Abstract
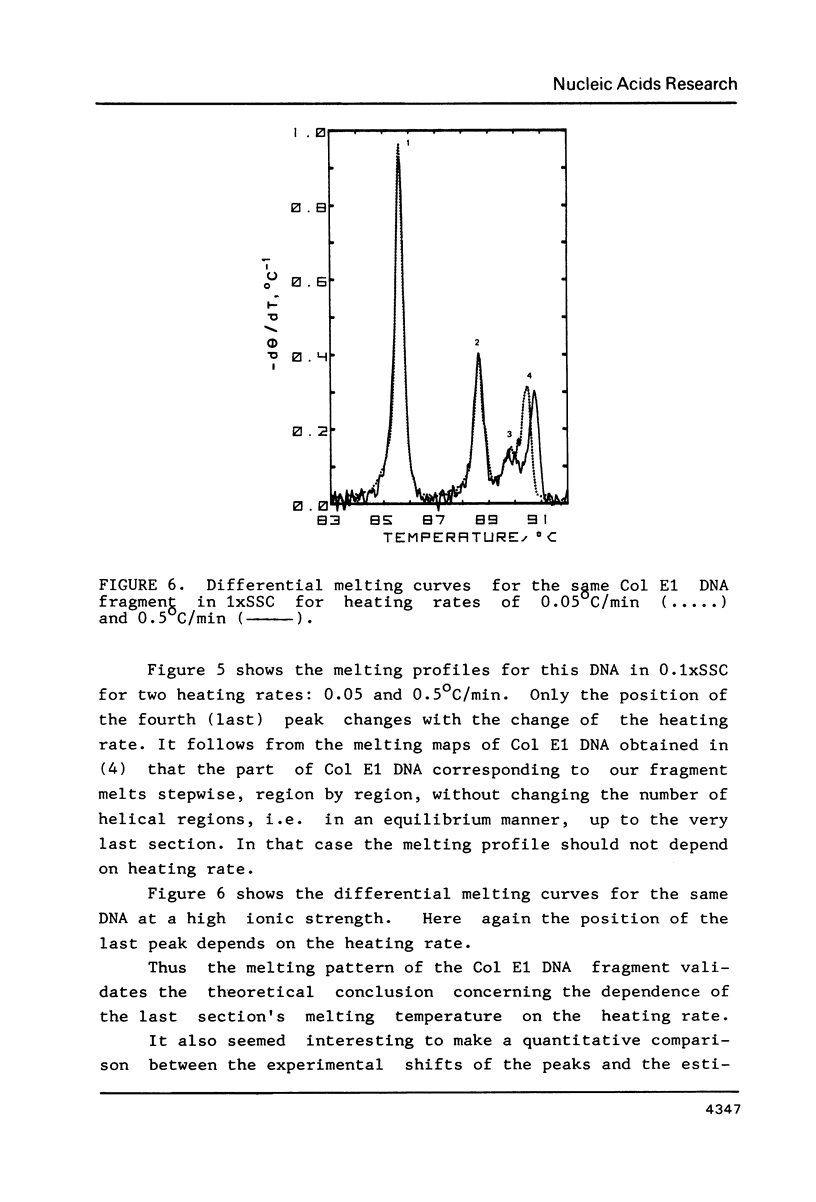
To investigate the effects of heating rate on the DNA melting profile and to test the predictions of the theory of slow relaxation processes in DNA melting (1) concerning these effects, we obtained differential melting curves for the Bsp I C1 fragment of T7 DNA (1461 bp) and the Sma I-Eco RI fragment of Col E1 DNA (1291 bp) at heating rates of 0.05 and 0.5 deg/min. At low ionic strength (0.02 M Na+) the heating rate has been shown to affect the position of the third peak in melting curve for C1 fragment. According to the melting maps (2), this peak corresponds to the unwinding of the section between the end of the molecule and the region already melted. At high ionic strength (0.2 M Na+), when the melting of this DNA is reversible (3), the position of the peaks does not depend on the heating rate. In the case of the Col E1 DNA fragment the heating rate affects, as might be expected from the melting maps (4), only the last peak, as the melting of the last section is always nonequilibrium. The results of the study are in good qualitative agreement and in satisfactory quantitative agreement with the theoretical predictions (1).
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amirikyan B. R., Vologodskii A. V., Lyubchenko YuL Determination of DNA cooperativity factor. Nucleic Acids Res. 1981 Oct 24;9(20):5469–5482. doi: 10.1093/nar/9.20.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anshelevich V. V., Vologodskii A. V., Lukashin A. V., Frank-Kamenetskii M. D. Slow relaxational processes in the melting of linear biopolymers: a theory and its application to nucleic acids. Biopolymers. 1984 Jan;23(1):39–58. doi: 10.1002/bip.360230105. [DOI] [PubMed] [Google Scholar]
- Azbel M. Y. DNA sequencing and helix-coil transition. I. Theory of DNA melting. Biopolymers. 1980 Jan;19(1):61–80. doi: 10.1002/bip.1980.360190105. [DOI] [PubMed] [Google Scholar]
- Blake R. D., Haydock P. V. Effect of sodium ion on the high-resolution melting of lambda DNA. Biopolymers. 1979 Dec;18(12):3089–3109. doi: 10.1002/bip.1979.360181214. [DOI] [PubMed] [Google Scholar]
- Borovik A. S., Kalambet Y. A., Lyubchenko Y. L., Shitov V. T., Golovanov E. I. Equilibrium melting of plasmid ColE1 DNA: electron-microscopic visualization. Nucleic Acids Res. 1980 Sep 25;8(18):4165–4184. doi: 10.1093/nar/8.18.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983 Jun 5;166(4):477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- Gotoh O., Wada A., Yabuki S. Salt-concentration dependence of melting profiles of lambda phage DNAs: evidence for long-range interactions and pronounced end effects. Biopolymers. 1979 Apr;18(4):805–824. doi: 10.1002/bip.1979.360180406. [DOI] [PubMed] [Google Scholar]
- Gravchev M. A., Zaichikov E. F., Kravchenko V. V., Pletnev A. G. Vydelenie promotornykh i terminatornogo fragmentov DNK faga T7 is gidrolizata, poluchennogo deivistviem éndonukleazy restrikstii BsuR. Dokl Akad Nauk SSSR. 1978;239(2):475–478. [PubMed] [Google Scholar]
- Hoff A. J., Roos A. L. Hysteresis of denaturation of DNA in the melting range. Biopolymers. 1972;11(6):1289–1294. doi: 10.1002/bip.1972.360110612. [DOI] [PubMed] [Google Scholar]
- Lyamichev V. I., Panyutin I. G., Cherny D. I., Lyubchenko YuL Localization of low-melting regions in phage T7 DNA. Nucleic Acids Res. 1983 Apr 11;11(7):2165–2176. doi: 10.1093/nar/11.7.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyamichev V. I., Panyutin I. G., Lyubchenko YuL Gel electrophoresis of partially denatured DNA. Retardation effect: its analysis and application. Nucleic Acids Res. 1982 Aug 11;10(15):4813–4826. doi: 10.1093/nar/10.15.4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Gabarro-Arpa J., Dujon B. The hierarchical approach to the DNA stability problem. II. Some applications and speculations with yeast mitochondrial DNA as an example. Biochimie. 1982 Feb;64(2):113–126. doi: 10.1016/s0300-9084(82)80413-6. [DOI] [PubMed] [Google Scholar]
- Michel F. Hysteresis and partial irreversibility of denaturation of DNA as a means of investigating the topology of base distribution constraints: application to a yeast rho- (petite) mitochondrial DNA. J Mol Biol. 1974 Oct 25;89(2):305–326. doi: 10.1016/0022-2836(74)90521-x. [DOI] [PubMed] [Google Scholar]
- Perelroyzen M. P., Lyamichev V. I., Kalambet YuA, Lyubchenko YuL, Vologodskii A. V. A study of the reversibility of helix-coil transition in DNA. Nucleic Acids Res. 1981 Aug 25;9(16):4043–4059. doi: 10.1093/nar/9.16.4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss C., Michel F., Gabarro J. An apparatus for studying the thermal transition of nucleic acids at high resolution. Anal Biochem. 1974 Dec;62(2):499–508. doi: 10.1016/0003-2697(74)90182-1. [DOI] [PubMed] [Google Scholar]
- Spatz H. C., Crothers D. M. The rate of DNA unwinding. J Mol Biol. 1969 Jun 14;42(2):191–219. doi: 10.1016/0022-2836(69)90038-2. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Simon M. N., Dunn J. J. Genetic and physical mapping in the early region of bacteriophage T7 DNA. J Mol Biol. 1979 Dec 25;135(4):917–937. doi: 10.1016/0022-2836(79)90520-5. [DOI] [PubMed] [Google Scholar]
- Sümegi J., Breedveld D., Hossenlopp P., Chambon P. A rapid procedure for purification of EcoRI endonuclease. Biochem Biophys Res Commun. 1977 May 9;76(1):78–85. doi: 10.1016/0006-291x(77)91670-9. [DOI] [PubMed] [Google Scholar]
- Tachibana H., Ueno-Nishio S., Gotoh O., Wada A. Salt-concentration dependence of thermal denaturation of restriction fragment DNAs from phi X174. J Biochem. 1982 Sep;92(3):623–635. doi: 10.1093/oxfordjournals.jbchem.a133973. [DOI] [PubMed] [Google Scholar]
- Vizard D. L., Ansevin A. T. High resolution thermal denaturation of DNA: thermalites of bacteriophage DNA. Biochemistry. 1976 Feb 24;15(4):741–750. doi: 10.1021/bi00649a004. [DOI] [PubMed] [Google Scholar]
- Yamada M., Ebina Y., Miyata T., Nakazawa T., Nakazawa A. Nucleotide sequence of the structural gene for colicin E1 and predicted structure of the protein. Proc Natl Acad Sci U S A. 1982 May;79(9):2827–2831. doi: 10.1073/pnas.79.9.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]


