Abstract
Synthetic analogs were designed to highlight the effect of the glycine moiety of bacillibactin on the overall stability of the ferric complex as compared to synthetic analogs of enterobactin. Insertion of a variety of amino acids to catecholamide analogs based on a Tren (tris(2-aminoethyl)amine) backbone increased the overall acidity of the ligands, causing an enhancement of the stability of the resulting ferric complex as compared to TRENCAM. Solution thermodynamic behavior of these siderophores and their synthetic analogs was investigated through potentiometric and spectrophotometric titrations. X-ray crystallography, circular dichroism, and molecular modeling were used to determine the chirality and geometry of the ferric complexes of bacillibactin and its analogs. In contrast to the Tren scaffold, addition of a glycine to the catechol chelating arms causes an inversion of the trilactone backbone, resulting in opposite chiralities of the two siderophores and a destabilization of the ferric complex of bacillibactin compared to ferric enterobactin.
Keywords: Bacillibactin, siderophore, Gram-positive bacteria, ferric stability constant
Introduction
The importance of siderophores for the acquisition of iron by microbes and the resulting bacterial pathogenicity is well established.3,4 Microorganisms use these low molecular weight compounds to overcome the insolubility of Fe3+ at pH 7 (~ 10−18 M). Specific receptor proteins on the cell membrane recognize the ferric complexes. The formation constants of siderophore complexes define the thermodynamic limits for the producing organisms to compete for iron, thus eventually providing superior propagation conditions for the organism producing the better chelating agent. The stability of an iron-siderophore complex must minimally be greater than that of iron hydroxide. Siderophores involved in virulence (which remove iron from transferrin or other iron transport storage proteins) must have a greater affinity for iron than the protein in order to be effective. The strength of iron binding may determine the mechanism of iron release (such as ligand exchange, reductive removal, or ligand destruction).5,6
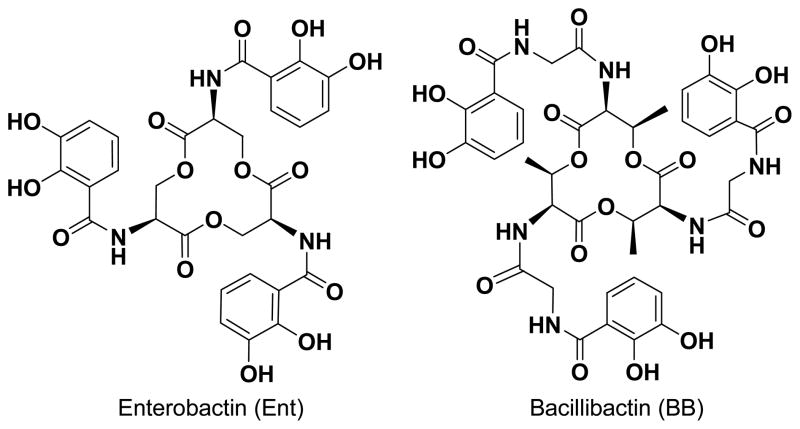
The siderophore with the highest known affinity for binding Fe3+ is enterobactin, produced by both Gram-positive7 and Gram-negative8,9 bacteria (Figure 1). Enterobactin has a high stability (Kf = 1049),10,11 with metal coordination at neutral pH through the six catecholate oxygens.12 This coordination leads to an iron center that has Δ or Λ chirality, and because the trilactone backbone is chiral, these conformations are diastereomers, with the lowest energy isomer being Δ for ferric enterobactin. This chirality is not essential for the receptor recognition and the transport through the membrane,13 but it is necessary for the iron utilization inside the microbial cell. The mirror image, Λferric enantioenterobactin does not promote microbial growth,14 indicating the importance of chirality for iron uptake.
Figure 1.
Enterobactin and Bacillibactin
By a standard convention,15 overall equilibria16 for metal stability constants are expressed as βmlh values for the reaction mM + lL + hH = MmLlHh. Deviations from the expected stability of a siderophore complex (based on comparison compounds) can indicate effects due to structural organization. Enterobactin is about 106 more stable than hexadentate synthetic analogs, but this extra stability disappears when the triserine ring is hydrolyzed.17 Many factors influence the stability of a metal complex;18–20 including overall ligand basicity and structural preorganization/predisposition. The intrinsic basicity of the donor atom also contributes to the overall stability of the metal complex.18–20 Although siderophores vary in their acidity, most are protonated at the pH experienced by the organism; H+ and Fe3+ compete with each other for binding to the siderophore. Thus the formal stability constant of a complex is not, by itself, a relative measure of a ligand’s ability to bind a metal. Variation in the protonation constants and the ligand denticity can lead to large differences in the magnitude of the overall formation constant among siderophores, which differ in pH dependence. To compare the true relative ability to bind a metal between differing ligands, a measure of the metal-ion free energy in the complex must be used. The pM value, analogous to pH, is defined as −log[M] where [M] represents the (usually very low) concentration of free aqueous metal ion.21 This is a direct measure of the chemical free energy of the metal ion at equilibrium with the chelating siderophore, which is why this is so direct and simple in application. The pM value is reported for a defined set of experimental conditions, at pH = 7.4, [M]tot = 1 μmol L−1, [L]tot = 10 μmol L−1. Successful siderophores are a balance between the basicity required for metal chelation and moieties that can effectively chelate at the environmental pH.
Bacillibactin (Figure 1), from Bacillus subtilis,22 is the second example of a siderophore with a trilactone backbone, but consists of L-threonine units in contrast to enterobactin, where L-serine units are incorporated. Also a glycine spacer separates the trilactone backbone from the catecholamide arms in bacillibactin, whereas in enterobactin the catecholamide is attached directly to the trilactone backbone. The relevance of individual components of the siderophore structure (such as methylation of the trilactone ring or addition of the glycine spacer) is difficult to accurately determine by studying only the natural siderophore. Synthetic analogs of naturally occurring siderophores provide a way to isolate key structural features in order to highlight the contribution of different components to the overall stability of the ferric complex. In addition, synthetic analogs can probe siderophore receptor recognition via alteration of specific structural features or chirality. Over the last few decades, synthetic siderophore analogs have provided information regarding stability, recognition and uptake.23 Synthetic analogs of bacillibactin can be used to measure indirectly the effect of the glycine spacer on the iron stability constant by comparison of these compounds to enterobactin and its analogs. Insertion of the spacer enhances the ability of the complex to chelate iron, through both lowering the overall acidity of the compounds and introduction of additional hydrogen bonds. Structural features of bacillibactin and synthetic analogs were investigated by circular dichroism spectroscopy and X-ray crystallography.
Materials and Methods
General
Unless otherwise noted, starting materials were obtained from commercial suppliers without further purification. Flash silica gel chromatography was performed using Merck silica gel (40–7 mesh). Microanalyses were performed by the Microanalytical Services Laboratory, College of Chemistry, University of California at Berkeley, Berkeley, CA 94720. Mass spectra were recorded at Mass Spectrometry Laboratory, College of Chemistry, University of California at Berkeley, Berkeley, CA 94720. All 1H NMR spectra were recorded at room temperature on the AVB-300/400 or DRX-500 Bruker FT spectrometer unless otherwise noted.
Syntheses
Bacillibactin was isolated from B. subtilis as described.24 Enterobactin and enantioenterobactin were synthesized by a published method.25 Dr. Martin Bluhm synthesized serine bacillibactin.26
2,3-Bis-benzyloxy-benzoyl chloride (Bncatcl) (1)
Bncatcl was prepared by established procedures from 2,3-dimethoxybenzoic acid, with purity established by 1H NMR spectroscopy.27
Ethyl-2,3-dibenzyl-benzamide glycylanoate (Bnglycamest) (2)
Glycine ethyl ester hydrochloride (45 mmol, 6.3 g) and Bncatcl (4.5 g, 0.013 mol) were dissolved in 100 mL of dry N, N-dimethylacetamide (DMAA). Triethylamine (TEA) was added in excess of three equivalents (0.135 mol, 19.0 mL) to yield a white precipitate of triethylamine hydrochloride. The reaction was stirred for 15 h, the solvent was removed with a rotary evaporator, and the orange solid was redissolved in CH2Cl2. The orange solution was washed with 1M of HCl (2×100 mL) and with brine (2×100 mL). The CH2Cl2 layer was applied to a silica gel column and eluted with a gradient from 1.5–3.5% methanol in CH2Cl2. The eluent was evaporated to yield a pale yellow solid. (Yield 3.57 g, 65.5%) 1H NMR (CDCl3): δ 8.50 (s, br, 1H, NH) 7.2–7.4 (m, 13H, ArH), 5.15 (s, 4H, CH2), 4.24 (q, J = 7.0, 2H CH2), 4.08 (d, J = 5.4, 2H CH2), 1.32 (t, J = 7.0, 3H, CH3). δ 13C NMR: δ 169.9, 165.5, 153.4, 147.5, 136.7, 129.5, 129.0, 128.9, 128.8, 128.6, 128.7, 128.1, 125.0, 124.7, 123.6, 117.6, 76.7, 71.6, 61.6, 42.2, 14.6. Anal. Calcd. (Found) for C25H25O5N: C 71.60 (71.50); H 5.96 (5.93); N 3.33 (3.05). MS (FAB+): m/z (MH+) calcd 420, obsd 420.
Glycyl-2,3-dibenzyl-benzamide (Bnglycam) (3)
Bnglycamest (3.0 g, 7.2 mmol) was dissolved in 50 mL of methanol. To this solution, KOH pellets (0.6 g, 0.01 mol) in 10 mL of MeOH was added and stirred for 20 h and monitored by TLC. The methanol was then evaporated, and the white solid was dissolved in 20 mL of water. The solution was extracted with 50 mL of ethyl acetate to remove any starting materials. Hydrochloric acid (1 mL, 6M) was added dropwise to the aqueous phase until a pH of 2 was reached. The solution was filtered, and the white solid was washed with water. (Yield 2.3 g, 82%) 1H NMR (DMSO-d6): δ 8.60 (t, J = 5.13, 1H, NH) 7.69 (dd, J = 2.73, 4.05, 1H, ArH), 7.30 – 7.40 (m, 12H, ArH), 5.15 (s, 2H OCH2), 5.12 (s, 2H OCH2), 4.01 (d, J = 5.40, 2H CH2). 13C NMR: δ 173.4, 166.1, 151.8, 147.1, 136.3, 136.1, 129.1, 128.8, 128.7, 128.6, 128.3, 127.8, 126.2, 124.5, 123.2, 117.6, 76.5, 71.33, 41.5. Anal. Calc. (Found) for C23H21O5N: C 70.58 (70.24); H 5.41 (5.36); N 3.58 (3.31). MS (FAB+): m/z (MH+) calcd 392 obsd 392.
Tren-glycyl-2,3-dibenzyl-benzamide (Bntrenglycam) (4)
Bnglycam (5.17 g, 13.1 mmol) was dissolved in dry THF (50 mL). To this solution, N-hydroxysuccinimide (NHS, 2.3 g, 0.020 mol) and 4-dimethylaminopyridine (DMAP, 0.24 g, 2.0 mmol) were added. When everything was dissolved, N, N′-dicyclohexylcarbodiimide (DCC, 4.1 g, 0.020 mol) was added and the mixture was stirred for 6 h and monitored by TLC. Tren (0.40 mL, 2.7 mmol) was then added and the solution was stirred for 15 h. Another portion of Tren (0.10 mL, 0.67 mmol) was added and the solution was stirred for 6 h. The mixture was filtered to remove the white precipitate, and the precipitate was washed with CH2Cl2. The solvent was evaporated off and the solid was redissolved in CH2Cl2 (250 mL), and washed with basic brine (0.5 M NaOH in saturated NaCl solution, 250 mL). The organic layer was collected and the solvent removed with a rotary evaporator. The resulting residue was applied to a silica gel column and eluted with MeOH (1–10 %) in CH2Cl2 to afford a pale yellow solid. (Yield: 4.06 g, 94%) 1H NMR (CDCl3): δ 8.59 (t, J = 4.4, 3H, NH), 7.67 (s, br, 3H, NH), 7.0–7.6 (m, 30H, ArH), 5.10 (s, 12H, CH2), 3.88 (d, J = 4.4, 6H CH2), 3.20 (s, br, 6H CH2), 2.52 (s, br, 6H, CH2). δ 13C NMR: δ 169.5, 166.0, 151.9, 136.7, 129.3, 128.7, 128.6, 128.3, 127.8, 126.9, 124.7, 124.4, 123.0, 117.6, 75.3, 71.3, 54.11, 43.8, 38.3, 30.9. Anal. Calcd. (Found) for C75H75N7O12·H2O: C 70.13 (70.28); H 6.04 (6.11); N 7.63 (7.44). MS (FAB+): m/z (MH+) calcd 1266.5, obsd 1266.5.
Tren-glycyl-2,3-dihydroxy-benzamide (TRENGlyCAM) (5)
BnTRENGlyCAM (0.323 g, 0.251 mmol) was dissolved in acetic acid/methanol solution (80/20 %, 75 mL). 10% Pd/C (0.30 g) was added and the mixture was hydrogenated for 15 h at atmospheric pressure. The catalyst was filtered, and 1 mL of HCl was added, and the solvent was evaporated. The product was then precipitated from methanol by ether as a pale purple solid. (Yield 0.147 g, 76%) 1H NMR (DMSO-d6): δ 9.20 (s, br, 3H, OH), δ 9.00 (s, br, 3H, NH), 7.92 (s, 3H, NH), 7.27 (d, J = 8.3, 3H, ArH), 6.88 (d, J = 7.8, 3H, ArH), 6.65 (t, J = 7.9, 3H, ArH), 3.85 (s, 6H, CH2), 3.11 (s, br, 6H CH2), 2.48 (s, br, 6H CH2). MS (ES+): m/z (MH+) calcd 726.3 obsd 726.3. Anal. Calc. (Found) for C33H39O12N7·HCl·H2O: C 47.61 (47.49); H 5.47 (5.07); N 11.50 (11.75).
2-Mercaptothiazolide 2,3-dibenzyl benzoic acid (Bncamthiaz)
Bncamthiaz was prepared by established procedures from 2,3-dimethoxybenzoic acid, with purity established by 1H NMR spectroscopy.28
Seryl-2,3-dibenzyl-benzamide (Bnserobutcam)
2-Mercaptothiazolide 2,3-dibenzyl benzoic acid (3.3 g, 7.6 mmol) and H-Ser-O-t-butyl-OH (1.2 g, 7.6 mmol) were dissolved in dry DMAA (100 mL). Triethylamine (14 mL) was added to the solution. The reaction was stirred for 60 h under N2 and monitored by TLC. The solvent was then evaporated with a rotary evaporator to yield a yellow residue. The residue was applied to a silica column and the product was eluted with a 95:5 methanol:dichloromethane. The solvent was removed to yield a white foam (2.7 g, 75 %). 1H NMR (CDCl3): δ 8.85 (d, 1H, J = 6.5, NH), 7.703 (dd, br, 1H, ArH), 7.1–7.4 (m, 12H, ArH), 5.15 (s, 2H, OCH2), 5.29 (m, br, 1H, CH), 5.13 (s, 4H, OCH2), 4.80 (s, 1H, CH2), 3.81 (s, 1H, CH2), 1.03 (s, 9H, CH3).
Tren-2,3-Dibenzoxy-benzamide-seryl-t-butyl-ester-cam (Bntrenserobutcam, Bntsc)
Bnserobutcam (2.7 g, 5.7 mmol) was dissolved in dry THF (20 mL). To this solution, NHS (0.83 g, 7.2 mmol) was added. When everything was dissolved, N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide (EDC, 1.6 g, 7.3 mmol) was added and the mixture was stirred for 6 h and monitored by TLC. Tren (0.28 mL, 1.9 mmol) was then added in 0.50 mL portions over 24 h. The mixture was filtered to remove the white precipitate, and the precipitate was washed with CH2Cl2. The solvent was evaporated, and the resulting residue was applied to a silica gel column and eluted with 2% MeOH in CH2Cl2. (Yield 1.1 g, 39 %). 1H NMR (DMSO-d6): δ 8.54 (d, 3H, J = 9.6, NH), 7.90 (m, 3H, NH), 7.5-7.1 (m, 39H, ArH), 5.16 (s, 6H, ArH), 5.04 (q, 6H, J = 13, OCH2), 4.48 (m, 3H, CH3), 3.42 (m, 6H, CH2), 3.15 (m, 12H, CH2), 0.93 (s, 27H, CH3).
Tren-2,3-Dihydroxy-benzamide-serylcam (TRENSerCAM)
Bntsc (0.68 g, 0.44 mmol) was dissolved in acetic acid (10 mL). 10% Pd/C (0.25 g) was added and the mixture was hydrogenated for 15 h at 400 psi. The catalyst was filtered, and 1 mL of concentrated HCl was added. The solvent was evaporated. The product was then precipitated from methanol by ether as a pale purple solid. (Yield 0.63 g, 100%) 1H NMR (DMSO-d6): δ 11.64 (s, 3H, OH), 9.38 (s, br, 3H, OH), δ 8.78 (d, J = 6.5, 3H, NH), 8.31 (m, 3H, NH), 7.40 (d, J = 8.5, 3H, ArH), 6.92 (d, J = 7.5, 3H, ArH), 6.69 (t, J = 8, 3H, ArH), 4.45 (m, 3H, CH), 3.72 (m, 6H, CH2), 3.46 (m, 6H, CH2), 3.26 (m, 6H, CH2). Anal. Calc. (Found) for C36H46O15N7Cl·H2O: C 49.69 (49.97); H 5.56 (5.56); N 11.27 (10.87). MS (FAB+): m/z (MH+) calcd 816, obsd 816.
Glutyl-2,3-dibenzyl-benzamide (Bngluobutcam)
2-Mercaptothiazolide 2,3-dibenzyl benzoic acid (3.1 g, 7.1 mmol) and H-Glu-O-t-Bu-OH (2.2 g, 10.8 mmol) were dissolved in dry DMAA (100 mL). Triethylamine (14 mL) was added to the solution. The reaction was stirred for 15 h under N2 and monitored by TLC. The solvent was then evaporated with a rotary evaporator to yield a yellow residue. The residue was applied to a silica column and the product was eluted with a 1:5:94 (acetic acid:methanol:dichloromethane). The solvents were removed to yield a yellow oil, which was redissolved in CH2Cl2 and the solution was washed with 0.5 M NaHCO3 to remove the remaining acetic acid. The organic layer was collected, and dried over Na2SO4, filtered, and the solvent was removed to yield a yellow oil (3.6 g, 97 %). 1H NMR (CDCl3): δ 8.55 (d, 1H, J = 7.0, NH), 7.66 (dd, J = 2, J = 7.5, 1H, ArH), 7.4-7.1 (m, 12H, ArH), 5.13 (m, 4H, OCH2), 4.54 (m, 1H, CH), 2.20 (m, 2H, CH2), 2.04 (m, 1H, CH2), 1.75 (m, 1H, CH2), 1.38 (s, 9H, OC(CH3)3). MS (FAB+): m/z (MNa+) calcd 542.4, obsd 542.4. Anal. Calc. (Found) for C30H33O7N·0.2CH3OH: C 68.96 (68.69); H 6.48 (6.66); N 2.66 (2.82).
Tren-2,3-Dibenzoxy-benzamide-glutamic-t-butyl-ester-cam (Bntrengluobutcam, Bntec)
Bngluobutcam (2.0 g, 3.8 mmol) was dissolved in dry DMF (50 mL). To this solution, 1-Hydroxybenzotriazole hydrate (HOBt, 0.62 g, 4.5 mmol) and was added. When everything was dissolved, EDC (0.88 g, 4.6 mmol) was added and the mixture was stirred for 6 h and monitored by TLC. Tren (0.17 mL, 1.1 mmol) was then added in 0.50 mL portions over 24 h. The mixture was filtered to remove the white precipitate, and the precipitate was washed with CH2Cl2. The solvent was evaporated, and the resulting residue was applied to a silica gel column and eluted with 2% MeOH in CH2Cl2. (Yield 1.9 g, 90 %). 1H NMR (DMSO-d6): δ 8.45 (d, 3H, J = 7.5, NH), 7.94 (m, 3H, NH), 7.5-7.1 (m, 39H, ArH), 5.17 (s, 6H, OCH2), 5.01 (d, 6H, OCH2), 4.46 (m, 3H, CH), 3.10 (m, 6H, CH2), 2.46 (m, 6H, CH2), 2.14 (m, 6H, CH2), 1.86 (m, 3H, CH2), 1.68 (m, 3H, CH2), 1.29 (m, 27H, CH3). Anal. Calc. (Found) for C96H111O18N7: C 69.84 (69.53); H 6.78 (6.97); N 5.94 (5.96). MS (FAB): m/z (MH+) calcd 1652, obsd 1652.
Tren-2,3-Dihydroxy-benzamide-glutamic-cam (TRENGluCAM)
Bntec (0.25 g, 0.15 mmol) was dissolved in acetic acid (10 mL). 10% Pd/C (0.1 g) was added and the mixture was hydrogenated for 15 h at 1000 psi. The catalyst was filtered, and 1 mL of concentrated HCl was added. The solvent was evaporated. The product was then precipitated from methanol by ether as a pale purple solid. (Yield 0.14 g, 100%) 1H NMR (DMSO-d6): δ 11.95 (s, 3H, OH), 10.01 (s, br, 1H, NH), 9.28 (s, br, 3H, OH), 8.82 (d, J = 6.5, 3H, NH), 8.36 (m, 3H, NH), 7.39 (d, J = 8, 3H, ArH), 6.92 (d, J = 8, 3H, ArH), 6.69 (t, J = 8, 3H, ArH), 4.45 (m, 3H, CH), 3.54 (m, 6H, CH2), 3.25 (m, 6H, CH2), 2.35 (m, 6H, CH2), 2.10 (m, 3H, CH2), 1.94 (m, 3H, CH2). Anal. Calc. (Found) for C36H46O15N7Cl·2CH3OH: C 50.70 (50.45); H 5.80 (5.65); N 9.40 (8.99). MS (ES−): m/z (MK2−) calcd 1018 obsd 1018.
Lysyl-2,3-dibenzyl-benzamide (Bnlysboccam)
2-Mercaptothiazolide 2,3-dibenzyl benzoic acid (3.0 g, 6.9 mmol) and H-Lys-BOC-OH (2.03 g, 8.24 mmol) were dissolved in dry DMAA (100 mL). Triethylamine (14 mL) was added to the solution. The reaction was stirred for 36 h under N2 and monitored by TLC. The solvent was then evaporated with a rotary evaporator to yield a yellow residue. The residue was applied to a silica column and the product was eluted with a 1:5:94 (acetic acid:methanol:dichloromethane). The solvents were removed to yield a yellow oil, which was redissolved in CH2Cl2 and the solution was washed with 0.5 M NaHCO3 to remove the remaining acetic acid. The organic layer was collected, and dried over Na2SO4. After filtration of the Na2SO4, the solvent was removed to yield a white foam (3.1 g, 80 %). 1H NMR (CDCl3): δ 8.40 (d, 1H, J = 6.0, NH), 7.52 (d, J = 7.5, 1H, ArH), 7.4 (m, 7H, ArH), 7.17 (m, 3H, ArH), 6.96 (d, J = 8, 1H, ArH), 6.88 (t, J = 7.5, 1H, ArH), 5.04 (d, 1H, J = 10.5, OCH2), 5.02 (s, 2H, OCH2), 4.98 (s, br, 1H, NH), 4.93 (d, J = 10, 1H, CH), 2.75 (m, 2H, CH2), 1.60 (m, 1H, CH2), 1.35 (m, 1H, CH2), 1.33 (s, 9H, C(CH3)3), 1.04-1.02 (m, 4H, CH2). Anal. Calc. (Found) for C32H37N2O7Na·0.5H2O: C 64.74 (64.45); H 6.45 (6.31); N 4.72 (4.71). MS (FAB+): m/z (MNa+) calcd 585.3, obsd 585.3.
Tren-2,3-Dibenzoxy-benzamide-lysylBoc-cam (Bntrenlysboccam, Bntkc)
Bnlysboccam (3.1 g, 5.5 mmol) was dissolved in dry DMF (50 mL). To this solution, HOBt (0.82 g, 6.1 mmol) and was added. When everything was dissolved, EDC (1.16 g, 6.1 mmol) was added and the mixture was stirred for 6 h and monitored by TLC. Tren (0.247 mL, 1.7 mmol) was then added in 0.50 mL portions over 24 h. The mixture was filtered to remove the white precipitate, and the precipitate was washed with CH2Cl2. The solvent was evaporated, and the resulting residue was applied to a silica gel column and eluted with 2% MeOH in CH2Cl2. (Yield 2.0 g, 63 %). 1H NMR (DMSO-d6): δ 8.39 (d, 3H, J = 8, NH), 7.92 (m, 3H, NH), 7.5-7.1 (m, 38H, ArH), 6.66 (m, 3H, ArH), 5.17 (s, 6H, OCH2), 5.01 (dd, 6H, OCH2), 4.38 (m, 3H, CH), 3.10 (m, 6H, CH2), 3.08 (m, 6H, CH2), 2.76 (m, 6H, CH2), 2.44 (m, 6H, CH2), 1.56 (m, 3H, CH2), 1.38 (m, 3H, CH2), 1.30 (s, 27H, C(CH3)3), 1.23 (m, 6H, CH2), 1.15 (m, 6H, CH2). Anal. Calc. (Found) for C102H126O18N10: C 68.82 (68.72); H 7.13 (7.23); N 7.87 (7.73). MS (FAB): m/z (MH+) calcd 1781, obsd 1781.
Tren-2,3-Dihydroxy-benzamide-lysine-cam (TRENLysCAM)
Bntkc (1.0 g, 0.5 mmol) was dissolved in acetic acid (10 mL). 10% Pd/C (0.3 g) was added and the mixture was hydrogenated for 24 h at 600 psi. The catalyst was filtered, and 4 mL concentrated HCl was added. The solvent was evaporated. The product was then precipitated from methanol by ether as a pale purple solid. (Yield 0.5 g, 92 %) 1H NMR (DMSO-d6): δ 12.05 (s, 3H, OH), 10.72 (s, br, 1H, OH), 9.33 (s, br, 3H, OH), 8.88 (d, J = 7.8, 3H, NH), 8.49 (m, 3H, NH), 7.93 (s, 9H, NH), 7.46 (d, J = 7.8, 3H, ArH), 6.93 (d, J = 7.8, 3H, ArH), 6.66 (t, J = 7.8, 3H, ArH), 4.44 (m, 3H, CH), 3.48 (m, 6H, CH2), 3.25 (m, 6H, CH2), 2.73 (m, 6H, CH2), 1.79 (m, 6H, CH2), 1.34 (m, 6H, CH2), 1.34 (m, 6H, CH2). Anal. Calc. (Found) for C45H70O12N10Cl4·1.5H2O: C 48.61 (48.79); H 6.62 (6.95); N 12.60 (12.29). MS (FAB+): m/z (MH+) calcd, 940 obsd 940.
β-alanine-2,3-dibenzyl-benzamide (Bnbalcamest)
To solution of Bncatcl (3.5 g, 9.9 mmol) in DMAA (50 mL), β-alanine ethyl ester (1.57 g, 10.2 mmol) was added. Triethylamine (10 mL) was added and the reaction was stirred under N2 for 15 h. The solvent was removed in vacuo and the residue was dissolved in CH2Cl2 and extracted with 1 M HCl and 1 M KOH successively. The organic fraction was dried over Na2SO4, filtered, and then the solvent was removed to yield a yellow oil (4.3 g, 100 %). 1H NMR (CDCl3): δ 8.23 (s, br, 1H, NH), 7.72 (m, 1H, ArH), 7.5-7.1 (m, 12H, ArH), 5.14 (s, 2H, OCH2), 5.08 (s, 2H, OCH2), 4.03 (q, 2H, J = 7.2, CH2), 3.55 (q, J = 6.4, 2H, CH2), 2.45 (t, J = 6.4, 1H, CH2), 1.18 (t, J = 7.2, 3H, CH3). Anal. Calc. (Found) for C26H27NO3: C 72.04 (72.38); H 6.28 (6.20); N 3.23 (2.98). MS (FAB+): m/z (MH+) calcd 434.2, obsd 434.2.
β-alanine-2,3-dibenzyl-benzamide (Bnbalcam)
Bnbalcamest (4.3g, 9.9 mmol) was dissolved in MeOH (50 mL). NaOH (0.48 g, 12 mmol) was dissolved in water (10 mL) and added to the MeOH solution. Water (20 mL) was added to the reaction until it became cloudy and then it was stirred under N2 for 15 h. The solvents were removed in vacuo and the resulting colorless oil was dissolved in water and the solution was filtered. The water was acidified to pH 2 with concentrated HCl and the product precipitated out of solution as a white solid. The solution was filtered and the solid was dried (3.4g, 85%). 1H NMR (CDCl3): δ 8.34 (s, br, 1H, NH), 7.72 (m, 1H, ArH), 7.5-7.1 (m, 12H, ArH), 5.15 (s, 2H, OCH2), 5.10 (s, 2H, OCH2), 3.52 (m, 2H, CH2), 2.51 (t, J = 6 1H, CH2). Anal. Calc. (Found) for C24H23NO5: C 71.10 (71.04); H 5.72 (5.80); N 3.45 (3.46). MS (FAB+): m/z (MH+) calcd 406.1, obsd 406.1.
Tren-2,3-Dibenzoxy-benzamide-β-alanine-cam (Bntrenbalcam, Bntac)
Bnbalcam (2.0 g, 4.9 mmol) was dissolved in dry DMF (50 mL). To this solution, HOBt (0.73 g, 5.4 mmol) and was added. When everything was dissolved, EDC (1.04 g, 5.43 mmol) was added and the mixture was stirred for 6 h and monitored by TLC. Tren (0.246 mL, 1.64 mmol) was then added in 0.50 mL portions over 24 h. The mixture was filtered to remove the white precipitate, and the precipitate was washed with CH2Cl2. The solvent was evaporated, and the resulting residue was applied to a silica gel column and eluted with 2% MeOH in CH2Cl2. (Yield 1.8 g, 86 %). 1H NMR (CDCl3): δ 8.25 (t, 3H, J = 6, NH), 7.55 (m, 3H, NH), 7.5-7.0 (m, 39H, ArH), 5.09 (s, 6H, OCH2), 5.05 (d, 6H, OCH2), 3.45 (m, 6H, CH), 3.10 (m, 6H, CH2), 2.38 (m, 6H, CH2), 2.28 (m, 6H, CH2). Anal. Calc. (Found) for C78H81O12N7·H2O: C 70.62 (70.29); H 6.31 (6.53); N 7.39 (7.30). MS (FAB): m/z (MH+) calcd 1309, obsd 1309.
Tren-2,3-Dihydroxy-benzamide-β-alanine-cam (TRENbAlaCAM)
Bntac (1.5 g, 1.1 mmol) was dissolved in acetic acid (10 mL). 10% Pd/C (0.25 g) was added and the mixture was hydrogenated for 24 h at ambient pressure. The catalyst was filtered, and 1 mL of concentrated HCl was added. The solvent was evaporated. The product was then precipitated from methanol by ether as a pale purple solid. (Yield 0.77 g, 87%) 1H NMR (DMSO-d6): δ 12.59 (s, 3H, OH), 10.17 (s, 1H, NH), 9.17 (s, br, 3H, OH), 8.78 (d, J = 5.5, 3H, NH), 8.31 (m, 3H, NH), 7.26 (dd, J = 1.5, J = 6.5, 3H, ArH), 6.89 (dd, J= 6.5, J = 1.5, 3H, ArH), 6.65 (t, J = 6.5, 3H, ArH), 3.46 (m, 6H, CH2), 3.43 (m, 6H, CH2), 3.23 (m, 6H, CH2), 2.44 (m, 6H, CH2). Anal. Calc. (Found) for C36H46O12N7Cl·2CH3OH: C 52.56 (51.60); H 6.27 (6.18); N 11.29 (11.05). MS (FAB+): m/z (MH+) calcd 768, obsd 768.
Ferric Tren-lysyl-2,3-dihydroxybenzamide (FeTRENLysCAM)
TRENLysCAM (287 mg, 0.258 mmol) was dissolved in degassed methanol (25 mL). Fe(acac)3 (87 mg, 0.246 mmol) was added and the solution turned a dark purple. After 30 min, KOH (7.28 mL, 0.1001 M) was added and the solution stirred for 6 h. The methanol was condensed (1 mL) and the product precipitated upon addition of acetone (30 mL). Centrifugation of the solution yielded a dark red product. X-ray quality crystals were obtained from a solution of FeTRENLysCAM complex in wet DMF diffused with ether over the course of one week.
Titration Solutions and Equipment
Experimental protocols and equipment followed closely to those of previous studies.29 The EDTA solution was standardized by potentiometric titration from pH 5 to 9 and back to 5 using 0.1 M HCl (standardized against tris(hydroxymethyl)aminomethane (TRIS) using bromocresol green (50 mg bromocresol green, 0.715 mL 0.1 M KOH in 120 mL water) as a visual end-point indicator) and 0.1 M KOH (standardized against potassium hydrogen phthalate using phenolphthalein (1 drop of a 1% solution in ethanol) as a visual indicator. A stock solution of iron was prepared by the dissolution of FeCl3·6H2O in 0.1 M HCl. This solution was standardized against the standardized EDTA solution using a literature method.30 Ligand acidity and Fe3+ coordination properties were examined by potentiometric (pH vs total proton concentration) and spectrophotometric (absorbance vs pH) titrations, with data analysis using Hyperquad31 and pHAB32 suite of computer programs. All experiments were performed at 25 °C and 0.1 M KCl. Equilibration times were 150 s for ligand only potentiometric titrations and 300 s for ligand-metal potentiometric titrations. The equilibration time for the metal-ligand batch titrations with EDTA was 48 h.
Ligand Protonation Constants
Protonation constants were determined by potentiometric titrations using ligand concentrations of approximately 0.4 mM with the average number of points collected in each titration to be 60, collected over a pH range of 4–10. The resulting solutions were titrated twice, first against 0.1 M KOH from pH 4 – 10 and then reverse against 0.1 M HCl back down to pH 4. For TRENGluCAM, the pH range was 5.5–10 and the protonation constants for the carboxylic acid side chains were fixed at 3.5, 4, and 4.5. For TRENLysCAM, the pH range was 4–10 and the protonation constants for the amine side chains were fixed at 11, 10.5, and 10. The results from each pair of titrations were combined for nonlinear least squares refinement with the program HYPERQUAD,31 giving the values for the protonation constants listed in Table 1.
Table 1.
Summary of solution thermodynamic behavior for Tren-based analogs
| TRENCAM37 | TRENbAlaCAM | TRENGlyCAM | TRENGluCAM | TRENSerCAM | TRENLysCAM | ||
|---|---|---|---|---|---|---|---|
| [LH7]+ | log Ka7 | 5.88 | 5.69(1) | 5.89(1) | 5.75(1) | 5.57(2) | 5.22(2) |
| [LH6] | log Ka6 | 6.71 | 6.95(1) | 6.84(1) | 6.87(1) | 6.61(2) | 6.24(2) |
| [LH5]− | log Ka5 | 8.61 | 7.54(1) | 7.36(1) | 7.32(1) | 6.98(1) | 6.72(1) |
| [LH4]−2 | log Ka4 | 8.75 | 8.33(1) | 8.23(1) | 8.19(1) | 7.91(2) | 7.37(2) |
| [LH3]−3 | log Ka3 | 11.3 | 11.3 | 11.3 | 11.3 | 11.3 | 11.3 |
| [LH2]−4 | log Ka2 | 12.1 | 12.1 | 12.1 | 12.1 | 12.1 | 12.1 |
| [LH]−5 | log Ka1 | 12.9 | 12.9 | 12.9 | 12.9 | 12.9 | 12.9 |
| ΣpKa1-7 | 65.25 | 64.81(1) | 64.60(1) | 64.43(1) | 63.37(2) | 61.85(1) | |
| FeL3− | log β110 | 43.6 | 42.6(1) | 45.2(1) | 45.1(2) | 45.2 (1) | 46.4(1) |
| FeLH2− | log β111 | 49.1 | 49.4 (1) | 50.1 | 51.5(2) | 51.3 (1) | 51.3(1) |
| FeL pKa | log KaFeL | 5.5 | 6.8 | 4.9 | 6.4 | 5.9 | 4.9 |
| pM | 27.8 | 28.0 | 30.9 | 30.9 | 31.6 | 32.8 |
FeTRENGlyCAM Complexation Constants
The iron stability constants were determined spectrophotometrically. In the titration cell (in the following order) 50.0 mL of electrolyte solution (0.1 M KCl) was combined with standardized EDTA (0.051 mmol, 0.50 mL) and Fe3+ (0.008 mmol, 0.351 mL) and TRENGlyCAM (approximately 7.5 mg, 0.009 mmol) previously dissolved in a MeOH (0.2 mL). The pH was adjusted to 5.5 with 0.1 M KOH. Each experiment consisted of two titrations (forward and reverse titrations) with ~24 points collected over ~48 h (an equilibration time of 2 h was used for each point) in a pH range of 5.5 to 6.5. Each data point consisted of a potentiometric measurement (pH) and an absorbance spectrum (400 to 700 nm). Equilibrium was not attainable under these conditions for the non-glycine analogs, so those constants were determined via a batch titration described below.
Ferric Stability Constants: Batch Titration with EDTA
The formation constants β110 and β111 were determined via a competition for Fe3+ between the ligand and EDTA. One stock solution was prepared containing approximately 8 × 10−5 M Fe3+, 9 × 10−4 M EDTA, and 8 × 10−5 M ligand (TRENGlyCAM, TRENGluCAM, TRENbAlaCAM, TRENLysCAM, TRENSerCAM) and the pH was adjusted to 5. The solution was then divided into 9 batches (10 mL each) and the pH of each batch was adjusted with ~0.1 M KOH to generate an equal distribution of pH values from 5 to 7.5. After an equilibration time of 48 h, both the spectra and pH values were recorded. Deconvolution of spectrophotometric and potentiometric data was accomplished with the refinement program pHAB.32
Ferric Complex Protonation Constants: Spectrophotometric Titrations
A stock solution containing 1.2 × 10−4 M ligand, 1.19 × 10−4 M Fe3+, 1.2 mM MES buffer, 1.2 mM HEPES buffer, and 1.2 mM sodium acetate was divided into 9 batches and the pH of each was adjusted to afford a pH range of 6 to 8. After 48 h, the spectra and pH of each sample was recorded and the data was refined in pHAB.32
Ferric Complex Protonation Constants: Potentiometric Titrations
Protonation constants for were determined by potentiometric titrations using ligand concentrations of approximately 0.22 mM and Fe3+ concentration of 0.2 mM with the average number of points collected in each titration to be 50, collected over a pH range of 2–6 (TRENLysCAM, TRENSerCAM) or 5.5–7.5 (TRENGluCAM, TRENbAlaCAM). The resulting solutions were titrated twice, first against 0.1 M KOH and then reverse against 0.1 M HCl. The results from each pair of titrations were combined for nonlinear least squares refinement with the program HYPERQUAD,31 giving the values for the protonation constants listed in Table 1.
Circular Dichroism Studies
Metal complexes studied were prepared in situ. An aliquot of a DMSO stock solution of the ligand (4 μM, 25 μL) and FeCl3 (0.0271 M, 3.7 μL) were combined and vigorously shaken in an eppendorf tube. Na2PO4 at pH 7.4 was added (0.1 M, 100 μL) and the solution was again shaken and equilibrated for 20 minutes. This solution was diluted with double distilled H2O to give a final volume of 1 mL and FeL concentration of 0.1 mM.
Results
Ligand Design
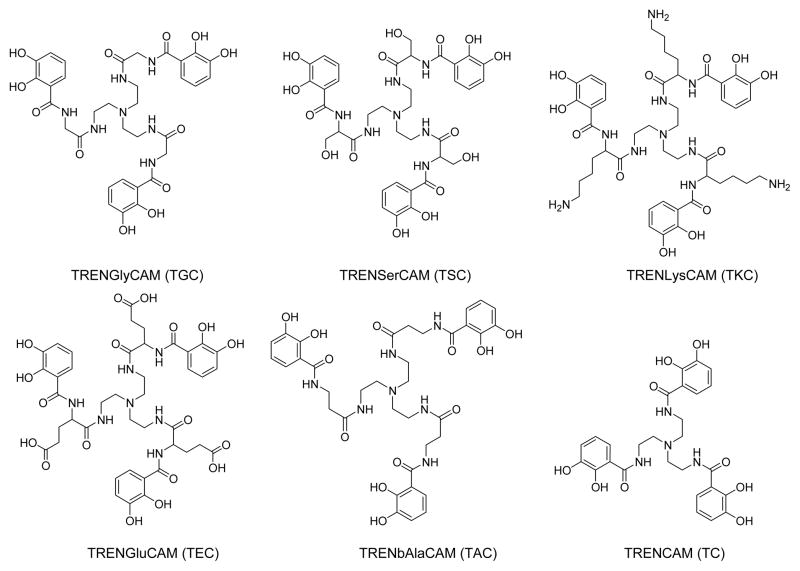
Several analogs of bacillibactin have been designed to probe the effect of the glycine spacer on iron complex stability of analogs employing a tris(2-aminoethyl)amine (Tren) scaffold (Figure 2). Tren-based ligands possess inherent organization due to the trigonal prismatic geometry of an amine, which is stabilized through hydrogen bonding to the pendent amine arms. The formation of five membered hydrogen bonded ring organizes the ligand to metal chelation in the catecholate derivatives. Both crystallographic and solution studies have confirmed that the backbone adapts this favorable geometry.33–36 Additionally, Tren does not hydrolyze, a major problem that would impede solution thermodynamic studies of similar series of trilactone-based ligands as utilized by the natural siderophores enterobactin and bacillibactin.
Figure 2.
Tren-based analogs
TRENCAM, an enterobactin analog, has previously been synthesized and characterized.37 Addition of glycine between the Tren backbone and the catecholate chelating moieties will serve as a comparison between enterobactin and bacillibactin. To expand upon this theme, new amino acid spacer analogs were also designed to explore the generality of any effect on stability. If extension of the chelating arms affects the thermodynamic stability of the complex, the reason can be determined by inserting a variety of amino acid spacers. If simply extending the arms changes the ferric ion chelating ability, then all amino acids should result in the same change. However, if the amino acid spacer itself becomes the source of the effect, the stability changes could be different for each spacer.
Water solubility is crucial for solution thermodynamic studies, requiring the amino acid analogs to be incorporated to possess hydrophilic side chains. Also, the amino acid selected should cause minimum changes to the overall structure of the analog. L-glycine, serine, glutamic acid, and lysine have these desired features, and synthesis and solution thermodynamic behavior was investigated. TRENSerCAM incorporates serine as the amino acid spacer unit in an attempt to preserve the overall charge of the compound, but to enhance the water solubility. TRENGluCAM (with a glutamic acid spacer) and TRENLysCAM (with a lysine spacer) are useful in determining the effect of the overall charge of the analog on the resulting stability of the ferric complex. These α-amino acid analogs should facilitate solution studies and determine the thermodynamic effect of elongation of the chelating arms through the addition of the glycine spacer.
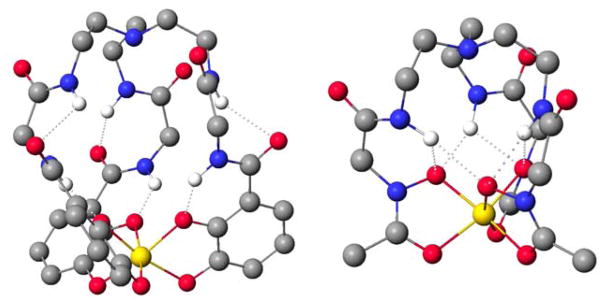
New hydrogen bonds formed within the chelating arms have been proposed as a result of the additional amino acid spacer as indicated by molecular models and crystallographic data in related structures.2 The presence of intrastrand hydrogen bonds in FeTRENGlyCAM was found in a MM3 molecular model by Dr. Ben Hay of the Pacific Northwest Laboratory (Figure 3). The additional hydrogen bond is seen between the carbonyl of the catecholamide and the second amide proton. The other set of hydrogen bonds is that seen typically of ferric catecholamide complexes, between the amide proton and the chelating phenolic oxygen. Masuda and coworkers also found inter- and intrastrand hydrogen bonding networks in a tripodal hydroxamic acid analog.2,38 Shanzer et al. also proposed a similar rationale for their Tren-based enterobactin analogs containing leucine and alanine, though with a different hydrogen-bonding scheme than seen in our model.39,40 Qualitative stability measurements indicated these analogs were often more stable than the enterobactin analog counterpart. Hydrogen bonding and increased ligand flexibility for better metal chelation were again proposed as reasons for the stability enhancement. In order to experimentally probe the possibility of the hydrogen bonding, another analog, using a β-alanine, was synthesized. In contrast to the common amino acids, β-alanine incorporates an extra methylene group between the amine and carboxylic acid. This lengthening of the spacer unit should disrupt any hydrogen bonding present, and the corresponding ferric complex should be less stable than the traditional amino acid analogs.
Figure 3.
(Left) Molecular model of Fe-TRENGlyCAM (Dr. Ben Hay). Intrastrand hydrogen bonding can be seen within the chelating arm (average hydrogen bond distances: Namide-Ocarbonyl: 3.09; Ocarbonyl-Hamide: 2.39; Namide-Ocatechol: 2.61; Ocatechol-Hamide 3.18). (Right) X-Ray crystal structure Fe-TAGE (average hydrogen bond distances: Namide-Ocatechol(intra): 2.88; Ocarbonyl-Hamide: 2.14; Namide-Ocatechol(inter): 3.2; Ocarbonyl-Hamide: 2.13).2
Synthesis of Analogs
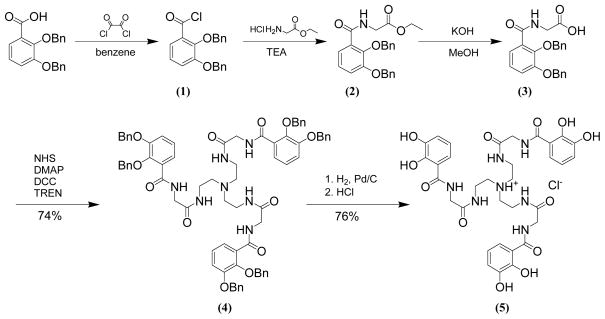
The five analogs were synthesized by a similar method; slight modifications were necessary when incorporating the amino acids with reactive side chains. The final ligand is an excellent iron chelator and can scavenge iron from glassware and the deprotected catecholate rings are sensitive to oxidation, so the deprotection step must require only the minimal amount of chemical manipulation. For these reasons, benzyl protection was used, since the deprotection conditions are mild. Different coupling techniques for the first amide bond formation were required under certain circumstances. The first approach directly couples the acid chloride of the benzyl-protected benzoic acid to the ethyl ester of the desired spacer (for glycine and β-alanine). Use of the ester form of the amino acid is necessary, since the zwitterionic form of the free acid is less reactive. The second approach converts the acid chloride the mercaptothiazolide activated benzyl-protected benzoic acid and then couples this to the amino acid (for serine, glutamic acid and lysine). Mercaptothiazolide activation provides control over the reaction by directing the coupling of the amino acid to the amine of the target amino acid. Though an additional step as compared to the first approach, this actually streamlines the procedure by allowing for coupling to the free acid of the amino acid, with protection only required for the side chain moiety. Standard amide formation coupling techniques yielded the second amide bond, which attached the arm to the backbone. Finally for serine, lysine and glutamic acid, protection of the reactive side chains is required. The acid-labile tert-butyl ester was selected over other esters (ethyl, benzyl) for its ease of deprotection. BOC protected the amine of lysine. A scheme for TRENGlyCAM is seen in Scheme 1, and the other four analogs follow a similar scheme and are detailed in the Materials and Methods section.
Scheme 1.
Synthetic scheme for TRENGlyCAM. The other analogs follow a similar strategy and are detailed in the Materials and Methods section.
Solution Thermodynamics
Formation of a ferric siderophore complex requires deprotonation of the phenolic hydrogen (catecholates). Therefore, determination of ligand pKa values is crucial to meaningfully interpret the stability of ferric complexes. The high pKa values of the catechol (9.2 and 13) make it the most basic ligand; addition of the amide moiety lowers these values substantially (6 and 11.1 for bidentate analog 2,3-dihydroxy-N, N′-diethylterephthalamide, ETA.) 41
With its high charge-to-radius ratio, the ferric ion is quite acidic. Formation of insoluble iron hydroxides is favored in pH ranges over 2, leaving free Fe3+ concentrations as low as 10−18 M at pH 7.4. However, iron is often solubilized through chelation by organic ligands or proteins. Siderophores must be thermodynamically able to remove the complexed iron from environmental sources such as transferrin, citrate, hydroxide, and minerals. Knowledge of the iron formation constant of a ligand indicates the strength of a siderophore to act as a ferric chelator. The stability constant for a ferric hexadentate catecholate siderophore complex is defined as:
| (1) |
However, the proton-independent ferric ion complex formation constants cannot be determined directly because the basicity of the ligand requires the measurement to be performed at unattainably high pH, precluding the direct measurement of the equilibrium of interest. Additionally, even at very low pH, the iron-siderophore complex does not fully dissociate and so the Fe3+ + LH6 = FeL3− + 6H+ equilibrium cannot be observed.
For the above reasons, direct measurement of the free Fe3+ after chelation is usually impossible. However, the stability of the complexes can be determined indirectly via a competition experiment with a chelator able to form strong iron complexes at lower pH ranges. Ethylenediamminetetraacetic acid (EDTA) is typically used for competition titrations because of its well-known stability constants and lack of absorbance in the visible region.42 By monitoring the LMCT (ligand to metal charge transfer) at 480–500 nm of a FeL3− tris-catecholate complex, and the amount of ferric siderophore complex formed can be determined by:
| (2) |
The resulting proton dependent formation constant is:
| (3) |
TRENGlyCAM pKa Values
Four of the seven protonation constants for TRENGlyCAM have been determined potentiometrically. Previous work with enterobactin and its analogs determined the protonation constants (pKa1 – pKa3 for TRENGlyCAM) of the m-hydroxy oxygen atoms are well separated from the o-hydroxy oxygen atoms (pKa4 – pKa6). Also, the capping amine nitrogen atom of the Tren backbone has been assigned the lowest protonation constant (pK7), evident from the lack of spectral change upon deprotonation and would be consistent with the model compound, Trenac where the only dissociable proton is that of the capping amine.43 Stepwise protonation (for n = 1–7) of the ligand could be seen by:
| (4) |
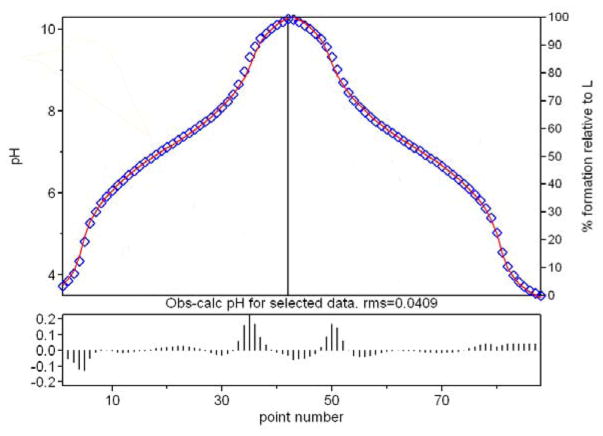
The forward and reverse potentiometric titration curves for TRENGlyCAM can be seen in Figures 4 and 5.
Figure 4.
Forward and reverse potentiometric titration curves for TRENGlyCAM. [TRENGlyCAM] = 0.4 mM, μ = 0.1 M KCl, 25 ° C.
Figure 5.
Theoretical (solid red line) and experimental (blue squares) curves for the potentiometric titration of TRENGlyCAM shown in Figure 4.
Ferric TRENGlyCAM Formation Constant
A solution of Fe, TRENGlyCAM, and EDTA (0.9:1:10) was titrated with base to pH 6.5 and then returned to pH 5.5 with acid to ascertain the ferric formation constant for TRENGlyCAM. A two hour delay period was sufficient to reach equilibration between each data point. The visible region was monitored for the appearance of the LMCT band at 490nm (Figure 6). As in equation 1, the stability constant for the ferric complex for TRENGlyCAM is defined as:
| (5) |
Figure 6.
Spectrophotometric EDTA competition to determine β110 for TRENGlyCAM. ([TRENGlyCAM] = 0.2 mM, [Fe3+] = 0.18 mM, [EDTA] = 1.0 mM, pH 4.6–7.6, 25°C, 2 hr eq. time, 0.1 M KCl, 25 °C).
Ferric TRENGlyCAM pKa Values
Since the Tren-based ligands contain a tertiary amine, which generally have pKa values around 10.5, determination of this pKa of the ferric complex is important. Although the intramolecular hydrogen bonding stabilizes the deprotonated form of this amine in the free ligand, as evident by the potentiometric protonation curve and the lack of spectral change in the spectrophotometric titration, the amino acid analogs may not lower the pKa as much and may yield two species at physiological pH, where the pM comparison is calculated.
In the spectrophotometric titration of a 1:1 TRENGlyCAM to Fe3+ solution, an isobestic point was seen at 540 nm (Figure 7), indicating the presence of two species in solution (equation 6). Deconvolution of the spectral data with pHab32 yielded a pKa value of 4.9, comparable to the pKa value for the enterobactin analog Fe-TRENCAM (pKa = 5.6).37
Figure 7.
Visible spectrum of FeTRENGlyCAM as a function of pH. [TRENGlyCAM] = 0.2 mM; [Fe3+]= 0.19 mM, 1 cm cell; μ= 0.1 M KCl; 25 ° C; pH
| (6) |
Since the pKa of ferric TRENGlyCAM is 2.5 orders of magnitude lower than physiological pH, the protonated ferric complex is not a major contributor to the solution equilibria occurring at pH 7.4.
At physiological pH, a high equilibrium constant (K7.4 = 1032.2) indicates TRENGlyCAM is a powerful iron chelator. The calculated pM value for TRENGlyCAM is 30.9, three log units higher than that reported for the enterobactin analog, TRENCAM (pM = 27.8). The only difference between these two ligands is the presence of the glycine spacer in the bacillibactin analog, TRENGlyCAM. One possible explanation for the enhanced stability of the ferric-TRENGlyCAM complex could be greater flexibility in the chelating arms, allowing the desired octahedral arrangement to be less strained. A second explanation could be the formation of additional hydrogen bonds that lead to a stabilization of the anionic form of the ligand, rendering it a more potent iron chelator at physiological pH.
Protonation Constants for Non-Glycine Amino Acid Analogs
Like TRENGlyCAM, TRENSerCAM has seven dissociable protons, thus has seven protonation constants. The protonation constants were determined in a similar manner as for TRENGlyCAM and lay within range of the TRENGlyCAM. The protonation constants for the other three analogs were determined in a similar manner. For TRENLysCAM, the three protonation constants for the amine side chain were fixed at 10, 10.5 and 11. For TRENGluCAM, the three protonation constants for the carboxylic acid side chain were fixed at 3.5,4, and 4.5. The pH range for potentiometric titration excluded these regions. The summary of all the potentiometric data is presented in Table 1.
Ferric Stability Constants for Non-Glycine Amino Acids
A solution of Fe3+, TRENSerCAM, and EDTA (0.9:1:10) was divided into 9 aliquots and base was added to each sample to give a pH range from 5 to 7. These samples were stored for 48 h before each spectrum and pH were recorded to ascertain the ferric formation constant for TRENSerCAM. (Note: In contrast to TRENGlyCAM, a two hour delay period was not sufficient to reach equilibration between each data point. TRENGlyCAM redone titrated in this manner yielded a similar result as above.) From equation 1, the stability constant for the ferric complex for TRENSerCAM is:
| (7) |
giving a pM value of 31.6. The formation constant of the other three analogs (TRENbAlaCAM, TRENGluCAM, TRENLysCAM) were determined in a similar fashion as TRENSerCAM and are summarized is Table 1.
Structure of Ferric TRENLysCAM
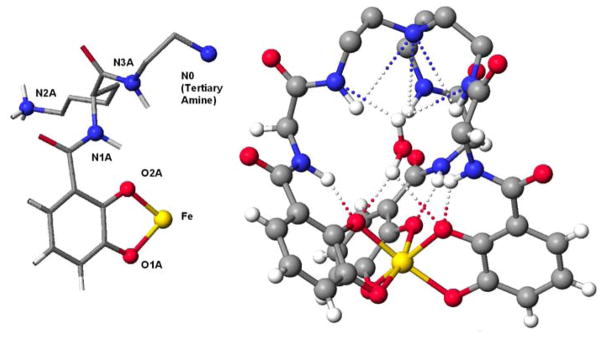
X-ray quality crystals were obtained from a solution of FeTRENLysCAM complex in wet DMF diffused with ether. One FeTRENLysCAM complex, two molecules of DMF, and five and half water molecules occupy each asymmetric unit. The compound crystallized as a neutral complex, which is unique for ferric tris-catecholamide compounds, since the overall charge at the metal center is −3. The three protonated ε-amino groups of pendent lysine arms serve as the counter ions to compensate the negative charge of the ferric tris-catecholamide center. This compound crystallized in the triclinic space group (P1̄). The ligand has three identical branches attached to a tertiary (Tren) amine nitrogen atom; a chiral carbon atom is on each branch of the ligand. In the crystal structure, the compound is a racemic mixture, although each complex molecule is homochiral. The two complex molecules in each cell are enantiomers (Δ-ferric TREN(LLL)LysCAM and Λ-ferric TREN(DDD)LysCAM). The FeTRENLysCAM complex contains a water molecule trapped within the chelating arms as seen in Figure 8, Left (the other solvent molecules have been omitted for clarity). The ferric complex has approximate C3 symmetry around the axis defined by the tertiary amine of the Tren backbone (N0) and the Fe center (Figure 8, Right).
Figure 8.
ORTEP (50% probability ellipsoid) of FeTRENLysCAM. (Left) Side view showing the trapped water molecule. (Right) Viewed down the pseudo-3-fold axis.
The Tren cap nitrogen atom (N0) has roughly equal distances to the corresponding atoms of each arm, as shown in Table 2 (the atom labels are correlated to the structure in Figure 9, Left).
Table 2.
Selected FeTRENLysCAM bond distances indicating the symmetric orientation of the three chelating arms relative to the tertiary amine (N0) of the Tren cap.
| Distances between N0 and Tren-amide nitrogens: | ||
| DN0-N3A (Å) | DN0-N3B (Å) | DN0-N3C (Å) |
| 3.024 | 3.098 | 3.036 |
| Distances between N0 and Lysine-α-amide nitrogens: | ||
| DN0-N1A (Å) | DN0-N1B (Å) | DN0-N1C (Å) |
| 5.527 | 5.576 | 5.296 |
| Distances between N0 and Lysine-ε-amide nitrogens: | ||
| DN0-N2A (Å) | DN0-N2B (Å) | DN0-N2C (Å) |
| 9.972 | 9.904 | 9.480 |
| Distances between N0 and catechol oxygens: | ||
| DN0-O2A (Å) | DN0-O2B (Å) | DN0-O2C (Å) |
| 6.551 | 6.466 | 6.447 |
| DN0-O1A | DN0-O1B | DN0-O1C |
| 8.715 | 8.680 | 8.638 |
Figure 9.
(Left) One arm of FeTRENLysCAM with the atom labeled to correspond to Tables 2 and 3. N0: Tren tertiary amine; N1A: α-lysine nitrogen; N2A: ε-lysine nitrogen; N3A: Tren-amide nitrogen; O1, O2: catecholate oxygen atoms. (Right) the intrastrand hydrogen bonding found in FeTRENLysCAM. The two amide nitrogen atoms are within hydrogen bonding distance of each other (these bonds and the lysine side chains are omitted for clarity). The intermolecular hydrogen bonding is evident between the complex and the trapped water
The FeTRENLysCAM is stabilized by an extensive hydrogen bonding network as shown in Figure 9, Right. The structural stabilization from both the intrastrand hydrogen-bonding and the hydrogen-bonding partners provided by the trapped water molecule contributes to the high thermodynamic stability of this complex. The tertiary amine (N0) of the Tren backbone is hydrogen-bonded to the three Tren-amide nitrogen atoms N3A, N3B and N3C, and is common feature found in related six-coordinate metal complexes including TRENCAM34 or Trenhopo.44 Similar to other catecholamide complexes, the amide of the α–nitrogen of the lysine form strong hydrogen bonds with their adjacent catechol oxygen atoms (Figure 9, Right; Table 2). The Tren-amide nitrogen and Tren-α-Lysine amide nitrogen are within hydrogen-bonding contact (i.e. N3A and N1A, Figure 9, Right; Table 2). A water guest (O1W) is trapped in the space of the Tren-Lysine cap of the complex and mediates the hydrogen bond stabilization. The water molecule is surrounded by three of the catechol oxygen donors and three of the Tren-α-Lysine amide nitrogen donors at relatively short distances (Figure 9, Right; Table 3). The three ε-Lysine amino atoms are all protonated, and form intermolecular hydrogen bonding with oxygen donors from solvent or neighboring complexes, as shown in the packing diagram (Figure 10).
Table 3.
FeTRENLysCAM intrastrand and intermolecular hydrogen bonding.
| Distances between O2 and Lysine-α-amide nitrogen atoms: | ||
| DN1A-O2A (Å) | DN1B-O2B (Å) | DN1C-O2C (Å) |
| 2.589 | 2.639 | 2.611 |
| Distances between Lysine-α-amide and Tren-amide nitrogen atoms: | ||
| DN1A-N3A (Å) | DN1B-N3B (Å) | DN1C-N3C (Å) |
| 2.876 | 2.866 | 2.751 |
| Distances between phenolate oxygen atoms and the trapped water molecule: | ||
| DO1w-O2A (Å) | DO1w-O2B (Å) | DO1w-O2C (Å) |
| 3.068 | 2.857 | 3.051 |
| Distances between Tren-amide nitrogens and the trapped water molecule: | ||
| DO1w-N3A (Å) | DO1w-N3B (Å) | DO1w-N3C (Å) |
| 3.272 | 3.097 | 3.526 |
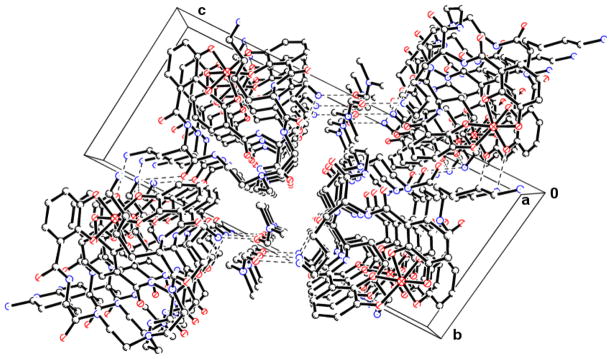
Figure 10.
Packing diagram of FeTRENLysCAM viewed down the ‘a’ axis. In the center of the cell is the tertiary amine of the Tren backbone, with the ferric tris-catecholate centers in opposite corners of the cell. Hydrogen bonding is evident between the lysine amine of the side chain and a neighboring FeTRENLysCAM molecule and solvent molecules.
A major contribution to the overall stability of ferric enterobactin is the optimal size of the trilactone backbone. Measurement of the two different V-O bond lengths in the crystal structure of V(Ent)3− revealed that the difference between the two (1.939(5) Å and 1.946(7) Å) is small (0.007 Å), indicating that the backbone is ideal for positioning the three arms to chelate the metal ion perfectly.12,45 The structure of [Fe(TRENCAM]3−, where the two Fe-O bond lengths differ significantly (by 0.038 Å) and indicate that the Tren cap is too small to accommodate the ferric ion.34 The average Fe-O2 bond length (the oxygen ortho to the amide) is 1.997(1) Å and is 2.013(1) Å for the Fe-O1 bond length (the oxygen meta to the amide) in FeTRENLysCAM, translating to a difference of 0.016 Å. Extension of the Tren cap by the amino acid spacer facilitates metal coordination, but still cannot match the perfect arrangement of the trilactone backbone. The stability enhancement of FeTRENLysCAM over FeTRENCAM is a result of two effects. The ligands incorporating the amino acid provide a better geometric fit for the ferric ion as compared to TRENCAM. The increased acidity also allows the ligand to compete for iron at a lower pH range than TRENCAM.
Circular Dichroism
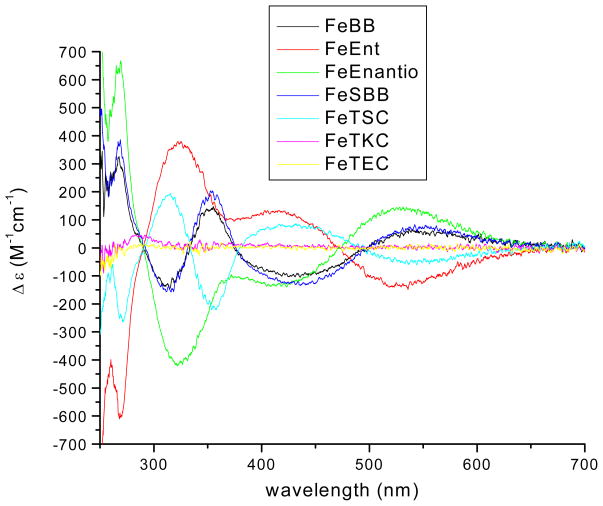
The solutions for circular dichroism were prepared in situ. The ferric complexes of the seven ligands (Figure 1 and 2, SBB is serine bacillibactin26) were prepared from the solutions of the free ligands in DMSO (4mM, 25 μL) with iron trichloride (27 mM, 3.7 μL). Sodium phosphate (pH = 7.4, 100 μL) was added to form the tris-catecholate complex, and the solutions were diluted with water to yield a final FeL concentration of 0.1 mM. The circular dichroism spectra were obtained within two h of making the complexes. The CD spectra are shown in Figure 11. All ferric complexes reveal intense CD bands at 270 nm corresponding to the carbonyl amide in the ligand. The bands of ferric bacillibactin and serine-bacillibactin (310 and 350 nm) and ferric enterobactin (320 nm) are due to the chiral trilactone scaffold. Two characteristic ferric catechol transitions are observed in the visible region at 420 nm and between 530 nm. These bands arise from ligand-to-metal charge transfer (LMCT) transitions and are therefore sensitive to the chirality at the metal center.46
Figure 11.
Circular dichroism spectra of ferric complexes studied. ([FeL] = 0.1 mM, path length = 1 cm, 22 °C) (Enantio = Enantioenterobactin).
The crystal structure of vanadium (IV) enterobactin established the absolute chirality as Δ in the solid state.12 Comparison of the ligand to metal charge transfer (LMCT) bands in the CD spectra of vanadium enterobactin to that of ferric enterobactin indicates that the two metal complexes have the same chirality. Using the CD spectrum of ferric enterobactin as a standard, ferric-bacillibactin appears to have the opposite chirality.26,47 A hybrid analog (serine-bacillibactin) containing the serine trilactone of enterobactin and the glycine catecholamide arms of bacillibactin also adopts the Λ configuration. Fe-TRENLysCAM is a racemate, evident in the absence of a CD spectrum and the presence of enantiomers in the crystal structure. The two enantiomers are homochiral, however, allowing for the comparison of the solid state chirality of the Fe-(LLL)TRENLysCAM and the chirality in solution of Fe-(LLL)TRENSerCAM. Both adopt the Δ conformation, the same as ferric enterobactin (Table 4). Similar chiralities were found in the tren-based amino acid containing analogs reported by Shanzer and coworkers.39
Table 4.
Circular dichroism results of ferric complexes.
| Ferric Complex | Diastereomer | λmax (nm) | Δ ε (M−1 cm−1) |
| Enterobactin | Δ | 527 | −133 |
| Bacillibactin | Λ | 546 | 58 |
| Enantioenterobactin | Λ | 526 | 138 |
| Serine Corynebactin | Λ | 546 | 70 |
| TRENSerCAM | Δ | 538 | −50 |
| TRENGluCAM | Racemic | ||
| TRENLysCAM | Racemic |
Molecular Modeling summary
The CD spectra of ferric complexes of enterobactin, bacillibactin, and the serine bacillibactin suggest differing chiralities at the metal centers for ferric bacillibactin and ferric serine-bacillibactin as compared to ferric enterobactin. This behavior has been confirmed by conformational analysis using an extended MM3 model.47 In full agreement with the CD spectroscopy and the X-ray crystallography, the modeling results predict Δ chirality for FeEnt and Λ chirality for FeBB and FeSBB. The calculations also reveal that the lowest energy forms of FeBB and FeSBB have an inverted macrocycle conformation. Calculations on the isolated macrocycle show that, in the absence of side chains, the inverted conformer is only 2 to 3 kcal/mol higher in energy than the normal conformer. The inverted macrocycle conformer seen in FeBB and FeSBB appears to be stabilized by hydrogen bonding between the macrocyclic CO groups and amide N-H hydrogen bond donors in the side chains.
Conclusion
The ferric ion affinity often scales with ligand basicity for chelates containing oxygen donor atoms.18,19 Thus, the basicity of the ligand is an important consideration in ligand design. However, many other considerations need to be made in order to design a ligand that meets the practical applications for which it will be used. Under basic conditions, hydrolysis of the metal ion begins to compete with complex formation with the desired ligand, resulting in a upper limit for ligand basicity. Under acidic conditions, protons compete with the metal ion for the ligand until the pH of the solution reaches a value above the highest pKa of the ligand. While lowering the acidity of the ligand lowers the pH at which the competition with protons occurs, it also diminishes the ligand’s affinity for not only proton, but also the metal ion as well. More acidic ligands do not bind the metal ion as strongly implying a lower limit in acidity for efficient iron chelation. Optimizing the basicity of the ligand was studied in depth for a series of Tren-based hydroxypyridonate and terephthalamide ligands designed as gadolinium chelators for use as MRI contrast agents.48
Comparison of the protonation and formation constants of the enterobactin analog, TRENCAM, to the bacillibactin analogs indicates that the amino acid spacer has a stabilizing effect and the magnitude of the effect depends on the spacer. The stability enhancement must not just be due to simply extending the arms, because all amino acids should result in the same change. The amino acid spacer must be the source of the effect, since the stability is different for each spacer.
The differing acidities of the Tren-based analogs are indicated by the sum of their protonation constants (ΣpKa), and are the likely cause for their different ferric stability constants. Comparing each proton dissociation constant for TRENCAM and TRENGlyCAM, the values are relatively the same, except for pKa5 (the second meta phenolic oxygen) where the two differ by 1.3 log units (Table 1). Possible stabilization from hydrogen bonding within and between the chelating arms may help stabilize the anionic species, making the amino acid spacer analog more acidic (Figure 12).
Figure 12.
Hydrogen bonding scheme for TRENCAM (left) and TRENGlyCAM (right). The glycine spacer analog is more acidic than that of the spacerless analog. The additional amide proton in bacillibactin analogs can facilitate the independent stabilization of two separate bases: the capping amine and the phenolic oxygen. The one amide proton is shared between three bases in TRENCAM. In TRENGlyCAM, each proton is shared by only two bases.
Deprotonation also induces a ligand conformational change, with rotation of 180° about the catechol-carbonyl bond. The rotation of the shorter arm TRENCAM may put these negatively charged oxygens in much closer proximity than in the longer armed TRENGlyCAM, leading to an increase basicity of TRENCAM to TRENGlyCAM. Since this conformational change is a major contributor to the overall stability of enterobactin, where having some arms pointing out, ‘prospecting’ for iron while the others are turned in, ready to chelate as soon as iron coordination induces the conformation change, it may be advantageous to lower the acidity of the ligand to have more arms pointing inward. (Figure 13).
Figure 13.
Difference in preorganization/predisposition of TRENCAM and TRENGlyCAM.
The lower acidity of TRENGlyCAM results in 2 of the 3 chelating arms to be directed inward at pH 7.4 (where pM is calculated). Two of the three chelating arms of the more basic TRENCAM are pointing outward. TRENGlyCAM is therefore more preorganized toward metal binding than TRENCAM, and may be the reason for the stability enhancement when forming the ferric complex.
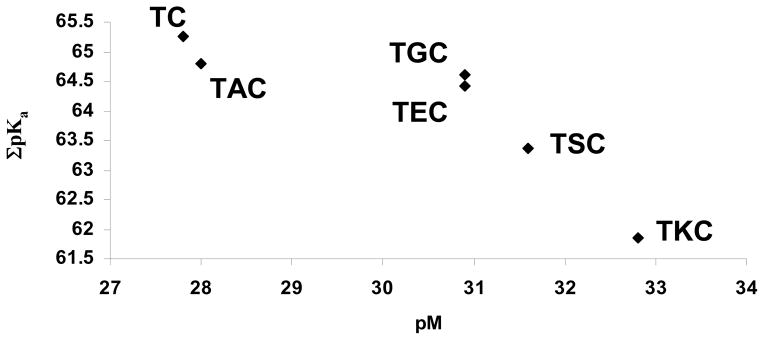
From these results we see that the enhancement of stability correlates to the acidity of the ligand. Had the glycine simply provided a more favorable geometry around the iron center, all the amino acid analogs (except TRENbAlaCAM) should provide the same amount of stabilization. However, TRENLysCAM is almost two orders of magnitude more stable than TRENGlyCAM, indicating that the enhancement is largely due to the greater iron chelating capacity at lower pH values (Figure 14).
Figure 14.
Increasing acidity of the ligand increases the pM.
The structural changes between bacillibactin and enterobactin result in these two siderophores having opposite chiralities and differing affinities for iron. The reason for different chiralities of the ferric complexes studied is complicated, since neither structural modification (methylation of the trilactone and addition of an amino acid spacer) has a consistent effect. Addition of the amino acid spacer cannot determine the chirality because FeTRENSerCAM and FeEnt adopt similar chiralities. Methylation of the trilactone cannot be the sole reason either because FeBB and FeSBB have virtually the same CD spectra. The reason must be a combination of the two. The trilactone backbone of enterobactin appears perfect for the size of the ferric ion, while the Tren cap is slightly smaller than optimal.12,45 The addition of the amino acid spacer for Tren-based ligands compensates for this size deficiency and allows the ligand to form highly stable metal complexes. The high ferric formation constant of FeTRENLysCAM can be explained by the stabilization seen in the X-ray crystal structure. First, the trapped water molecule extends the intrastrand hydrogen bonding network in the metal complex. Second, addition of a spacer to the Tren cap (previously determined to be too small for optimal metal coordination) allows for a less strained metal center. These two effects work in concert to stabilize the amino acid containing Tren-based analogs over the parent TRENCAM. However, addition of the glycine spacer to the already perfectly sized trilactone is detrimental to the overall stability evident in the lowered thermodynamic stability and increased basicity of ferric bacillibactin compared to ferric enterobactin.49 The trilactone of bacillibactin contorts to the less favored ‘inverted’ conformation in order to bind the ferric ion.47
Supplementary Material
Acknowledgments
We are grateful for grant support from NIH AI 11744. We thank Dr. Ben Hay from Pacific Northwest Laboratories, Dr. Marco Melchior and Rebecca Abergel for helpful discussions.
Footnotes
Supporting Information Available. Crystallographic information files (CIF) and data collection and refinement methods for Ferric TRENLysCAM. This information is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Paper number 77 in the series Microbial Iron Transport. For the previous paper in the series see: Dertz EA, Xu J, Stintzi A, Raymond KN. J Am Chem Soc. 2006;128:22–23. doi: 10.1021/ja055898c.
- 2.Matsumoto K, Ozawa T, Jitsukawa K, Einaga H, Masuda H. Inorg Chem. 2001;40:190–191. doi: 10.1021/ic0007411. [DOI] [PubMed] [Google Scholar]
- 3.Braun V. In J Med Microbiol. 2001;291:67–79. doi: 10.1078/1438-4221-00103. [DOI] [PubMed] [Google Scholar]
- 4.Andrews SC, Robinson AK, Rodriguez-Quinones F. Fems Microbiol Rev. 2003;27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 5.Boukhalfa H, Crumbliss AL. BioMetals. 2002;15:325–339. doi: 10.1023/a:1020218608266. [DOI] [PubMed] [Google Scholar]
- 6.Winkelmann G. Biochem Soc Trans. 2002;30:691–696. doi: 10.1042/bst0300691. [DOI] [PubMed] [Google Scholar]
- 7.Fiedler HP, Krastel P, Muller J, Gebhardt K, Zeeck A. Fems Microbiol Lett. 2001;196:147–151. doi: 10.1111/j.1574-6968.2001.tb10556.x. [DOI] [PubMed] [Google Scholar]
- 8.O’Brien IG, Gibson F. Biochim Biophys Acta. 1970;215:393–402. doi: 10.1016/0304-4165(70)90038-3. [DOI] [PubMed] [Google Scholar]
- 9.Pollack JR, Neilands JB. Biochim Biophys Res Commun. 1970;38:989–992. doi: 10.1016/0006-291x(70)90819-3. [DOI] [PubMed] [Google Scholar]
- 10.Hou Z, Stack TDP, Sunderland CJ, Raymond KN. Inorg Chim Acta. 1997;263:341–355. [Google Scholar]
- 11.Loomis LD, Raymond KN. Inorg Chem. 1991;30:906–911. [Google Scholar]
- 12.Karpishin TB, Dewey TM, Raymond KN. J Am Chem Soc. 1993;115:1842–1851. [Google Scholar]
- 13.Sprencel C, Cao ZH, Qi ZB, Scott DC, Montague MA, Ivanoff N, Xu JD, Raymond KN, Newton SMC, Klebba PE. J Bacteriol. 2000;182:5359–5364. doi: 10.1128/jb.182.19.5359-5364.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neilands JB, Erikson TJ, Rastetter WH. J Biol Chem. 1981;256:3831. [PubMed] [Google Scholar]
- 15.Martell AE, Motekaitis RJ. The Determination and Use of Stability Constants. VCH; New York: 1988. [Google Scholar]
- 16.Leggett DJ. Computational Methods for the Determination of Formation Constants. Plenum Press; New York: 1985. [Google Scholar]
- 17.Scarrow RC, Ecker DJ, Ng C, Liu S, Raymond KN. Inorg Chem. 1991;30:900–906. [Google Scholar]
- 18.Martell AE, Hancock RD, Motekaitis RJ. Coord Chem Rev. 1994;133:39–65. [Google Scholar]
- 19.Hancock RD, Martell AE. Chemical Reviews. 1989;89:1875–1914. [Google Scholar]
- 20.Evers A, Hancock RD, Martell AE, Motekaitis RJ. Inorg Chem. 1989;28:2189–2195. [Google Scholar]
- 21.Avdeef A, Sofen SR, Bregante TL, Raymond KN. J Am Chem Soc. 1978;100:5362–5370. [Google Scholar]
- 22.May JJ, Wendrich TM, Marahiel MA. J Biol Chem. 2001;276:7209–7217. doi: 10.1074/jbc.M009140200. [DOI] [PubMed] [Google Scholar]
- 23.Dertz EA, Raymond KN. In: Comprehensive Coordination Chemistry - II. McCleverty J, Meyer T, editors. Vol. 8. Pergamon; Oxford: 2003. pp. 141–168. [Google Scholar]
- 24.Dertz EA, Stintzi A, Raymond KN. 2006 In preparation. [Google Scholar]
- 25.Meyer M, Telford JR, Cohen SM, White DJ, Xu J, Raymond KN. J Am Chem Soc. 1997;119:10093–10103. [Google Scholar]
- 26.Bluhm ME, Kim SS, Dertz EA, Raymond KN. J Am Chem Soc. 2002;124:2436–2437. doi: 10.1021/ja016651s. [DOI] [PubMed] [Google Scholar]
- 27.Schuda P, Botti C, Vanti MC. OPPI Briefs. 1984;16:119–125. [Google Scholar]
- 28.Nagao Y, Seno K, Kawabata K, Miyasaka T, Takao S, Fujita E. Tetrahedron Let. 1980;21:841–844. [Google Scholar]
- 29.Johnson AR, O’Sullivan B, Raymond KN. Inorg Chem. 2000;39:2652–2660. doi: 10.1021/ic991471t. [DOI] [PubMed] [Google Scholar]
- 30.Ueno K, Imamura T, Cheng KL. Handbook of organic analytical reagents. 2. CRC Press; Boca Raton, FL: 1992. pp. 520–521. [Google Scholar]
- 31.Gans P, Sabatini A, Vacca A. Talanta. 1996;43:1739–1753. doi: 10.1016/0039-9140(96)01958-3. [DOI] [PubMed] [Google Scholar]
- 32.Gans P, Sabatini A, Vacca A. Ann Chim (Rome) 1999;89:45–49. [Google Scholar]
- 33.Bulls AR, Pippin CG, Hahn FE, Raymond KN. J Am Chem Soc. 1990;112:2627–2632. [Google Scholar]
- 34.Stack TDP, Karpishin TB, Raymond KN. J Am Chem Soc. 1992;114:1512–1514. [Google Scholar]
- 35.Xiao GY, Vanderhelm D, Hider RC, Dobbin PS. Inorg Chem. 1995;34:1268–1270. [Google Scholar]
- 36.Xu J, Franklin SJ, Whisenhunt DW, Raymond KN. J Am Chem Soc. 1995;117:7245–7246. [Google Scholar]
- 37.Rodgers SJ, Lee CW, Ng CY, Raymond KN. Inorg Chem. 1987;26:1622–1625. [Google Scholar]
- 38.Matsumoto K, Ozawa T, Jitsukawa K, Einaga H, Masuda H. Chem Commun. 2001:978–979. doi: 10.1021/ic0007411. [DOI] [PubMed] [Google Scholar]
- 39.Tor Y, Libman J, Shanzer A, Felder CE, Lifson S. J Am Chem Soc. 1992;114:6661–6671. [Google Scholar]
- 40.Tor Y, Libman J, Shanzer A, Lifson S. J Am Chem Soc. 1987;109:6517–6518. [Google Scholar]
- 41.Garrett TM, Miller PW, Raymond KN. Inorg Chem. 1989;28:128–133. [Google Scholar]
- 42.Martell AE, Smith RM. Critical Stability Constants. Vol. 1 Plenum Press; New York: 1974. [Google Scholar]
- 43.Cohen SM, Meyer M, Raymond KN. J Am Chem Soc. 1998;120:6277–6286. [Google Scholar]
- 44.Xu JD, O’Sullivan B, Raymond KN. Inorg Chem. 2002;41:6731–6742. doi: 10.1021/ic025610+. [DOI] [PubMed] [Google Scholar]
- 45.Karpishin TB, Raymond KN. Angew Chemie Int Ed. 1992;31:466–468. [Google Scholar]
- 46.Karpishin TB, Gebhard MS, Solomon EI, Raymond KN. J Am Chem Soc. 1991;113:2977–2984. [Google Scholar]
- 47.Bluhm ME, Hay BP, Kim SS, Dertz EA, Raymond KN. Inorg Chem. 2002;41:5475–5478. doi: 10.1021/ic025531y. [DOI] [PubMed] [Google Scholar]
- 48.Doble DMJ, Melchior M, O’Sullivan B, Siering C, Xu JD, Pierre VC, Raymond KN. Inorg Chem. 2003;42:4930–4937. doi: 10.1021/ic026240s. [DOI] [PubMed] [Google Scholar]
- 49.Dertz EA, Xu J, Stintzi A, Raymond KN. J Am Chem Soc. 2006;128:22–23. doi: 10.1021/ja055898c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.