Abstract
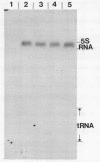
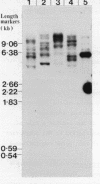
In Saccharomyces cerevisiae the majority of the genes for 5S rRNA lie within a 9kb rDNA sequence that is present as 100-200 tandemly-repeated copies on Chromosome XII. Following our observations that about 10% of yeast 5S rRNA exists as minor variant sequences, we screened a collection of yeast DNA fragments cloned in lambda gt for 5S rRNA genes whose flanking sequences differed from those adjacent to 5S rRNA genes of the rDNA repeat. Three variant 5S rRNA genes were isolated on the basis of such dissimilarity to rDNA repeat sequences. They display a remarkable conservation of their DNA in the vicinity of the 5S coding region, and are examples of a minor form of 5S rRNA coding sequence present in a small number of copies in the yeast genome. These variant sequences appear to be transcribed as efficiently as 5S rRNA genes of the rDNA repeat. In one of our isolates of the variant sequence a Ty transposable element is inserted 145bp upstream of the initiation point for 5S rRNA synthesis.
Full text
PDF













Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bayev A., Georgiev O. I., Hadjiolov A. A., Nikolaev N., Skryabin K. G., Zakharyev V. M. The structure of the yeast ribosomal RNA genes. 3. Precise mapping of the 18 S and 25 S rRNA genes and structure of the adjacent regions. Nucleic Acids Res. 1981 Feb 25;9(4):789–799. doi: 10.1093/nar/9.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G. I., DeGennaro L. J., Gelfand D. H., Bishop R. J., Valenzuela P., Rutter W. J. Ribosomal RNA genes of Saccharomyces cerevisiae. I. Physical map of the repeating unit and location of the regions coding for 5 S, 5.8 S, 18 S, and 25 S ribosomal RNAs. J Biol Chem. 1977 Nov 25;252(22):8118–8125. [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Bowen B. A., Fulton A. M., Tuite M. F., Kingsman S. M., Kingsman A. J. Expression of Ty-lacZ fusions in Saccharomyces cerevisiae. Nucleic Acids Res. 1984 Feb 10;12(3):1627–1640. doi: 10.1093/nar/12.3.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutlag D. L. Molecular arrangement and evolution of heterochromatic DNA. Annu Rev Genet. 1980;14:121–144. doi: 10.1146/annurev.ge.14.120180.001005. [DOI] [PubMed] [Google Scholar]
- Cameron J. R., Loh E. Y., Davis R. W. Evidence for transposition of dispersed repetitive DNA families in yeast. Cell. 1979 Apr;16(4):739–751. doi: 10.1016/0092-8674(79)90090-4. [DOI] [PubMed] [Google Scholar]
- Cameron J. R., Philippsen P., Davis R. W. Analysis of chromosomal integration and deletions of yeast plasmids. Nucleic Acids Res. 1977;4(5):1429–1448. doi: 10.1093/nar/4.5.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer J. H., Farrelly F. W., Barnitz J. T., Rownd R. H. Construction and restriction endonuclease mapping of hybrid plasmids containing Saccharomyces cerevisiae ribosomal DNA. Mol Gen Genet. 1977 Mar 16;151(3):229–244. doi: 10.1007/BF00268786. [DOI] [PubMed] [Google Scholar]
- DeFranco D., Schmidt O., Söll D. Two control regions for eukaryotic tRNA gene transcription. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3365–3368. doi: 10.1073/pnas.77.6.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eibel H., Philippsen P. Preferential integration of yeast transposable element Ty into a promoter region. 1984 Jan 26-Feb 1Nature. 307(5949):386–388. doi: 10.1038/307386a0. [DOI] [PubMed] [Google Scholar]
- Elder R. T., Loh E. Y., Davis R. W. RNA from the yeast transposable element Ty1 has both ends in the direct repeats, a structure similar to retrovirus RNA. Proc Natl Acad Sci U S A. 1983 May;80(9):2432–2436. doi: 10.1073/pnas.80.9.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh P. J., Fink G. R. Insertion of the eukaryotic transposable element Ty1 creates a 5-base pair duplication. Nature. 1980 Jul 24;286(5771):352–356. doi: 10.1038/286352a0. [DOI] [PubMed] [Google Scholar]
- Gafner J., Philippsen P. The yeast transposon Ty1 generates duplications of target DNA on insertion. Nature. 1980 Jul 24;286(5771):414–418. doi: 10.1038/286414a0. [DOI] [PubMed] [Google Scholar]
- Gafner J., Robertis E. M., Philippsen P. Delta sequences in the 5' non-coding region of yeast tRNA genes. EMBO J. 1983;2(4):583–591. doi: 10.1002/j.1460-2075.1983.tb01467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard J. P., Squire M., Bienz M., Smith J. D. A human tRNAGlu gene of high transcriptional activity. Nucleic Acids Res. 1983 May 11;11(9):2551–2562. doi: 10.1093/nar/11.9.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafatos F. C., Jones C. W., Efstratiadis A. Determination of nucleic acid sequence homologies and relative concentrations by a dot hybridization procedure. Nucleic Acids Res. 1979 Nov 24;7(6):1541–1552. doi: 10.1093/nar/7.6.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsman A. J., Gimlich R. L., Clarke L., Chinault A. C., Carbon J. Sequence variation in dispersed repetitive sequences in Saccharomyces cerevisiae. J Mol Biol. 1981 Feb 5;145(4):619–632. doi: 10.1016/0022-2836(81)90306-5. [DOI] [PubMed] [Google Scholar]
- Korn L. J. Transcription of Xenopus 5S ribosomal RNA genes. Nature. 1982 Jan 14;295(5845):101–105. doi: 10.1038/295101a0. [DOI] [PubMed] [Google Scholar]
- Koski R. A., Allison D. S., Worthington M., Hall B. D. An in vitro RNA polymerase III system from S. cerevisiae: effects of deletions and point mutations upon SUP4 gene transcription. Nucleic Acids Res. 1982 Dec 20;10(24):8127–8143. doi: 10.1093/nar/10.24.8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer R. A., Philippsen P., Davis R. W. Divergent transcription in the yeast ribosomal RNA coding region as shown by hybridization to separated strands and sequence analysis of cloned DNA. J Mol Biol. 1978 Aug 15;123(3):405–416. doi: 10.1016/0022-2836(78)90087-6. [DOI] [PubMed] [Google Scholar]
- Mao J., Appel B., Schaack J., Sharp S., Yamada H., Söll D. The 5S RNA genes of Schizosaccharomyces pombe. Nucleic Acids Res. 1982 Jan 22;10(2):487–500. doi: 10.1093/nar/10.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Tizard R., Skryabin K. G., Gilbert W. Promotor region for yeast 5S ribosomal RNA. Nature. 1977 Jun 16;267(5612):643–645. doi: 10.1038/267643a0. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Nath K., Bollon A. P. Organization of the yeast ribosomal RNA gene cluster via cloning and restriction analysis. J Biol Chem. 1977 Sep 25;252(18):6562–6571. [PubMed] [Google Scholar]
- Olson M. V., Hall B. D., Cameron J. R., Davis R. W. Cloning of the yeast tyrosine transfer RNA genes in bacteriophage lambda. J Mol Biol. 1979 Jan 25;127(3):285–295. doi: 10.1016/0022-2836(79)90330-9. [DOI] [PubMed] [Google Scholar]
- Olson M. V., Loughney K., Hall B. D. Identification of the yeast DNA sequences that correspond to specific tyrosine-inserting nonsense suppressor loci. J Mol Biol. 1979 Aug 15;132(3):387–410. doi: 10.1016/0022-2836(79)90267-5. [DOI] [PubMed] [Google Scholar]
- Petes T. D., Hereford L. M., Skryabin K. G. Characterization of two types of yeast ribosomal DNA genes. J Bacteriol. 1978 Apr;134(1):295–305. doi: 10.1128/jb.134.1.295-305.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes T. D. Unequal meiotic recombination within tandem arrays of yeast ribosomal DNA genes. Cell. 1980 Mar;19(3):765–774. doi: 10.1016/s0092-8674(80)80052-3. [DOI] [PubMed] [Google Scholar]
- Petes T. D. Yeast ribosomal DNA genes are located on chromosome XII. Proc Natl Acad Sci U S A. 1979 Jan;76(1):410–414. doi: 10.1073/pnas.76.1.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippsen P., Thomas M., Kramer R. A., Davis R. W. Unique arrangement of coding sequences for 5 S, 5.8 S, 18 S and 25 S ribosomal RNA in Saccharomyces cerevisiae as determined by R-loop and hybridization analysis. J Mol Biol. 1978 Aug 15;123(3):387–404. doi: 10.1016/0022-2836(78)90086-4. [DOI] [PubMed] [Google Scholar]
- Piper P. W., Bellatin J. A., Lockheart A. Altered maturation of sequences at the 3' terminus of 5S gene transcripts in a Saccharomyces cerevisiae mutant that lacks a RNA processing endonuclease. EMBO J. 1983;2(3):353–359. doi: 10.1002/j.1460-2075.1983.tb01430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Roeder G. S., Farabaugh P. J., Chaleff D. T., Fink G. R. The origins of gene instability in yeast. Science. 1980 Sep 19;209(4463):1375–1380. doi: 10.1126/science.6251544. [DOI] [PubMed] [Google Scholar]
- Rubin G. M. Preparation of RNA and ribosomes from yeast. Methods Cell Biol. 1975;12:45–64. doi: 10.1016/s0091-679x(08)60951-6. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Selker E. U., Yanofsky C., Driftmier K., Metzenberg R. L., Alzner-DeWeerd B., RajBhandary U. L. Dispersed 5S RNA genes in N. crassa: structure, expression and evolution. Cell. 1981 Jun;24(3):819–828. doi: 10.1016/0092-8674(81)90107-0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Squires C., Lee F., Bertrand K., Squires C. L., Bronson M. J., Yanofsky C. Nucleotide sequence of the 5' end of tryptophan messenger RNA of Escherichia coli. J Mol Biol. 1976 May 15;103(2):351–381. doi: 10.1016/0022-2836(76)90317-x. [DOI] [PubMed] [Google Scholar]
- Szostak J. W., Wu R. Insertion of a genetic marker into the ribosomal DNA of yeast. Plasmid. 1979 Oct;2(4):536–554. doi: 10.1016/0147-619x(79)90053-2. [DOI] [PubMed] [Google Scholar]
- Thomas M., Davis R. W. Studies on the cleavage of bacteriophage lambda DNA with EcoRI Restriction endonuclease. J Mol Biol. 1975 Jan 25;91(3):315–328. doi: 10.1016/0022-2836(75)90383-6. [DOI] [PubMed] [Google Scholar]
- Valenzuela P., Bell G. I., Masiarz F. R., DeGennaro L. J., Rutter W. J. Nucleotide sequence of the yeast 5S ribosomal RNA gene and adjacent putative control regions. Nature. 1977 Jun 16;267(5612):641–643. doi: 10.1038/267641a0. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Williamson V. M., Cox D., Young E. T., Russell D. W., Smith M. Characterization of transposable element-associated mutations that alter yeast alcohol dehydrogenase II expression. Mol Cell Biol. 1983 Jan;3(1):20–31. doi: 10.1128/mcb.3.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamb T. J., Petes T. D. Analysis of the junction between ribosomal RNA genes and single-copy chromosomal sequences in the yeast Saccharomyces cerevisiae. Cell. 1982 Feb;28(2):355–364. doi: 10.1016/0092-8674(82)90353-1. [DOI] [PubMed] [Google Scholar]