Abstract
The N-hydroxylating flavoprotein monooxygenases are siderophore biosynthetic enzymes that catalyze the hydroxylation of the sidechain amino-group of ornithine or lysine or the primary amino-group of putrescine. This hydroxylated product is subsequently formylated or acylated and incorporated into the siderophore. Importantly, the modified amino-group is a hydroxamate and serves as an iron chelating moiety in the siderophore. This review describes recent work to characterize the ornithine hydroxylases from Pseudomonas aeruginosa (PvdA) and Aspergillus fumigatus (SidA) and the lysine hydroxylase from Escherichia coli (IucD). This includes summaries of steady and transient state kinetic data for all three enzymes and the X-ray crystallographic structure of PvdA.
Keywords: Ornithine, lysine, putrescine, hydroxylase, monooxygenase, N-hydroxylating, flavoprotein, siderophore, hydroxamate, PvdA, SidA, IucD
1. Introduction
Since iron serves as a cofactor in enzymes that catalyze essential biological reactions, bacteria, fungi and plants have developed several mechanisms for scavenging iron from their environments [1, 2]. One such mechanism is the production and secretion of low molecular weight iron chelators called siderophores, followed by the import of the iron-loaded form [3, 4]. Some species of bacteria and fungi generate hydroxamate siderophores, which are associated with pathogen virulence [5–7] and are derived from amino acids and other metabolic intermediates [8–10], with the iron chelating moiety derived from a hydroxylated and formylated or acylated lysine, ornithine or putrescine. The enzymes that hydroxylate the primary amino-group of the sidechain belong a group of functionally-related enzymes called the N-hydroxylating flavoprotein monooxygenases (NMO). NMOs are part of the class B flavoprotein monooxygenases, which also contains the flavin-containing monooxygneases (FMOs) and Baeyer Villiger monooxygenase (BVMOs) [11]. The focus of this review is on the recent advances in the study of the NMO enzymes.
2. Study of NMOs hampered by the inability to produce soluble enzyme for experimentation
The most studied of the N-hydroxylating flavoprotein monooxygenase proteins are the ornithine hydroxylases from Pseudomonas aeruginosa (PvdA) and Aspergillus fumigatus (SidA) and the lysine hydroxylase from Escherichia coli (IucD). All three have been produced as soluble, recombinant proteins and purified for steady state and/or transient state analyses. PvdA has been produced with an N-terminal histidine tag [12–14]. Other researchers had difficulty producing a soluble form of PvdA; however, this may be due to problems with mutations introduced by cloning. The original sequence reported for PvdA [15] is inconsistent with that from the Pseudomonas aeruginosa PAO1 sequencing project [16]. The inconsistent sequence is due to a deletion of a single adenine nucleotide at position 1242 that resulted in a frameshift at amino acid 385. The resulting protein has 41 incorrect C-terminal amino acids followed by a premature stop (426 amino acids total, not 443). This construct appears to have been used to promote the idea that PvdA is a membrane associated protein in later work (clones producing full-length and deletions of a 426 amino acid protein), although this is unclear as the sequence alignment published in the same article uses the correct sequence (443 amino acids) [17]. The production of SidA, the fungal ornithine hydroxylase, also met with solubility problems. SidA has been produced with an N-terminal histidine tag [18] or as an maltose binding protein fusion [19].
Several truncation mutants were generated of IucD, a bacterial lysine hydroxylase, to produce a soluble form of this enzyme [7]. The final result was a form of the enzyme with the first 48 amino acids replaced with an N-terminal segment of β-galactosidase. These data were also used to substantiate the idea that IucD is generally membrane associated [20], most likely through the N-terminus [21]. However, active enzyme was purified from E. coli strain EN222 (not recombinant protein) [22] and also with an N-terminal histidine tag [23]. As we will see in the discussion of the structure of PvdA later, the IucD-βGal fusion removes a significant portion of the FAD binding motif, and therefore will not be used herein for discussion of mechanism.
3. Oligomerization and FAD binding of purified enzymes
Two interesting differences are noted upon purification of these three enzymes. First, they are reported to have differing oligomeric states: PvdA is observed to be monomeric in solution [14], SidA is reported to be dimeric [18] or tetrameric [19], and the βGal-IucD fusion is active as a tetramer at high salt but the removal of salt led to an inactive monomer [24]. Finally, PvdA [12, 14] and recombinant IucD [24, 25] purified without flavin and do not bind flavin tightly, whereas ~60% of SidA has FAD bound following purification [18, 19].
4. Steady state analyses of PvdA, SidA and IucD
Steady state analyses of PvdA, SidA and IucD have produced results that are consistent among the three. The enzymes are FAD-dependent: FMN was tested for PvdA and found to be insufficient as a flavin co-factor [12, 14], and IucD purified from E. coli strain EN222 (not recombinant protein) had FAD bound [22]. PvdA [12, 14] and SidA [19] are specific for NADPH. NADH does not serve as an efficient electron donor. PvdA [12, 14], SidA [19] and IucD [25] were found to be highly specific for their substrates. In terms of hydroxylation, PvdA [14] and SidA [19] are specific for ornithine, whereas lysine is a non-substrate effector that promotes NADPH oxidase activity without subsequent N-hydroxylation. Putrescine, among other analogues, shows no activity by either the NADPH oxidation or N-hydroxylation assay [14]. IucD is specific for lysine, showed no catalytic activity for ornithine, and homolysine was a non-substrate effector [25]. Steady state experiments also provided the first mechanistic information for these proteins. First, FAD reduction is substrate independent (PvdA [14], SidA [19]). Second, a long lived flavin-intermediate is observed without substrate (PvdA [14] and SidA [19]). Finally, NADP+ remains bound throughout the oxidative half reaction (PvdA [26] and SidA [18, 19]).
5. Kinetic mechanism as determined by transient kinetic analyses of PvdA and SidA
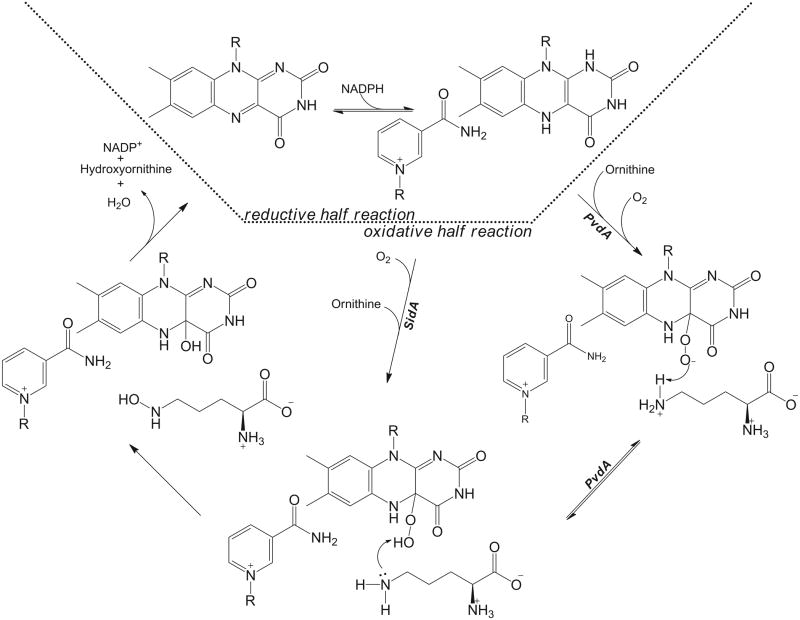
Transient kinetic analyses of PvdA [14] and SidA [18] have provided similar reaction mechanisms for the two enzymes with only one difference, and can be considered the basis for all of this class (Figure 1). Substrate is not required for the reduction of flavin by NADPH, nor does it accelerate the process [13, 18]. The reduction of the flavin is observed as a decrease in absorbance at 450 nm [13, 18]. The biphasic nature of the reduction curves has been attributed to reduction and NADP+ release [13] or to binding and reduction [18], with the latter correct in light of the more recent observations that NADP+ remains bound throughout the oxidative half reaction as cited in the previous section. After NADPH binds and reduces the flavin, oxygen binds and adds to the C4a-position of the flavin forming a long-lived flavin intermediate. For PvdA, this intermediate is hypothesized to be a peroxyflavin (absorbs at ~360), which is converted to a hydroperoxyflavin (~380 nm) by donation of a proton from the substrate amino-group [13]. The evidence for this comes from very early timepoints at which a spectral shift from 361 to 376 nm is observed [13], as seen previously for cyclohexanone monooxygenase [27]. For SidA, the long-lived intermediate is proposed to be the hydroperoxyflavin (~380 nm) and stabilized by the presence of NADP+ with no initial peroxyflavin intermediate observed [18]. The rate of the formation of the intermediate is enhanced by the addition of substrate, 80-fold for PvdA [13] and 5-fold for SidA [18]. The hydroperoxyflavin is the reactive intermediate, which donates the distal oxygen to the substrate thereby forming hydroxyornithine and the hydroxyflavin intermediate (increase in absorbance at both 390 nm [13, 18]). The hydroxyflavin intermediate dehydrates to regenerate the oxidized flavin (decrease in absorbance at 390 nm [13] and increase in 450 nm [13, 18]). The hydroxyornithine and NADP+ dissociate and the cycle continues.
Figure 1.
Kinetic mechanism for N-hydroxylating flavoprotein monooxygenases based on data for PvdA [13, 14, 26] and SidA [18, 19].
6. Structure determination of PvdA
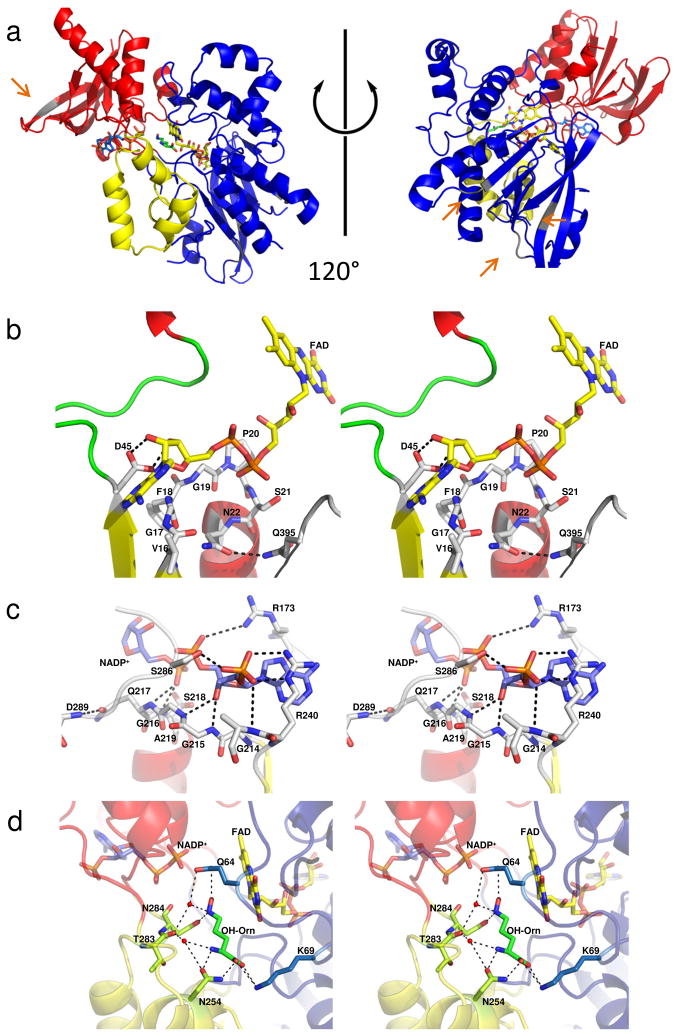
The first structure of an N-hydroxylating flavoprotein monooxygenase was determined by seleno-methionine multiwavelength anomalous dispersion to 1.9 Å [26]. The structure revealed a three domain enzyme with an FAD binding domain comprising residues 1 – 171, 356 – 396 and 405 – 443 (blue in Figure 2A). The NADPH binding domain is inserted into a loop of the FAD binding domain and includes residues 170 – 245 and 285 – 355 (red in Figure 2A). Both of these domains are characteristic /β Rossman-like nucleotide binding folds. The third domain is a substrate binding domain composed of residues 248–285 and 398 – 404 (yellow in Figure 2A). This smallest domain is helical in nature. This structure, along with the 3.03 Å reduced structure published in the same article [26], have FAD(H2), NADP(H) and (hydroxy)ornithine bound in the active site (sticks in Figure 2).
Figure 2.
Structure of the ornithine hydroxylase from Pseudomonas aeruginosa, PvdA. A) Cartoon of the oxidized structure of PvdA (PDB code 3S5W) [26]. The FAD binding domain is shown in blue, the NADPH binding domain in red and the ornithine binding domain in yellow. The FAD is yellow sticks, the NADP+ is blue sticks, and the ornithine is green sticks. The grey elements of secondary structure and loops regions represent areas of sequence insertion for SidA and are highlighted with orange arrows. B) Stereo image of the FAD binding site and C) Stereo image of the NADP+ binding site. In parts B and C, the cartoon is colored by secondary structure with α-helices red, β-strands yellow and loop regions green. D) Stereo image of the ornithine binding site. Here the cartoon is colored as in part A. Hydrogen bonding and salt links are shown as dashed lines.
6.1. FAD binding
Considerable speculation on the structural composition of this class of enzymes, especially related to substrate and coenzyme binding motifs, predated the completion of the PvdA structure. Sequence analysis led to the proposal that the FAD binding motif was GXGXXP (residues 15 – 20 in PvdA, Figure 3) [28]. Due to the previously mentioned difficulty in expression both PvdA and IucD in a soluble form, it was hypothesized that the N-terminal region of these proteins constituted a membrane binding region, but the overlap with the FAD binding sequence led to some debate about the assignment of substrate, co-substrate and FAD binding motifs [21, 29]. Admittedly, by hydropathy plot the region (residues 10–30) is quite hydrophobic and appears to be transmembrane, leading the authors to suggest that the “N-terminal hydrophobic domain interacts with the lipid bilayer by forming a U-shaped or re-entrant loop aided by contiguous G19–P20 residues without actually crossing the membrane” [30]. The structure of PvdA [26] allows for the assignment of the FAD binding motif as two amino acids C-terminal to that proposed: 17GXGXXN22 (Figure 2B). Furthermore, the structure clearly shows that the residues of the putative membrane binding motif form the first strand and helix of the classical βαβ dinucleotide binding motif involved in binding of the ribose and phosphates of the FAD [26].
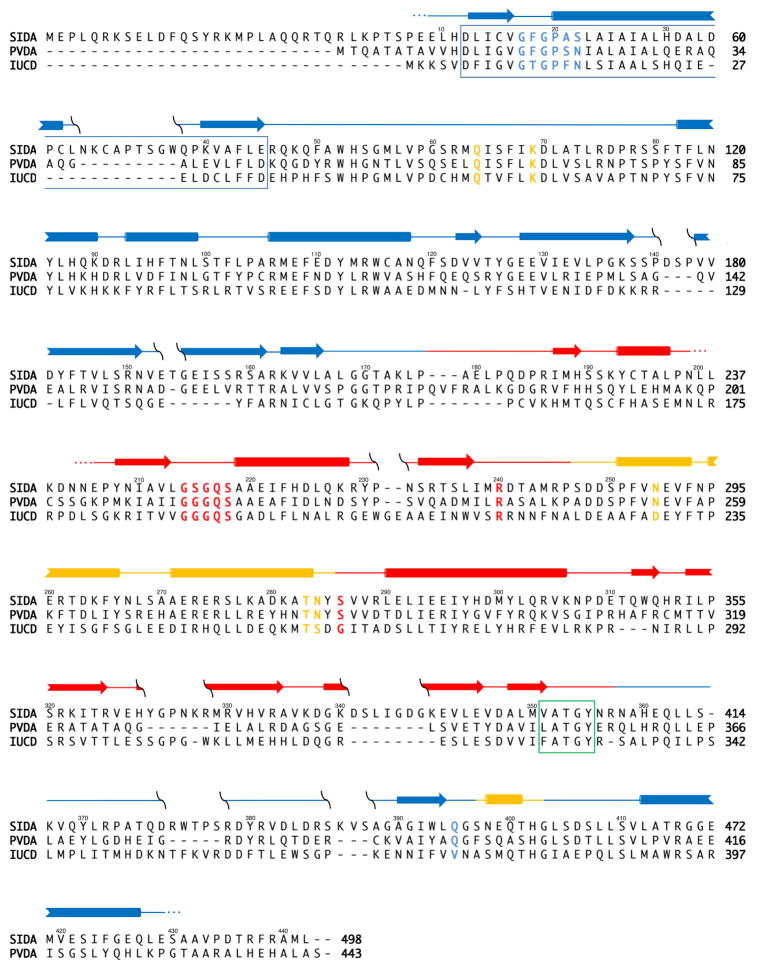
Figure 3.
Sequence alignment for SidA, PvdA, and IucD. Secondary structure elements of PvdA shown above the sequences as arrows (β-strands) and cylinders (α-helices) [26]. Blue elements are found in the FAD binding domain, red in the NADPH binding domain and yellow in the ornithine binding domain. Residues colored blue are involved in FAD binding, red in NADPH binding and yellow in ornithine binding. The blue box represents the region of the protein previously predicted to be both involved in FAD and membrane binding [21, 30]. The green box highlights the ‘FATGY’ sequence hypothesized to be involved in substrate binding [21]. The numbers above the sequences correspond to the amino acid numbers for PvdA. Primary sequence alignments indicate an overall identity of ~15% and 46% similarity among the three enzymes. There is a 40% identity (73% similarity) between PvdA and SidA, the two ornithine hydroxylases [44]. Each ornithine hydroxylase shares similar percentages when compared to IucD, the lysine hydroxylase, with 25% identity (59% similarity) between PvdA and IucD, and 20% identity (53% similarity) between IucD and SidA.
As noted previously, PvdA and IucD bind FAD weakly, whereas SidA can be purified with FAD bound. Comparing the structure of PvdA to the available structures for flavin-containing monooxygenases and Baeyer Villiger monooxygeanses led to the proposal that PvdA may not bind FAD as tightly because the FAD binding cleft is much shallower in PvdA [26]. Indeed, most of the FAD is solvent exposed in PvdA, whereas in FMOs and BVMOs, the FAD is buried and solvent inaccessible [31–35]. An exception is phenylacetone monooxygenase, a BVMO that also does not bind FAD stably, and indeed has the adenine moiety of the FAD exposed to solvent [36]. Based on the sequence alignment in Figure 3, we propose that IucD, which is smaller than PvdA by 46 amino acids, will have a similarly open FAD binding site. On the other hand, SidA is 45 residues longer than PvdA with insertions in loops and strands. An initial hypothesis is that the additional residues are at least partially responsible for shielding the FAD from competing solvent interactions when bound to SidA, promoting more stable FAD binding and less uncoupling of NADPH oxidiation and product formation. However, this hypothesis seems unlikely since the insertions are not in close proximity to the FAD (grey sections of structure highlighted by orange arrows in Figure 2A).
6.2. NADPH binding
NADPH binding has been hypothesized to be a function of the consensus sequence GGG(Q/N)S(G/A) [28]. Indeed, in PvdA, these residues (214–219) form a turn between the first strand and helix of the dinucleotide binding βαβ motif analogous to the GXGXXN for FAD binding (Figure 2C) [26]. The glutamine hydrogen bonds with the backbone of Asp289 thereby stabilizing the loop conformation, and the serine hydrogen bonds with the phosphate of the NADPH molecule. The co-substrate specificity (NADPH instead of NADH) is conferred by two residues: Arg240 and Ser286. These two residues form hydrogen bonds to the phosphate of the adenine ribose of the NADPH, which is not present in NADH. The arginine is conserved in both SidA (Arg276) and IucD (Arg216), whereas the serine is only conserved in SidA (Ser322 in SidA, Gly262 in IucD; Figure 3).
6.3. Substrate binding
Sequence alignment of the N-hydroxylating monooxygenases led to the proposal that the ‘FATGY’ motif (residues 351–355 in PvdA) is involved in substrate binding [28]. The structure of PvdA reveals this sequence to be located in a loop which forms part of the NADPH binding pocket [26]. The substrate binding domain is instead composed of a three helix insertion within the NADPH binding domain and one helix derived from the C-terminal portion of the FAD binding domain (Figure 2A). The ornithine is stabilized in the structure by five residues: Gln64, Lys69, Asn254, Thr283 and Asn284 (Figure 2C). Whereas Gln64 and Asn284 align the N5-amino-group for hydroxylation, Lys69, Asn254, and the backbone carbonyl of Thr283 stabilize the ornithine backbone amine and carboxylate, conferring substrate and stereochemical specificity. As seen in Figure 3, these residues are conserved in both SidA and IucD, with the exception of Asn284 (Ser260 in IucD) and Asn254 (Asp230 in IucD), which are conservative changes that would provide similar hydrogen binding capabilities. In the case of Ser260 of IucD, this might be an artifact of the sequence alignment, and the corresponding residue at that position may be Asp261.
7. Other NMOs identified to date
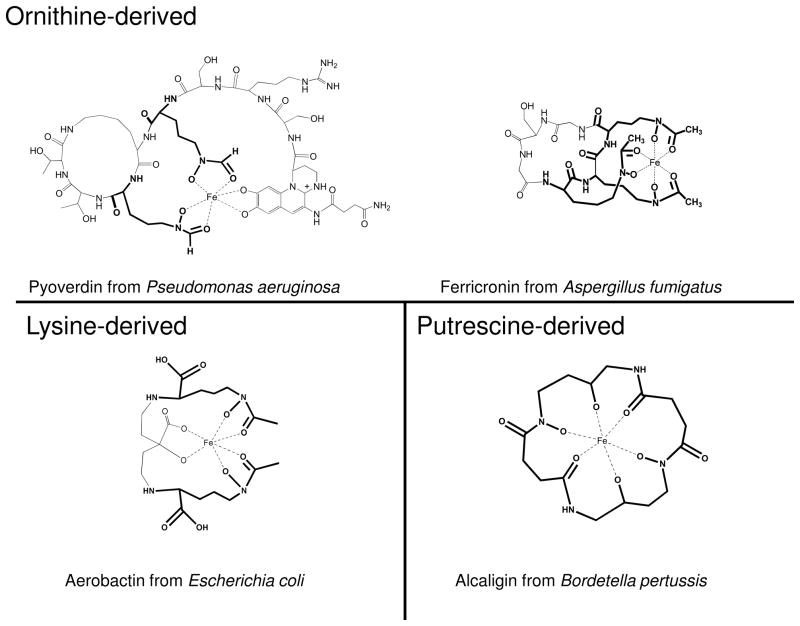
The siderophores into which the products of catalysis are incorporated are shown for PvdA (pyoverdin), SidA (ferricronin) and IucD (aerobactin) in Figure 4. At least nine other ornithine hydroxylases have been identified, all involved in siderophore production (Table 1). Along with IucD, at least one other lysine hydroxylase has been identified, with mycobactin assembly being unusual. The accepted norm is that the ornithine or lysine sidechain is hydroxylated as a first step in biosynthesis of the siderophore, followed by formyl- or acylation and finally incorporation into the siderophore [15, 37–39]. The MtbG enzyme from Mycobacterium tuberculosis appears to be a tailoring enzyme, hydroxylating the previously incorporated and acylated sidechain amino-group as a last step in siderophore biosynthesis [40, 41]. Finally, a putrescine (or 1,4-diaminobutane) hydroxylase has been identified from Bordetella sp siderophore production [42, 43].
Figure 4.
Examples of hydroxamate siderophores that include products of the N-hydroxylating flavoprotein monooxygenases. Pyoverdin is the siderophore produced with the assistance of PvdA. Ferrocronin production involves SidA, aerobactin production requires IucD, and alcaligin requires AlcA catalysis. The portion of the compound that is derived from ornithine, lysine or putrescine is shown in bold.
Table 1.
N-hydroxylating flavoprotein monooxygenases.
| Substrate | Enzyme | Organism | Siderophore | Reference |
|---|---|---|---|---|
| Lysine
| ||||
| IucD | Escherichia coli | aerobactin | [7, 21–25, 28, 29, 45–47] | |
|
| ||||
|
N-acyl-lysine
| ||||
| MbtG | Mycobacterium tuberculosis | mycobactin | [40, 41] | |
|
| ||||
| Ornithine
| ||||
| CchB | Streptomyces coelicolor | coelichelin | [48] | |
| OMO1 | Magnaporthe grisea | coprogen, ferricrocin | [49, 50] | |
| OMO1 | Omphalotus alearius | ferrichrome | [51] | |
| OrnOH | Aureobasidium pullulans | fusigen | [52] | |
| PsbA | Pseudomonas aeruginosa | pseudobactin | [53] | |
| PvdA | Pseudomonas aeruginosa | pyoverdin | [12–15, 17, 30, 54] | |
| PvdA | Burkholderia cepacia | ornibactin | [55] | |
| Sid1 | Ustilago maydis | ferrichrome | [56] | |
| SidA | Aspergillus fumigatus | fusarinine, ferricronin | [6, 18, 19, 39] | |
| SidA | Aspergillus nidulans | fusarinine, ferricronin | [57] | |
| VbsO | Rhizobium leguminosarum | vicibactin | [58] | |
|
| ||||
| Putrescine
| ||||
| AlcA | Bordetella bronchiseptica | alcaligin | [42] | |
| AlcA | Bordetella pertussis | alcaligin | [43] | |
8. Future work
The mechanism of catalysis for the N-hydroxylating monooxygenases has begun to be elucidated and the first structure has been reported for this functionally-related class of enzymes. While the controversy over membrane-binding seems to be decided in favor of these being soluble enzymes, new questions arise for further investigation. For example, further transient kinetic work is required to validate the presence of a peroxyflavin intermediate and continued crystallographic work may identify structural determinants of flavin-intermediate stabilization.
N-hydroxylating flavoprotein monooxygenases in hydroxamate siderophore biosynthesis
Review of kinetic studies revealing mechanism
Review of structural studies revealing FAD, NADPH and substrate binding/specificity
Acknowledgments
This publication was made possible by the Graduate Training Program in Dynamic Aspects of Chemical Biology NIH grant number T32 GM08545 (J.O.) from the National Institute of General Medical Sciences and by NIH grant number K02 AI093675 from the National Institute for Allergy and Infectious Disease (A.L.L.). We are grateful to Drs. R. J. Hondal and R. L. Schowen for critically reading this manuscript.
Abbreviations
The abbreviations used are:
- BVMO
Baeyer-Villiger monooxygenase
- FMO
flavin-containing monooxygenase
- IucD
lysine hydroxylase from Escherichia coli
- MbtG
N-acyl-lysine hydroxylase from Mycobacterium tuberculosis
- NMO
microbial N-hydroxylating monooxygenases
- PvdA
ornithine hydroxylase from Pseudomonas aeruginosa
- SidA
ornithine hydroxylase from Aspergillus fumigatus.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Crichton RR, Ward RJ. Biochemistry. 1992;31:11255–64. doi: 10.1021/bi00161a001. [DOI] [PubMed] [Google Scholar]
- 2.Crichton RR, Ward RJ. Met Ions Biol Syst. 1998;35:633–65. [PubMed] [Google Scholar]
- 3.Sandy M, Butler A. Chem Rev. 2009;109:4580–95. doi: 10.1021/cr9002787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schalk IJ. J Inorg Biochem. 2008;102:1159–69. doi: 10.1016/j.jinorgbio.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 5.Lamont IL, Beare PA, Ochsner U, Vasil AI, Vasil ML. Proc Natl Acad Sci U S A. 2002;99:7072–7. doi: 10.1073/pnas.092016999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schrettl M, Bignell E, Kragl C, Joechl C, Rogers T, Arst HN, Jr, Haynes K, Haas H. J Exp Med. 2004;200:1213–9. doi: 10.1084/jem.20041242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thariath A, Socha D, Valvano MA, Viswanatha T. J Bacteriol. 1993;175:589–96. doi: 10.1128/jb.175.3.589-596.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cornelis P, Matthijs S. Environ Microbiol. 2002;4:787–98. doi: 10.1046/j.1462-2920.2002.00369.x. [DOI] [PubMed] [Google Scholar]
- 9.Crosa JH, Walsh CT. Microbiol Mol Biol Rev. 2002;66:223–49. doi: 10.1128/MMBR.66.2.223-249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marahiel MA, Stachelhaus T, Mootz HD. Chem Rev. 1997;97:2651–2674. doi: 10.1021/cr960029e. [DOI] [PubMed] [Google Scholar]
- 11.van Berkel WJH, Kamerbeek NM, Fraaije MW. J Biotech. 2006;124:670–89. doi: 10.1016/j.jbiotec.2006.03.044. [DOI] [PubMed] [Google Scholar]
- 12.Ge L, Seah SY. J Bacteriol. 2006;188:7205–10. doi: 10.1128/JB.00949-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meneely KM, Barr EW, Bollinger JM, Jr, Lamb AL. Biochemistry. 2009;48:4371–4376. doi: 10.1021/bi900442z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meneely KM, Lamb AL. Biochemistry. 2007;46:11930–7. doi: 10.1021/bi700932q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Visca P, Ciervo A, Orsi N. J Bacteriol. 1994;176:1128–40. doi: 10.1128/jb.176.4.1128-1140.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Winsor GL, Van Rossum T, Lo R, Khaira B, Whiteside MD, Hancock RE, Brinkman FS. Nucleic Acids Res. 2009;37:D483–8. doi: 10.1093/nar/gkn861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Putignani L, Ambrosi C, Ascenzi P, Visca P. Biochem Biophys Res Commun. 2004;313:245–57. doi: 10.1016/j.bbrc.2003.11.116. [DOI] [PubMed] [Google Scholar]
- 18.Mayfield JA, Frederick RE, Streit BR, Wencewicz TA, Ballou DP, DuBois JL. J Biol Chem. 2010;285:30375–88. doi: 10.1074/jbc.M110.157578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chocklett SW, Sobrado P. Biochemistry. 2010;49:6777–83. doi: 10.1021/bi100291n. [DOI] [PubMed] [Google Scholar]
- 20.Goh CJ, Szczepan EW, Menhart N, Viswanatha T. Biochim Biophys Acta. 1989;990:240–5. doi: 10.1016/s0304-4165(89)80040-6. [DOI] [PubMed] [Google Scholar]
- 21.Dick S, Marrone L, Thariath AM, Valvano MA, Viswanatha T. Trends Biochem Sci. 1998;23:414–5. doi: 10.1016/s0968-0004(98)01271-7. [DOI] [PubMed] [Google Scholar]
- 22.Plattner HJ, Pfefferle P, Romaguera A, Waschutza S, Diekmann H. Biol Met. 1989;2:1–5. doi: 10.1007/BF01116193. [DOI] [PubMed] [Google Scholar]
- 23.Stehr M, Smau L, Singh M, Seth O, Macheroux P, Ghisla S, Diekmann H. Biol Chem. 1999;380:47–54. doi: 10.1515/BC.1999.006. [DOI] [PubMed] [Google Scholar]
- 24.Thariath AM, Fatum KL, Valvano MA, Viswanatha T. Biochim Biophys Acta. 1993;1203:27–35. doi: 10.1016/0167-4838(93)90032-m. [DOI] [PubMed] [Google Scholar]
- 25.Macheroux P, Plattner HJ, Romaguera A, Diekmann H. Eur J Biochem. 1993;213:995–1002. doi: 10.1111/j.1432-1033.1993.tb17846.x. [DOI] [PubMed] [Google Scholar]
- 26.Olucha J, Meneely KM, Chilton AS, Lamb AL. J Biol Chem. 2011 doi: 10.1074/jbc.M111.265876. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheng D, Ballou DP, Massey V. Biochemistry. 2001;40:11156–67. doi: 10.1021/bi011153h. [DOI] [PubMed] [Google Scholar]
- 28.Stehr M, Diekmann H, Smau L, Seth O, Ghisla S, Singh M, Macheroux P. Trends Biochem Sci. 1998;23:56–7. doi: 10.1016/s0968-0004(97)01166-3. [DOI] [PubMed] [Google Scholar]
- 29.Seth O, Smau L, Welte W, Ghisla S, Stehr M, Diekmann H, Macheroux P. TIBS. 1998;23:414–415. doi: 10.1016/s0968-0004(97)01166-3. [DOI] [PubMed] [Google Scholar]
- 30.Imperi F, Putignani L, Tiburzi F, Ambrosi C, Cipollone R, Ascenzi P, Visca P. Microbiology. 2008;154:2804–13. doi: 10.1099/mic.0.2008/018804-0. [DOI] [PubMed] [Google Scholar]
- 31.Alfieri A, Malito E, Orru R, Fraaije MW, Mattevi A. Proc Natl Acad Sci USA. 2008;105:6572–7. doi: 10.1073/pnas.0800859105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cho HJ, Cho HY, Kim KJ, Kim MH, Kim SW, Kang BS. J Struct Biol. 2011;175:39–48. doi: 10.1016/j.jsb.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 33.Eswaramoorthy S, Bonanno JB, Burley SK, Swaminathan S. Proc Natl Acad Sci U S A. 2006;103:9832–7. doi: 10.1073/pnas.0602398103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mirza IA, Yachnin BJ, Wang S, Grosse S, Bergeron Hln, Imura A, Iwaki H, Hasegawa Y, Lau PCK, Berghuis AM. J Amer Chem Soc. 2009;131:8848–54. doi: 10.1021/ja9010578. [DOI] [PubMed] [Google Scholar]
- 35.Orru R, Pazmino DE, Fraaije MW, Mattevi A. J Biol Chem. 2010;285:35021–8. doi: 10.1074/jbc.M110.161372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malito E, Alfieri A, Fraaije MW, Mattevi A. Proc Natl Acad Sci U S A. 2004;101:13157–62. doi: 10.1073/pnas.0404538101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Lorenzo V, Bindereif A, Paw BH, Neilands JB. J Bacteriol. 1986;165:570–8. doi: 10.1128/jb.165.2.570-578.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gross R, Engelbrecht F, Braun V. Mol Gen Genet. 1985;201:204–12. doi: 10.1007/BF00425661. [DOI] [PubMed] [Google Scholar]
- 39.Hissen AH, Wan AN, Warwas ML, Pinto LJ, Moore MM. Infect Immun. 2005;73:5493–503. doi: 10.1128/IAI.73.9.5493-5503.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krithika R, Marathe U, Saxena P, Ansari MZ, Mohanty D, Gokhale RS. Proc Natl Acad Sci U S A. 2006;103:2069–74. doi: 10.1073/pnas.0507924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quadri LE, Sello J, Keating TA, Weinreb PH, Walsh CT. Chem Biol. 1998;5:631–45. doi: 10.1016/s1074-5521(98)90291-5. [DOI] [PubMed] [Google Scholar]
- 42.Giardina PC, Foster LA, Toth SI, Roe BA, Dyer DW. Gene. 1995;167:133–6. doi: 10.1016/0378-1119(95)00659-1. [DOI] [PubMed] [Google Scholar]
- 43.Kang HY, Brickman TJ, Beaumont FC, Armstrong SK. J Bacteriol. 1996;178:4877–84. doi: 10.1128/jb.178.16.4877-4884.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uniprot_Consortium. Nucleic Acids Res. 39:D214–9. [Google Scholar]
- 45.Dick S, Marrone L, Duewel H, Beecroft M, McCourt J, Viswanatha T. J Protein Chem. 1999;18:893–903. doi: 10.1023/a:1020639514998. [DOI] [PubMed] [Google Scholar]
- 46.Marrone L, Beecroft M, Viswanatha T. Bioorg Chem. 1996;24:304–317. [Google Scholar]
- 47.Marrone L, Viswanatha T. Biochim Biophys Acta. 1997;1343:263–77. doi: 10.1016/s0167-4838(97)00129-5. [DOI] [PubMed] [Google Scholar]
- 48.Pohlmann V, Marahiel MA. Org Biomol Chem. 2008;6:1843–8. doi: 10.1039/b801016a. [DOI] [PubMed] [Google Scholar]
- 49.Hof C, Eisfeld K, Antelo L, Foster AJ, Anke H. Fungal Genet Biol. 2009;46:321–32. doi: 10.1016/j.fgb.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 50.Hof C, Eisfeld K, Welzel K, Antelo L, Foster AJ, Anke H. Mol Plant Pathol. 2007;8:163–72. doi: 10.1111/j.1364-3703.2007.00380.x. [DOI] [PubMed] [Google Scholar]
- 51.Welzel K, Eisfeld K, Antelo L, Anke T, Anke H. FEMS Microbiol Lett. 2005;249:157–63. doi: 10.1016/j.femsle.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 52.Wang W, Chi Z, Liu G, Buzdar MA, Chi Z, Gu Q. Biometals. 2009;22:965–72. doi: 10.1007/s10534-009-9248-x. [DOI] [PubMed] [Google Scholar]
- 53.Ambrosi C, Leoni L, Putignani L, Orsi N, Visca P. J Bacteriol. 2000;182:6233–8. doi: 10.1128/jb.182.21.6233-6238.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Visca P, Serino L, Orsi N. J Bacteriol. 1992;174:5727–31. doi: 10.1128/jb.174.17.5727-5731.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sokol PA, Darling P, Woods DE, Mahenthiralingam E, Kooi C. Infect Immun. 1999;67:4443–55. doi: 10.1128/iai.67.9.4443-4455.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mei B, Budde AD, Leong SA. Proc Natl Acad Sci U S A. 1993;90:903–7. doi: 10.1073/pnas.90.3.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eisendle M, Oberegger H, Zadra I, Haas H. Mol Microbiol. 2003;49:359–75. doi: 10.1046/j.1365-2958.2003.03586.x. [DOI] [PubMed] [Google Scholar]
- 58.Heemstra JR, Jr, Walsh CT, Sattely ES. J Am Chem Soc. 2009;131:15317–29. doi: 10.1021/ja9056008. [DOI] [PMC free article] [PubMed] [Google Scholar]