhandary U. L. Sequence analysis of 5'[32P] labeled mRNA and tRNA using polyacrylamide gel electrophoresis. Nucleic Acids Res. 1978 Jan;5(1):37–56. doi: 10.1093/nar/5.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki M. Studies on the nucleotide sequence of pseudouridine-containing 5S RNA from Saccharomyces cerevisiae. J Biochem. 1974 Jun;75(6):1407–1410. doi: 10.1093/oxfordjournals.jbchem.a130532. [DOI] [PubMed] [Google Scholar]
- Nishikawa K., Takemura S. Structure and function of 5S ribosomal ribonucleic acid from Torulopsis utilis. II. Partial digestion with ribonucleases and derivation of the complete sequence. J Biochem. 1974 Nov;76(5):935–947. [PubMed] [Google Scholar]
- Piechulla B., Hahn U., McLaughlin L. W., Küntzel H. Nucleotide sequence of 5S ribosomal RNA from Aspergillus nidulans and Neurospora crassa. Nucleic Acids Res. 1981 Mar 25;9(6):1445–1450. doi: 10.1093/nar/9.6.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker E. U., Yanofsky C., Driftmier K., Metzenberg R. L., Alzner-DeWeerd B., RajBhandary U. L. Dispersed 5S RNA genes in N. crassa: structure, expression and evolution. Cell. 1981 Jun;24(3):819–828. doi: 10.1016/0092-8674(81)90107-0. [DOI] [PubMed] [Google Scholar]
- Silberklang M., Gillum A. M., RajBhandary U. L. The use of nuclease P1 in sequence analysis of end group labeled RNA. Nucleic Acids Res. 1977 Dec;4(12):4091–4108. doi: 10.1093/nar/4.12.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata S. Structure of the 5-S ribosomal RNA gene and its adjacent regions in Torulopsis utilis. Eur J Biochem. 1980 Sep;110(1):107–114. doi: 10.1111/j.1432-1033.1980.tb04845.x. [DOI] [PubMed] [Google Scholar]
- Walker W. F., Doolittle W. F. 5S rRNA sequences from eight basidiomycetes and fungi imperfecti. Nucleic Acids Res. 1983 Nov 11;11(21):7625–7630. doi: 10.1093/nar/11.21.7625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker W. F., Doolittle W. F. Redividing the basidiomycetes on the basis of 5S rRNA sequences. Nature. 1982 Oct 21;299(5885):723–724. doi: 10.1038/299723a0. [DOI] [PubMed] [Google Scholar]



