Abstract
DNA sequence variations in individual genomes within the same species give rise to different phenotypes. One mechanism in this process is alteration of chromatin structure due to sequence variation that impacts gene regulation downstream. In this study, we compose a high-confidence collection of human indels and SNPs based on the analysis of a large set of publicly available sequencing data and investigate whether the DNA loci associated with stable nucleosome positions are protected against sequence mutations. We address how the sequence variation is reflected in the occupancy profiles of nucleosomes of different types at regulatory sequences and genome-wide. We find that indels are depleted around nucleosome positions of all considered types; SNPs, on the other hand, are enriched around the positions of bulk nucleosomes but depleted around the positions preferentially occupied by epigenetically modified nucleosomes. Such a behavior indicates an increased level of conservation for the sequences associated with epigenetically modified nucleosomes and highlights complex organization of the human chromatin.
Introduction
Growing evidence indicates that structural organization of chromatin, and the presence of regular nucleosome-positioning patterns in particular, are crucial for faithful gene regulation1–3. There is an on-going debate in the literature about the role of DNA sequence in establishing such patterns in vivo in different organisms4–6. For this question, analysis of the variability of the genomic sequences associated with stable nucleosome positions within the population can provide important clues. Mutations in genomic DNA can disrupt nucleosome positioning signal encoded in DNA and/or binding targets of transcription factors which are often coordinated with the stable nucleosomes. Therefore, functionally significant positions of stable nucleosomes are likely to be reflected in the density of sequence variations.
Two types of genomic sequence variation are the most relevant in this context: single nucleotide polymorphism (SNP) and short insertions and deletions (indels). Recent analysis of sequence variability in the yeast genome has shown that SNP density is higher by 10–15% in the DNA fragments associated with nucleosome cores than in linkers7. The analysis of the SNP distribution in the human genome revealed presence of a periodic signal close to the nucleosomal length in the promoter proximal regions and increased SNP density in the closed chromatin enriched with nucleosomes8,9. Association of both SNPs and indels with chromatin structure was recently characterized for regions around gene starts in the fish genome10. It was shown that unlike SNP density, the density of indels is decreased within stable nucleosome positions as compared to linker DNA.
Association of sequence variation with nucleosome organization in the human genome has not been studied comprehensively yet. Earlier studies focused exclusively on SNPs8, and the direct comparison of the genome variability and nucleosome occupancy profile was not possible for the human genome due to the lack of genome-scale nucleosome profiles. However, recent advances in high-throughput sequencing technology have made it possible to map nucleosomes and to accurately identify sequence variants on a genome-scale in humans11–14.
To this end, we collected sequencing data available from the NCBI Trace Archive and composed high-confidence non-redundant data sets of SNPs and indels from 1 to 100 bp in length (see Methods for detail). These sets comprise data from different sequencing centers obtained for multiple unrelated genomes and thus any biases due to possible experimental artifacts or genome sampling should be significantly reduced. We also use the data from a recently published set of genome variations based on the analysis of 8 individual genomes for the validation of our findings14 (results for the ‘8-genome’ set are presented as Supplementary material). Nucleosome occupancy has been profiled in the human genome for several types of epigenetically modified nucleosomes and for ‘bulk’ nucleosomes not selected for any histone variant or modification11,12. Based on these data we have recently identified with high resolution the stable positions for bulk nucleosomes and for the nucleosomes containing the H2A.Z histone variant and the histone H3 tri-methylated at lysine 46. The H2A.Z and H3K4me3 nucleosomes are associated with transcription activation and are enriched at gene starts, while the bulk nucleosomes are distributed throughout the genome more evenly, making the combination of these sets a convenient tool for our analysis.
Availability of these data allows us to address the question of how sequence variations are distributed relative to nucleosome positions on genome scale. For the first time, we consider nucleosomes of different types, both epigenetically modified and bulk, which play different roles in gene regulation. We also compare patterns of sequence variability and their association with chromatin structure in different regulatory genome regions such as transcription start and end sites (TSS and TES) and splicing sites.
Results
1. Distribution of indels and SNPs around stable nucleosome positions genome-wide
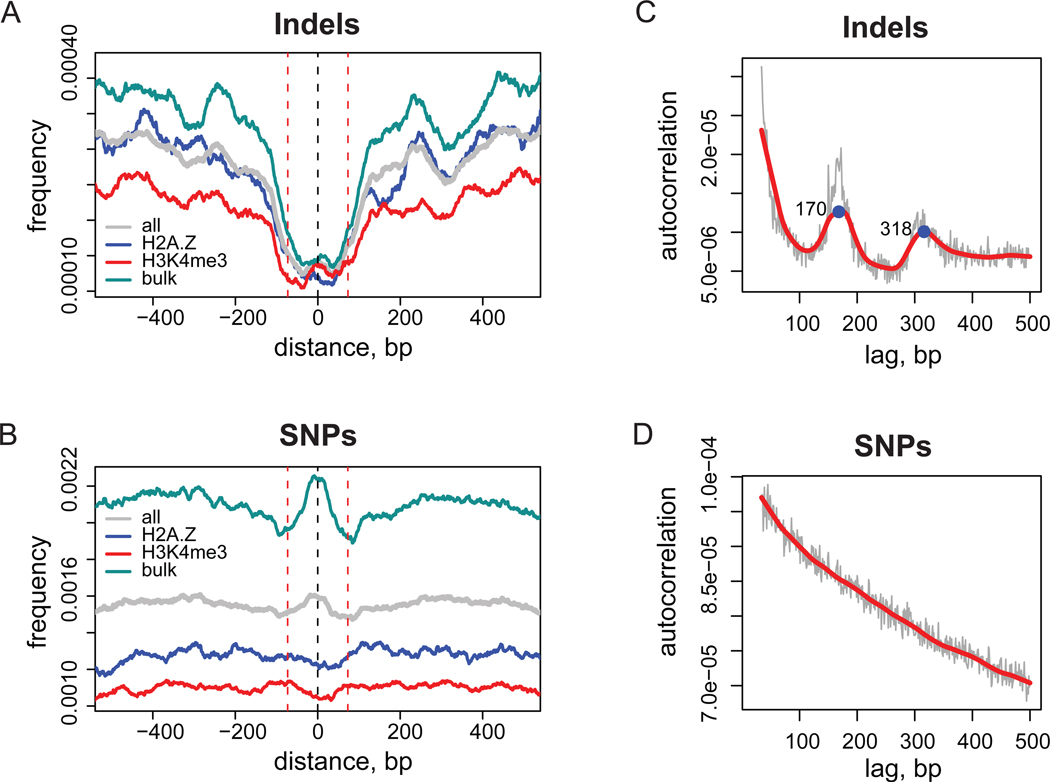
Distributions of the genome variation instances around stable nucleosome positions follow different patterns for indels and SNPs (Figure 1). Frequencies of indels are decreased inside core sequences for all types of nucleosomes as compared to the frequencies in linker DNA (Figure 1A). The distribution of SNP frequency is more complex: the SNP frequency is higher inside bulk nucleosomes, while it does not show significant variation around H2A.Z and H3K4me3 nucleosomes (Figure 1B).
Figure 1.
Genome-wide distributions of indel and SNP events. (A,B) Distributions of indel (A) and SNP (B) frequencies around stable nucleosome positions. Results are shown for a combined set of nucleosome positions (grey) and for individual nucleosome sets: bulk (cyan), H2A.Z (blue), and H3K4me3 (red). The frequency profiles were normalized and smoothed as described in Methods. Black dashed line at position zero corresponds to the center of nucleosome position and red dashed lines at positions ±73 bp give reference of nucleosomal size. (C,D) Auto-correlation profiles for indel (C) and SNP (D) occurrences. Thin grey lines correspond to the initial profile calculated with one base-pair lag increments and thick red line represents loess smoothing of the initial data. Two local maxima in the indel profile corresponding to mono- and di-nucleosomal sizes are indicated with numbers.
The coordination of indel and SNP distributions with nucleosome positioning is further illustrated in Figures 1C and 1D, where genome-wide autocorrelations of indel and SNP occurrences are shown. Autocorrelation is a measure of probability to find two and more instances of a variation separated by the specific distance in the genome. Therefore, if a coordinated pattern exists in the distribution of the variations, it should be reflected in the autocorrelation function. Unlike the monotonic autocorrelation plot for SNPs, the plot for indels features two pronounced maxima at distances of 170 bp and 318 bp, which agrees reasonably well with nucleosomal repeat length in the human chromatin15.
We performed a number of further analyses to confirm our results. We checked that the frequency profiles around stable nucleosome positions share the same features when indels were split into insertions and deletions and SNPs were split into transitions and transversions and analyzed independently (Supplementary Figure 1A–D). Since nucleosomes are known to favor GC-rich sequences16,17 we stratified nucleosome positions by GC-content and verified that the distribution of indels and SNPs is similar for GC-rich and GC-poor sequences (Supplementary Figure 1E–G).
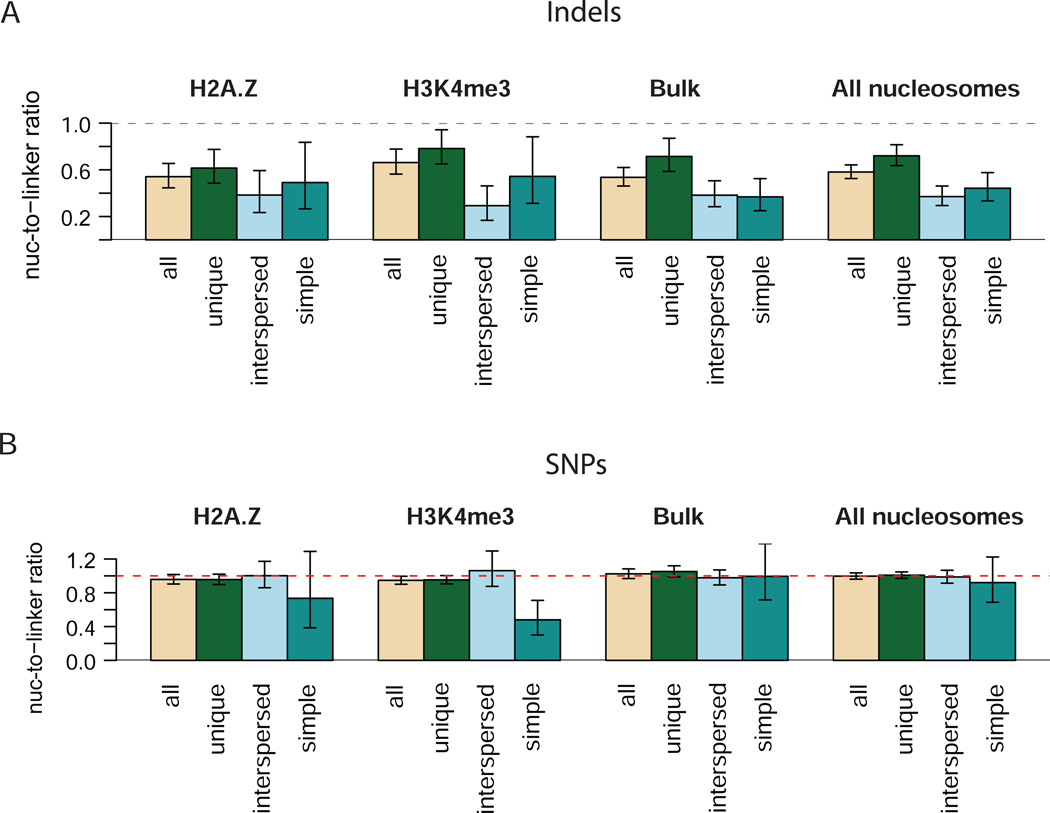
A recent study showed that tandem repeats can interfere with nucleosome positioning in yeast18. To check how the presence of repeats affects our findings, we examined the association of the genome variations located in the unique and in the repetitive regions of different classes with nucleosome positioning (Figure 2). This analysis reveals that the indel frequencies in core nucleosomes are lower than the indel frequencies in linker DNA for all classes of repeats (Figure 2). At the same time, the indels associated with repeats are further excluded from the nucleosome cores than the indels from unique regions. We also note that the indels from interspersed repeats contribute the most to the maxima in the autocorrelation function shown in Figure 1C (data not shown). In contrast, SNPs of all classes show nucleosome-to-linker occurrence ratio close to one, with the exception of the SNPs associated with the simple repeats in epigenetic nucleosomes which are depleted in nucleosome cores (Figure 2B). It is noteworthy that although the profile of SNP occurrences around bulk nucleosomes is not flat, the overall nucleosome-to-linker ratio is close to one for the linker DNA, defined in this study as 50-bp fragments flanking the core nucleosome.
Figure 2.
Relative occurrences of indels (A) and SNPs (B) of different classes inside nucleosome core sequences and linkers. Two 50-bp linker sequences flanking the core nucleosome sequence of 147-bp in length were considered and correction for different lengths of the core nucleosome and linker sequences was performed. The 95% confidence intervals are shown with thin arrows. Dashed horizontal line corresponding to the ratio of one is shown for reference. The nucleosome type for which the data are shown is indicated above each group of bars.
Since a noticeably weaker coordination between nucleosome positioning and frequency of the 1-bp indels was reported for the fish genome10, we compared the relative occurrences of indels inside and outside the nucleosome core for different indel lengths (Supplementary Figure 2). In our dataset, the nucleosome-to-linker ratio is about 0.6 for all the indel lengths for which such a ratio could be determined with statistical significance. We note that for the ‘8-genome’ set, such a ratio shows greater variability for different indel lengths and is close to 1 for the 1-bp indels. Nonetheless, the DNA sequences around stable nucleosome positions are still moderately depleted of the 1-bp indels from the ‘8-genome’ set compared to the fragment located 100–200 bp away from the nucleosome center (Supplementary Figure 2C,D).
One may expect that the 5-bp indels are excluded from the core nucleosome sequences to a higher extent than the 10-bp indels since the 5-bp shift would disrupt the sequence patterns determining rotational phasing of nucleosomes, while the 10-bp shift would preserve them19–21. However, we do not observe such dependence in the ratio of indel occurrences (Supplementary Figure 2A,C). The absolute number of occurrences of indels longer than 5 bp is relatively small in our dataset, therefore we do not completely rule out the stronger exclusion of the 5-bp indels than that of the 10-bp ones. A more likely explanation for the lack of the noticeable difference, however, is a relatively low number of the sequences that encode the 10-bp signal in the human genome6.
2. Nucleosome positioning and genome variation density at splicing sites
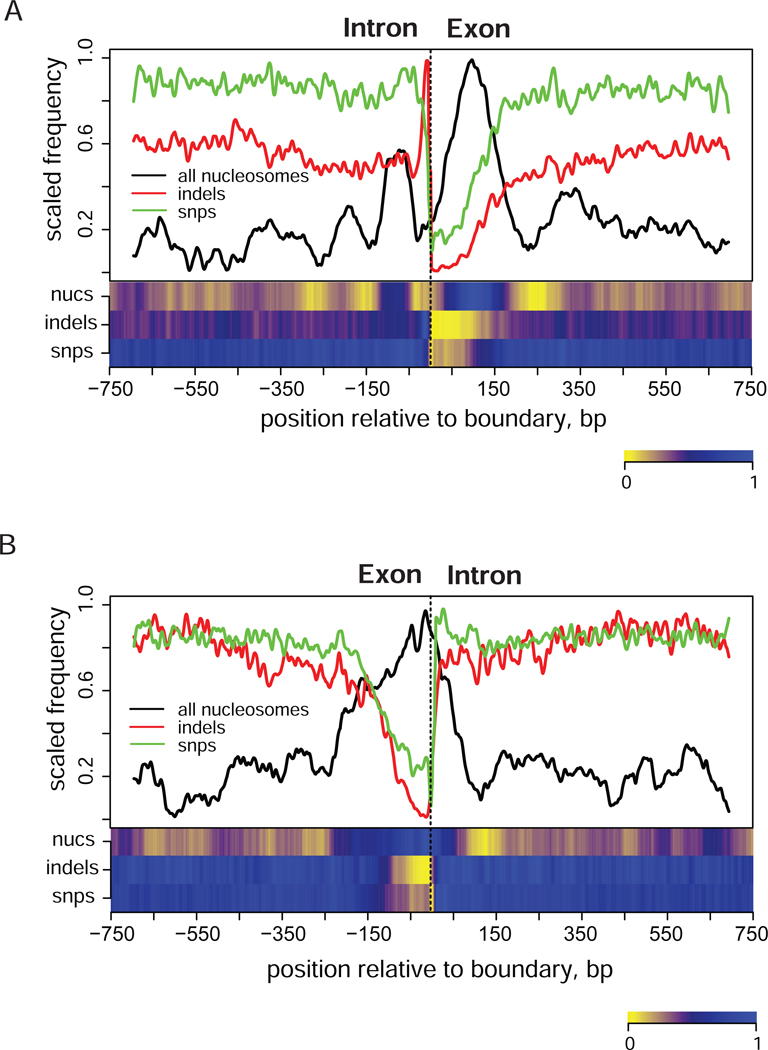
Intron-exon and exon-intron boundaries are among the mostly conserved genomic regions. Nucleosome positioning in these regions was recently studied22,23. Our analysis reveals that the nucleosome density is different at intron-exon and exon-intron junctions, while the pattern of genome variation is similar (Figure 3). We observe a pronounced stable nucleosome position at the exon-intron junction, while the intron-exon boundary seems to align with a trough in the nucleosome density, flanked by two positioned nucleosomes. This difference in the profile of stable nucleosome positions at the two boundaries is consistent with the distribution of nucleosome-disfavoring sequences, which has a stronger and wider peak at the intron-exon junctions than at the exon-intron junctions22.
Figure 3.
Distribution of indels (red), SNPs (green), and stable nucleosome positions from combined set (black) around intron-exon (A) and exon-intron (B) boundaries. Zero position in each plot corresponds to the position of boundary. Exonic coordinates were taken from the USCS track RefGene that reports known protein-coding genes from the NCBI mRNA sequences collection (RefSeq)34,35. First and last exons were excluded from the analysis. Only genes for which no alternative start site was reported we considered (14,946 genes). The combined nucleosome set (‘all nucleosomes’) was used to produce this plot. The frequency profiles were calculated as described in Methods. Heatmaps shown at the bottom panels represent de-trended profiles where large-scale variations were removed.
Distributions of SNPs and indels reach minima at both intron-exon and exon-intron junctions and feature wide trough on the exon side of the splice site. The appearance of the trough inside exons is consistent with their coding function and the presence of the conserved regulatory elements such as exonic splicing enhancers and silencers located in these regions24. Overall the distribution of genome variation around splice sites seems to be driven by the requirement on the conservation of splicing signals rather than by the nucleosomal patterns. Nevertheless, indels follow the trends observed on the genome scale (Figure 1) and are excluded from such positions at the splicing sites. On the other hand, the distribution of SNPs differs from that expected from the genome-wide analysis since we do not observe any increase in the SNP frequency at the nucleosome positions, even though more than half of the nucleosomes at the splicing sites are bulk. These observations suggest that strong selective pressure at specific genomic locations can overcome the traits in the genome variation profile imposed by nucleosome positioning.
3. Nucleosome positioning is coordinated with indel and SNP distribution at transcription start and end sites
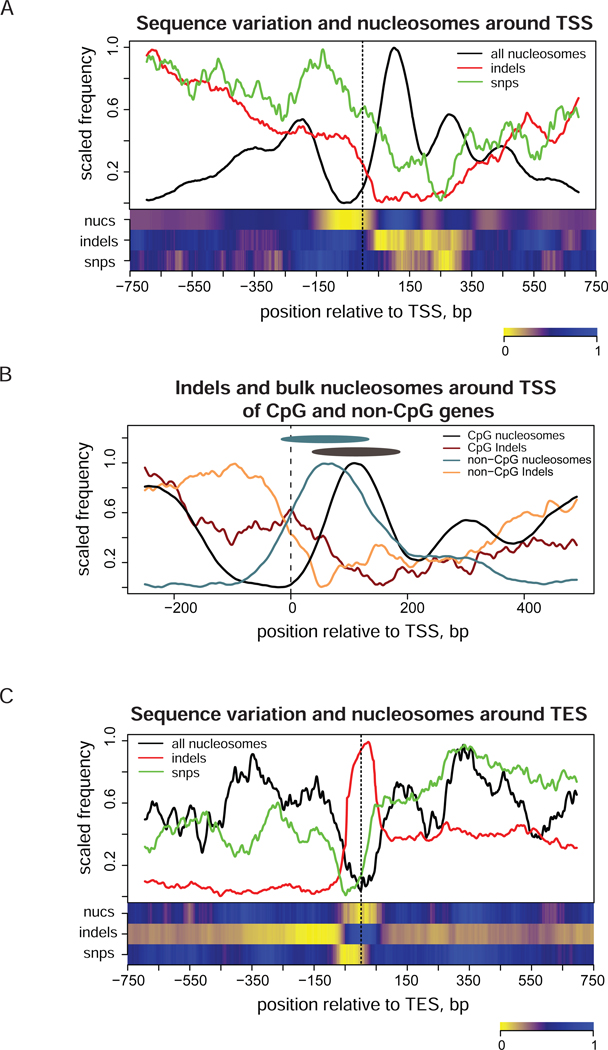
Comparison of the distributions of SNPs, indels, and stable nucleosome positions around transcription starts reveals two levels of coordination between genome variability and nucleosome positioning (Figure 4A). First, the overall increase in the nucleosome density around TSS is correlated with the decrease in density of both SNPs and indels in this region. It should be noted that the increase in the nucleosome density corresponds to stable positions only and may not represent the overall density of nucleosomes. Also, higher accessibility of open chromatin at TSS for nuclease digestion used to produce mono-nucleosome fragments for sequencing can contribute to the appearance of such an increase.
Figure 4.
Distribution of indels (red), SNPs (green), and stable nucleosome positions (black) around TSS and TES of human genes. Profiles were calculated as described in Methods. Heatmaps shown at the bottom panels represent de-trended profiles where large-scale variations were removed. (A) Profiles around TSS (position zero). The combined nucleosome set (‘all nucleosomes’) was used to produce this plot. Genes were oriented in the direction of transcription in such a way that the up-stream region is shown on the left and the downstream region is shown on the right of TSS. (B) Profiles shown separately for the frequencies of indels (dark red and orange lines) and bulk nucleosomes (black and cyan lines) for the subsets of genes associated and not associated with CpG islands at TSS. Black and cyan ovals represent nucleosomes at position +1 in CpG and non-CpG genes and are shown for a nucleosome size reference. Coordinates of CpG islands were taken from USCS genome browser annotation34. (C) Profiles computed around TES (position zero) for all genes. The combined nucleosome set was used.
Second, genome variation and nucleosome profiles are negatively correlated at the level of individual nucleosome positions, specifically at the nucleosome-free region and at the positions +1,+2, and +3 downstream of TSS. The Pearson correlations calculated inside the 1.5 kb region around TSS between genome variations and nucleosome occupancy indicate that both SNPs and indels are depleted at stable nucleosome positions at gene starts (Supplementary Table 1). Of the two, indels are anti-correlated with nucleosome occupancy to a higher degree than SNPs for all types of nucleosomes, in agreement with the results shown in Figure 1. Here, the profiles were de-trended before the calculation of correlation coefficients (see Methods for detail), and therefore our results are not influenced by the ‘overall’ coordination described above.
The exact location of the position +1 for bulk nucleosomes has been shown to depend on the transcription status of the gene12. The genes that are highly transcribed in a broad range of tissues often have their TSS encompassed by CpG islands25,26, implying that the transcription status of those genes is reflected in the underlying DNA sequences. In this context, it is interesting to compare the profiles of the sequence variation and nucleosome positioning around TSS for the CpG and non-CpG genes. We focus this analysis on bulk nucleosomes because most of the epigenetic nucleosomes considered in the current study are associated with the transcriptionally active genes and the nucleosome occupancy profiles are nearly identical around TSS of CpG and non-CpG genes for these nucleosomes6.
Since the number of stable nucleosome positions determined from the experimental data for bulk nucleosomes is not sufficient to obtain a reliable average profile around TSS for each gene group, we treat all sequenced tags as independent nucleosome fragments (Figures 3B). This approach allows increased statistical power to detect small changes in average profiles although it may reduce accuracy at the level of individual nucleosomes. This comparison shows that position +1 of bulk nucleosomes is shifted downstream in CpG genes as compared to non-CpG genes, as expected for the genes with increased expression level12. The minimum in the indel distribution for non-CpG nucleosomes aligns well with the bulk nucleosome position +1 for this group and the indel frequency minimum moves in the direction of the nucleosome position +1 for CpG genes (Figure 4B). This shift of the minimum in the indel profile indicates that the nucleosome positioning at TSS of CpG genes has evolved together with DNA sequence, presumably to accommodate high levels of transcription in a broad range of tissues26. The distribution of SNPs does not exhibit the same level of coordination with nucleosome occupancy at TSS of CpG and non-CpG genes as indel distribution does (Supplementary Figure 3), in accordance with the lower correlation between SNP and nucleosome density observed previously (Supplementary Table 1).
Around TES, indel density is negatively correlated with stable nucleosome positions, while SNP density is positively correlated (Figure 4C, Supplementary Table 1). We note that most nucleosomes at TES are bulk, and thus the positive correlation between SNPs and nucleosome density agrees with our finding that the SNP occurrence is higher on average inside the core sequences of the bulk nucleosomes (Figure 1B).
4. Different distribution of SNPs around bulk and epigenetic nucleosomes
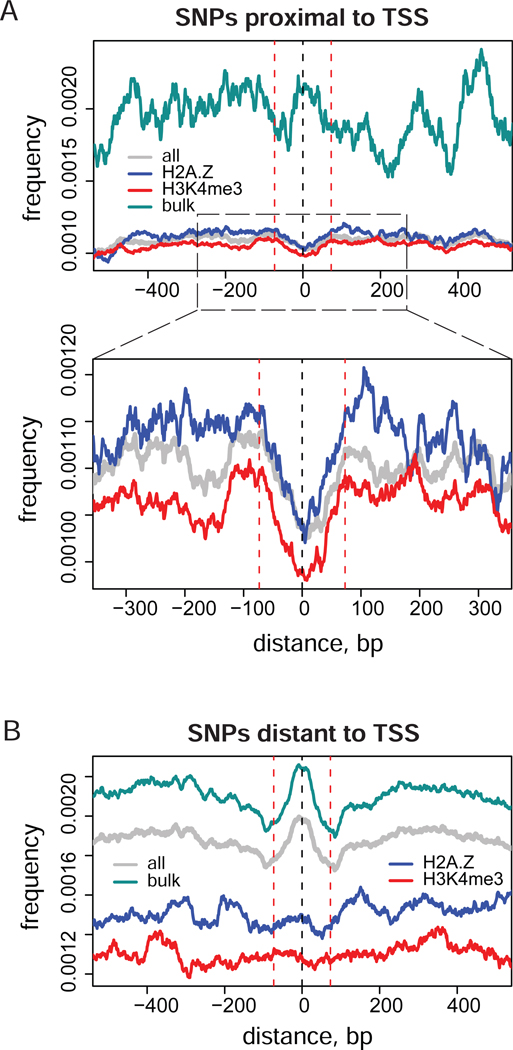
There are two possible explanations of the differences in SNP occurrence profiles around bulk and epigenetic nucleosomes observed in our analysis. One possibility is that the sequences associated with epigenetic nucleosomes of the types considered in this study are themselves conserved to a higher extent than the positions of ‘less important’ bulk nucleosomes. Another possibility for the lower frequency of SNPs detected for epigenetic nucleosome positions is simply the result of higher conservation of the TSS region, where most of such nucleosomes are located. To clarify this issue, we calculated the distributions of genome variations around nucleosome positions of each type in the regions that are proximal to and distant from TSS (Figure 5).
Figure 5.
Distribution of SNP frequencies around stable nucleosome positions in the regions that are proximal (A) and distant (B) to the TSS of human genes. TSS proximal and distant nucleosome positions were identified as those located less than 1 kb and more than 2 kb from the closest TSS respectively. Normalized profiles are shown for the positions from the combined nucleosome set (grey) and for the individual nucleosome sets: bulk (cyan), H2A.Z (blue), and H3K4me3 (red). Vertical dashed lines at zero and ±73 bp give reference of the nucleosome position and size.
We observed a clear decrease in SNP density for the epigenetic positions proximal to TSS (Figure 5A). Even far from TSS, the epigenetic positions are not associated with an increase in SNP rate, which is characteristic of bulk nucleosomes (Figure 5B). In the TSS proximal regions, most of the stable nucleosome positions in our set are epigenetic and only a small number of bulk positions are available. Despite the resulting jaggedness of the SNP density profile for bulk nucleosomes, it is obvious that there is no clear dip at TSS proximal regions, consistent with the first explanation given above.
Discussion
Our results indicate that while indels are depleted on average in all types of nucleosomes at TSS, TES, and genome-wide, the density of SNPs exhibits a more intricate behavior. In particular, SNPs are negatively correlated with nucleosome occupancy at TSS and positively correlated with nucleosome occupancy at TES (Figure 4 and Supplementary Table 1). In agreement with this observation, the density of SNPs is increased in the core sequences associated with bulk but not epigenetic nucleosomes, which constitute a majority of the nucleosomes in our set at TES and TSS respectively (Figure 1). The positive correlation between SNP density and nucleosome occupancy was reported earlier for the fish and yeast genomes7,10. We note that the nucleosome positions used in earlier studies correspond to bulk positions in our notation; thus, the results between the previous studies and ours are consistent. However, in the current paper we show that this rule does not hold in a number of important cases in the human genome. At least some classes of epigenetic nucleosomes are associated with a decrease rather than increase in the SNP density (Figure 5). We also report that SNPs may be negatively correlated with the nucleosome density in the DNA regions that are under strong selective pressure, such as exon-intron boundaries (Figure 3). Thus, our findings highlight the complexities in the interplay between the mechanisms that control SNP appearance and nucleosome positioning in humans.
It is interesting to consider why nucleosomal sequences in bulk are strongly depleted of one type of mutations, indels, while they are either only moderately depleted or even enriched in another type of mutations, SNPs. In general, two mechanisms are potentially responsible for the difference in the density of genome variations inside and outside nucleosomes27. One is connected to the alteration of the mutation rate in nucleosomal DNA, e.g. due to physical interaction the nucleosomal DNA with histones7,10,28. Another assumes that the DNA sequences that contain nucleosome positioning signals and/or binding sites of transcription factors are evolutionarily conserved to a higher extent than the adjacent DNA fragments29. These mechanisms are not mutually exclusive and can both contribute as discussed below.
Our observation of roughly the same frequency of indels inside nucleosomes of different types (Figure 1) suggests alteration of mutation rate rather than action of purifying selection for indels. Indeed, our results provide little support for the hypothesis that the selection pressure excludes indels from nucleosomes. We did not detect a dependence of the nucleosome-to-linker ratio of the indel occurrences on indel length (Supplementary Figure 2), which would be suggestive of this mechanism. Overall, our results indicate that the stable nucleosome positions are reflected in the indel frequency profile regardless of the local base composition or details of regulatory pathways in which a specific DNA locus is involved. This is illustrated by a shift of the nucleosome position +1 at starts of the CpG genes relative to the corresponding position at starts of the non-CpG genes in the indel frequency profile (Figure 4B). The sequence composition of the TSS proximal regions of CpG and non-CpG genes is quite different and CpG genes are actively transcribed in a broader range of cell types than the non-CpG genes26, yet the nucleosome position +1 is reflected in the indel frequency profile in each of these groups.
On the other hand, the density of SNPs appears to be affected by natural selection. A single nucleotide mutation can disrupt a transcription factor binding site to interfere with regulatory pathways. It is less likely that such a mutation would significantly alter the positioning properties of a 147-bp sequence associated with a nucleosome. Furthermore, even if a mutation changes the position of a bulk nucleosome by several base pairs, this may not have any biological effect. As a result, mutations would be tolerated in the core sequence of bulk nucleosomes but would be excluded from the linkers where many transcription factors bind3,30,31. In contrast, correct placement of epigenetically modified nucleosomes is important for gene regulation, and the positions preferentially occupied by these nucleosomes are likely to be conserved to the same or greater extent compared to the linker sequences. It should be emphasized that our results do not imply a complete absence of selective pressure on the bulk nucleosome sequences but rather that the pressure is stronger in linkers than in the nucleosomes of this type. Neither do we suggest that the SNP occurrence rate is not changed in nucleosome core sequences at all. Rather, our results provide information about the mechanisms contributing the most to the observed features of sequence variation profiles.
The interpretation of a stronger conservation of epigenetic nucleosome positions, rather than the difference in mutation rates in the bulk and epigenetic nucleosomes is further supported by two lines of evidence. The fraction of SNPs rarely occurring in population, in particular those associated with only one genome in our data set, is higher for the epigenetic than for bulk nucleosomes (Supplementary Figure 4). This indicates a stronger selection against SNPs from the epigenetic nucleosomes. As discussed above, we also observe a clear drop in SNP density at the nucleosome positions coinciding with exon-intron boundaries (Figure 3), which is likely to result from the strong selection pressure acting on the splicing sites. Since the greater part of the nucleosomes proximal to exon-intron junctions are bulk, the anti-correlation of SNP frequency with nucleosome occupancy argues against the idea that the presence of nucleosomes of this type necessarily increases the SNP accumulation rate.
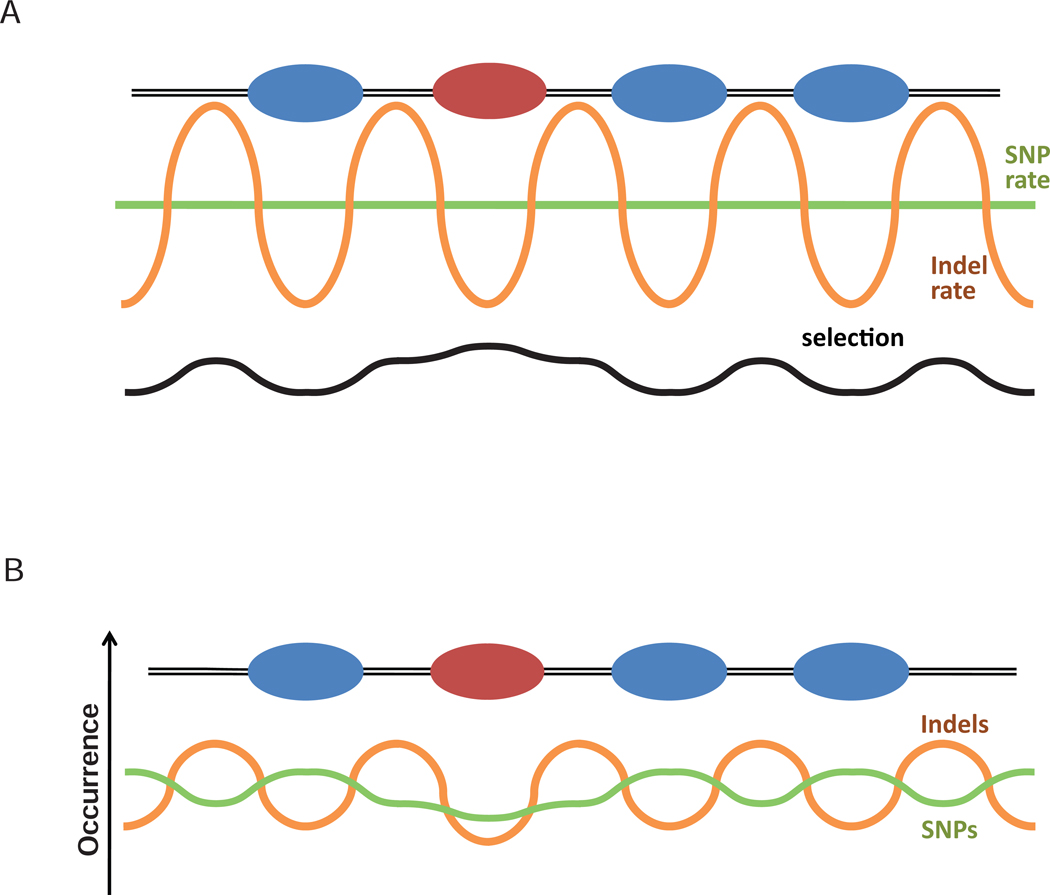
Taken together, our results suggest that a combination of purifying selection acting on biologically important sequences and the alteration of the mutation rate in nucleosomal DNA determine the pattern of sequence variation in the human genome (Figure 6). Further studies are required, however, to unambiguously prove or disprove the involvement of the above mechanisms in the evolution of nucleosome positioning sequences in the human genome. In particular, characterization of molecular mechanisms that can underlie chromatin-directed mutational bias will undoubtedly advance our understanding of the principles of genome evolution.
Figure 6.
Interplay of chromatin-mediated mutation bias and selection can shape sequence variation profile (cf. to schematic illustration in Ref. 27). (A) Bulk and epigenetically modified nucleosomes are represented with blue and red ovals. Green and orange lines represent mutation rate of SNPs and indels respectively, and black line represents selection pressure acting on the DNA sequence. (B) The significant difference in the indel rate inside and outside nucleosomes mainly determines the indel density profile observed in the genome (orange), while SNP density profile (green) is mainly affected by selection. Our results do not exclude the possibility that natural selection can affect the distribution of indels and that alteration of the mutation rate affects the distribution of SNPs. Rather, they indicate that these mechanisms are not the major factors shaping the resulting profiles.
Methods
Data
We identified SNPs and indels by comparing trace sequences with the sequence of the reference human genome (NCBI version 36.2). The trace data from the human libraries produced in 8 different sequencing centers (Agencourt Biosciences (ABC), Baylor College of Medicine (BCM), Celera (CRA), Cold Spring Harbor Laboratory –Watson Genome (CSHL), J. Craig Venter Institute (JCVI), Santa Cruz Genome Center (SC), Whitehead Center for Biomedical Research (WIBR), Washington University Sequencing Center (WUGSC) – referred to as sources) were downloaded from the Trace Archive (NCBI). The traces were mapped to the genomic reference DNA using GMAP32 software and the high score alignments were detected by the previously described procedure33. The GMAP alignments were parsed using the following parameters (i) distance of the reported variation from the end of the alignment – more than 20 bp; (ii) perfect alignment of 5 bp of flanking regions on both sides of the variations. All SNPs and indels of lengths from 1 bp to 100 bp were taken for analysis. The repeats content of the indels/SNPs loci was analyzed by comparing variations positions with the RepeatMasker annotation of Human genome. The indels that have lengths of more than 5 bp and contain mono- and dinucleotide repeats were filtered out from the final set. All variations were reported on the positive strand, so each chromosomal position represents a separate event of specific length, type (SNP, insertion or deletion) and allele. The events corresponding to SNPs and indels were clustered separately by the 5'-end for each source and for all sources together. The final data set includes 907,324 indel and 4,068,654 SNP events that were supported by at least 3 traces covering the variation from at least 2 sources (Supplementary Table 2). The histogram showing the frequency of SNP/indel events relative to the reference sequence is shown in Supplementary Figure 5A,B. The distribution of the indel lengths is shown in Supplementary Figure 5C.
We also used a recently published set14 of indels and SNPs based on analysis of 8 human genomes for comparison and validation of the results (the results of the analysis obtained for this dataset, shown in Supplementary Figures 6–8, support our findings described above). The genomic positions of the sequence variation events in the ‘8-genome’ set originally are presented in the coordinates that correspond to the human genome build hg17. The coordinates were converted to the hg18 coordinate frame with UCSC utility liftOver. Both data sets were generated by the analyses of the sequence alignments of the trace sequences to the reference Human genome. We found that the overlaps between the genome loci from both data sets constitute ~50–60% (Supplementary Figure 9). The difference between the sets is explained by the differences in the original traces data used for variation analysis and the definition of the indels/SNPs calling parameters. The first data set was produced based on the libraries from 8 centers including the ABC, which was the only source of the second data set (list of all libraries is given in Supplementary Table 3). The distinction in the indel/SNP calling procedure includes different alignment tools (GMAP and ssahaSNP) applied in the analysis and different classification of the alignments (see Supplementary Material for detail).
Stable positions for nucleosomes bearing H3K4me3 mark (28,976 positions), H2A.Z variant (17,667 positions), and bulk nucleosomes not selected for a specific epigenetic mark or histone variant (27,486 positions) were taken from a recent analysis of ChIP-Seq and MNase-Seq data6. In addition we composed an aggregative set of nucleosome positions that comprises all three individual sets. In the cases of two or more positions located closer than 150 bp to each other only the position that is associated with the largest number of sequenced tags was retained. The final set included 63,554 nucleosome positions.
The autocorrelations in the sequence variation positioning were computed for different lag distances for each chromosome separately and then averaged genome-wide accounting for chromosome sizes, similar to our previous analysis of nucleosome positions6. The frequency profiles of genome variations around stable nucleosome positions represent the indel and SNP occurrences normalized to the number of nucleosome positions in the corresponding set and smoothed in the 75-bp running window. The frequency profiles of the nucleosome positions and sequence variations around TSS, TES, and splicing sites were normalized to the number of genes or exons in the corresponding sets. The genes were oriented in the direction of transcription prior to averaging. Smoothing in the 100-bp running window was used for TSS, TES profiles and for nucleosome frequency in the splicing site profiles. The smaller running window of 11-bp was used in case of the genome variation profiles around splicing sites to allow for a better resolution in this case. The profiles were scaled to the interval from zero to one for easier comparison. Additional loess smoothing in 11-bp window, which does not affect positions of the major minima and maxima on the plots, was applied to reduce the jaggedness in the TSS, TES, and splicing site profiles. For the calculation of Pearson correlations between nucleosome and sequence variation frequencies and creating heatmaps, the profiles were de-trended by subtracting the same profile smoothed in the 750-bp running window.
Supplementary Material
References
- 1.Jiang C, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet. 2009;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schones DE, Zhao K. Genome-wide approaches to studying chromatin modifications. Nat Rev Genet. 2008;9:179–191. doi: 10.1038/nrg2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Segal E, Widom J. From DNA sequence to transcriptional behaviour: a quantitative approach. Nat Rev Genet. 2009;10:443–456. doi: 10.1038/nrg2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaplan N, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y, et al. Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo. Nat Struct Mol Biol. 2009;16:847–852. doi: 10.1038/nsmb.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tolstorukov MY, Kharchenko PV, Goldman JA, Kingston RE, Park PJ. Comparative analysis of H2A.Z nucleosome organization in the human and yeast genomes. Genome Res. 2009;19:967–977. doi: 10.1101/gr.084830.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Washietl S, Machne R, Goldman N. Evolutionary footprints of nucleosome positions in yeast. Trends Genet. 2008;24:583–587. doi: 10.1016/j.tig.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 8.Higasa K, Hayashi K. Periodicity of SNP distribution around transcription start sites. BMC Genomics. 2006;7:66. doi: 10.1186/1471-2164-7-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prendergast JG, et al. Chromatin structure and evolution in the human genome. BMC Evol Biol. 2007;7:72. doi: 10.1186/1471-2148-7-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sasaki S, et al. Chromatin-associated periodicity in genetic variation downstream of transcriptional start sites. Science. 2009;323:401–404. doi: 10.1126/science.1163183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 12.Schones DE, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin C, et al. H3.3/H2A.Z double variant-containing nucleosomes mark 'nucleosome-free regions' of active promoters and other regulatory regions. Nat Genet. 2009;41:941–945. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kidd JM, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lohr D, Corden J, Tatchell K, Kovacic RT, Van Holde KE. Comparative subunit structure of HeLa, yeast, and chicken erythrocyte chromatin. Proc Natl Acad Sci U S A. 1977;74:79–83. doi: 10.1073/pnas.74.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peckham HE, et al. Nucleosome positioning signals in genomic DNA. Genome Res. 2007;17:1170–1177. doi: 10.1101/gr.6101007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kharchenko PV, Woo CJ, Tolstorukov MY, Kingston RE, Park PJ. Nucleosome positioning in human HOX gene clusters. Genome Res. 2008;18:1554–1561. doi: 10.1101/gr.075952.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vinces MD, Legendre M, Caldara M, Hagihara M, Verstrepen KJ. Unstable tandem repeats in promoters confer transcriptional evolvability. Science. 2009;324:1213–1216. doi: 10.1126/science.1170097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trifonov EN, Sussman JL. The pitch of chromatin DNA is reflected in its nucleotide sequence. Proc Natl Acad Sci U S A. 1980;77:3816–3820. doi: 10.1073/pnas.77.7.3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Satchwell SC, Drew HR, Travers AA. Sequence periodicities in chicken nucleosome core DNA. J Mol Biol. 1986;191:659–675. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- 21.Segal E, et al. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwartz S, Meshorer E, Ast G. Chromatin organization marks exon-intron structure. Nat Struct Mol Biol. 2009;16:990–995. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- 23.Tilgner H, et al. Nucleosome positioning as a determinant of exon recognition. Nat Struct Mol Biol. 2009;16:996–1001. doi: 10.1038/nsmb.1658. [DOI] [PubMed] [Google Scholar]
- 24.Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. Rna. 2008;14:802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 26.Zhu J, He F, Hu S, Yu J. On the nature of human housekeeping genes. Trends Genet. 2008;24:481–484. doi: 10.1016/j.tig.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 27.Semple CA, Taylor MS. Molecular biology. The structure of change. Science. 2009;323:347–348. doi: 10.1126/science.1169408. [DOI] [PubMed] [Google Scholar]
- 28.Kogan S, Trifonov EN. Gene splice sites correlate with nucleosome positions. Gene. 2005;352:57–62. doi: 10.1016/j.gene.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 29.Warnecke T, Batada NN, Hurst LD. The impact of the nucleosome code on protein-coding sequence evolution in yeast. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000250. e1000250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Albert I, et al. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- 31.Lee W, et al. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 32.Akagi K, Li J, Stephens RM, Volfovsky N, Symer DE. Extensive variation between inbred mouse strains due to endogenous L1 retrotransposition. Genome Res. 2008;18:869–880. doi: 10.1101/gr.075770.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Volfovsky N, et al. Genome and gene alterations by insertions and deletions in the evolution of human and chimpanzee chromosome 22. BMC Genomics. 2009;10:51. doi: 10.1186/1471-2164-10-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuhn RM, et al. The UCSC Genome Browser Database: update 2009. Nucleic Acids Res. 2009;37:D755–D761. doi: 10.1093/nar/gkn875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.