Abstract
In humans, genetic variation and dietary factors may alter the biologic effects of exposure to 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP), one of the major heterocyclic amines generated from cooking meats at high temperatures that has carcinogenic potential through the formation of DNA adducts. Previously, we reported grilled red meat consumption associated with PhIP-DNA adduct levels in human prostate. In the present study, we expanded our investigation to estimate the associations between beverage consumption and PhIP-DNA adduct levels in prostate for 391 prostate cancer cases. Of the 15 beverages analyzed, red wine consumption had the strongest association with PhIP-DNA adduct levels showing an inverse correlation in both tumor (p=0.006) and non-tumor (p=0.002) prostate cells. Red wine consumption differed significantly between African-American and white cases, but PhIP-DNA adduct levels in prostate did not vary by race. In African Americans compared with whites, however, associations between red wine consumption and PhIP-DNA adduct levels were not as strong as associations with specific (e.g., SULT1A1 and UGT1A10 genotypes) and non-specific (e.g., African ancestry) genetic variation. In a multivariable model, the covariate for red wine consumption explained a comparable percentage (13-16%) of the variation in PhIP-DNA adduct levels in prostate across the two racial groups, but the aforementioned genetic factors explained 33% of the PhIP-DNA adduct variation in African-American cases, while only 19% of the PhIPDNA adduct variation in whites. We conclude that red wine consumption may counteract biologic effects of PhIP exposure in human prostate, but genetic factors may play an even larger role, particularly in African Americans.
Keywords: compounds, heterocyclic; resveratrol; UDP-glucuronosyltransferase; sulfotransferases; African Americans; chemoprevention
Introduction
Prostate cancer occurs more often and with a greater loss of life in men of African descent in the United States compared with whites. The most recent figures show a 60 percent higher prostate cancer incidence rate in African-American men and 140 percent higher mortality rate (1). In the Cancer Prevention Study II Nutrition Cohort, total red meat intake was associated with higher risk of prostate cancer only in African-American men and was mainly due to higher consumption levels of cooked processed meats (2). Heterocyclic amines are present in higher amounts in fried or grilled meats with the predominant heterocyclic amine compound being amino-1-methyl-6-phenylimidazo [4,5-b] pyridine (PhIP). PhIP has been shown to be carcinogenic in rat prostate (3, 4) and may derive its carcinogenic potential through the formation of DNA adducts. Based on dietary survey (5) and urinary excretion data (6), PhIP intake is higher in African Americans than Whites. PhIP dietary consumption has also been shown to be associated with higher PSA levels in healthy African-American men (7), and we have previously reported a positive association between PSA level and PhIP-DNA adducts in African Americans that was not observed in whites (8).
Beverages such as coffee, beer and wine may diminish PhIP mutagenicity by altering the PhIP metabolism pathway (9-11). Survey data suggests that whites consume more of these three beverages compared with African Americans (12). The antioxidant polyphenol, resveratrol, found in red wines has also been shown to inhibit PhIP-DNA adduct formation presumably by O-acetyltransferase and sulfotransferase catalysis (11). In rodents, beer inhibits PhIP-induced tumorigenicity and DNA adduct formation most likely by inhibiting enzymes involved in PhIP metabolism (9).
In addition to racial differences in PhIP metabolism, inherited genetic differences in enzymes that metabolize PhIP may also contribute to variation in the biologic effects of this compound across populations. Among the nine sulfotransferases (SULTs) known to be expressed in human tissues, SULT1A1, 1A2, and 1A3 catalyze the sulfate conjugation of many phenolic compounds and other xenobiotics, such as PhIP (13). A common single nucleotide polymorphism (SNP) has been observed in the SULT1A1 gene that results in an arginine-tohistidine amino acid change in codon 213. Individuals with two His213 alleles have only 15% of the SULT1A1 activity compared with the carriers of the Arg213 allele (14). Dietary polyphenols, such as resveratrol, may compete with PhIP as substrates and/or inhibitors for human SULT1A1. Since SULT1A1 is involved in the chemical activation of PhIP by forming highly reactive ester compounds that form DNA adducts, resveratrol could potentially reduce cancer risk through this competitive inhibition. The SULT1A1 codon 213 variant, which has been linked with increased risk to some cancers (15), has lower activity toward both resveratrol (16) and PhIP (13). PhIP and its carcinogenic metabolite N-hydroxy-PhIP (N-OH-PhIP) are extensively conjugated by UDP-glucuronosyltransferase (UGTs). UGT1A1 is the predominant UGT involved in PhIP metabolism, and genetic variation in UGT1A1 may affect the rate of PhIP metabolism (17, 18).
Based on the potential for PhIP metabolism to be affected by the chemical compounds in common dietary beverages, we hypothesized that beverage consumption patterns may influence prostatic levels of PhIP-DNA adducts. Therefore, we evaluated potential associations between beverage intake and PhIP-DNA adduct levels (as quantified by immunohistochemistry) in 391 prostate cancer patients who underwent radical prostatectomy, taking into account variation in the SULT1A1 and UGT1A10 genes and the level of African-ancestry.
Methods
Study Population
The study population consisted of men who were part of the Henry Ford Health System. Details concerning the ascertainment and recruitment of study cases can be found in a previous publication (19). The present study includes 391 of the 419 prostate cancer cases that underwent radical prostatectomy and had tissue specimens with sufficient areas of tumor and nontumor cells for immunohistochemical DNA adduct studies. The mean age at diagnosis was 60.8 ± 6.6 years with an average of 2.9 ± 2.4 months between diagnosis and surgery. The 391 study participants that comprised the analytic sample self-identified as 56.5% white and 43.5% African American. Cases had a mean age at time of diagnosis of 60.8 ± 6.7 years with 42.7% of patients diagnosed under the age of 60 years. The majority of patients (80.1%) had PSA levels greater than 4 ng/ml at diagnosis with a median PSA level at 5.2 ng/ml. A Gleason grade of 7 was the predominant tumor grade (45.9%) of cases with 19% of cases having a Gleason grade of 8 or higher. Most cases (80%) had stage 2 tumors with a median tumor volume of 15% of the prostate gland.
Food Questionnaire
Beverage consumption was measured by self-report using the SELECT food frequency questionnaire (FFQ) developed from FFQs used in the Women's Health Initiative (20) and the Prostate Cancer Prevention Trial (21). The FFQ asked about usual beverage consumption over the year preceding prostate cancer diagnosis and included portion and frequency information for seventeen beverages or beverage additives, such as milk in coffee or tea. The distribution of amount consumed for any of the specific beverages was not uniform; therefore we limited our analysis to a yes/no variable as to whether a beverage was consumed for the 15 beverage items with consumption frequencies. Since the SELECT questionnaire does not include food preparation questions, supplemental questions adapted from a validated questionnaire were added to assess grilled meat intake (22) . To account for dietary PhIP exposure relevant to PhIPDNA adduct levels in prostate, as previously reported by our group (23), in multivariate analyses we adjusted for combined red and white grilled meat consumption using a dichotomous dummy variable defined as 0 to 2 servings and >2 servings per month.
Pathology and Immunohistochemistry
Hematoxylin-eosin stained slides of study cases were reviewed by the study uropathologist (ATS) to confirm the diagnosis and identify a paraffin block with sufficient tumor and non-tumor prostatic tissue for staining. Using a microtome, five consecutive sections (5 micron thick) were cut from the tissue block of each patient sample. One slide was hematoxylin and eosin stained and examined by the study uropathologist who circled separate areas of tumor and non-tumor cell populations to be used for adduct scoring. The paraffin-embedded sections were baked at 50°C one hour, deparaffinized in xylene, and rehydrated in serial alcohol. After treatment using RNase and proteinase K, the sections were blocked using 3% BSA and normal goat serum. The primary anti-PhIP-DNA adduct polyclonal antibody (24, 25) was incubated with the sections at 4°C overnight in a humid chamber at a dilution of 1:500. Also, the biotinylated secondary antibody was incubated with the sections at room temperature for 30 minutes, at a dilution of 1: 200. Endogenous peroxidase activity was blocked using 0.3% hydrogen peroxide in methanol for 20 minutes. The antibody complex was detected using an avidin-biotin-peroxidase complex solution and visualized using 3,3′-diaminobenzidine (Zymed Laboratories, Inc., San Francisco, CA). A negative control was included in each experiment by omitting the primary antibody. The staining specificity was confirmed using the primary antibody that had been preabsorbed with 2 or 20 μg/ml DNA extract from MCF-7 cells treated with 150 μM N-hydroxy-PhIP. A cytospin sample of MCF-7 cells without PhIP treatment was included in each batch of staining. Staining was measured by absorbance image analysis using a Cell Analysis System 200 microscope. Absorbance of light at a wavelength of 500 nmol/L was measured because methyl green does not absorb light at this wavelength, whereas diaminobenzidine does. For each prostate specimen, two technicians independently scored 50 epithelial cells (five fields with 10 cells per field scored) in the two areas (tumor and non-tumor) circumscribed by the study pathologist. The final score was the mean of the two technicians’ scores. Scored cells were selected to be representative, in terms of intensity, of the cells in the field. Staining intensity was represented by the absorbance unit of optical density (OD).
Genotyping
Genotype data for this study were generated using several different methods. Most genotypes came from two separate OPAs (Oligo Pool All) run on the Illumina GoldenGate® genotyping platform. The first OPA was a panel of 1,509 SNPs informative for West African versus European ancestry that was a precursor to the current Illumina SNP panel for determining ancestry (illumina.com/products/african_american_admixture_panel.ilmn). The second OPA was a custom panel of 1,473 tag SNPs for 172 candidate genes drawn from the phase II version of the International HapMap project (hapmap.org). To optimally select a minimal number of tagSNPs that capture variation in both of the European and African samples, we used the multiple population tagging method TAGster (26). For the purposes of the present study, we used genotype data on three tag SNPs (genotyping of four tagSNPs attempted) for the SULT1A1 gene and 59 (successfully genotyped 60 of 66 attempted SNPs with one SNP removed because it had an exact HWE p-value < 0.01) tag SNPs for the UGT1A10 gene that tagged approximately 80/98 percent of the common (10% frequency or greater) variation present in these two genes in the HapMap YRI/CEU samples (see the method supplement for more details). We also genotyped the known functional exon 7 SNP of the SULT1A1 gene (rs9282861) that results in an amino acid change (Arg to His) at codon 213 of the translated protein. The PCR reactions were based on a previous report (27) and performed in a total volume of 25 μL of a solution containing 5X Colorless GoTaq Flexi Buffer, 8% DMSO, 0.2mM dNTP, 2mM MgCl2, 0.8μM of each primer (upstream primer 5”-GGTTGAGGAGTTGGCTCTGC-3' and downstream primer 5'-ATGAACTCCTGGGGGACGGT-3') and 1.25 U of GoTaq DNA Polymerase. The reaction started with 95°C for 2 min, followed by 40 cycles consisting of denaturation (94°C for 1 min), annealing (60°C for 1 min) and extension (72°C for 2 min). The 289bp PCR product was digested with the restriction enzyme HhaI at 37°C for 4 hours.
Statistical Analysis
Chi square tests were used to evaluate differences in beverage intake by race with a statistical correction made for multiple comparisons (28). Potential batch effects in the PhIPDNA adduct assay were taken into account by computing a batch correction factor that was the difference between the adduct level of the positive control slide in a single batch and the mean adduct level of the positive control slides across all batches. The batch-adjusted adduct level was the crude adduct level minus the batch correction factor. To account for correlation of observations within the same experimental batch, we used a mixed linear model implemented as PROC MIXED in SAS (29). In this modeling framework, the correlation of observations within each experimental batch were treated as random effects whereas other model covariates were treated as fixed effects.
Principal-components analysis (PCA) was used to derive linear transformations of the original tagSNPs data, in which eigenvectors are chosen to maximize the variance of each PC relative to the overall variation in the region (30). The number of principal components needed for each gene was determined using an 80% explained variance criteria, reducing the number of parameters to be tested. Once the necessary PCs were determined, generalized linear models were used to assess the significance of the gene. Please see the supplementary methods section for a more detailed description of how PCs were determined and loaded on individual SNPs. For a subset of the African-American cases that were genotyped with ancestry informative markers, estimates of African ancestry were generated with the ADMIXMAP statistical program (http://homepages.ed.ac.uk/pmckeigu/admixmap/), which uses a hybrid of Bayesian and traditional modeling to compare observed vs. expected ancestry across the genome (31).
Results
Types of Beverages Consumed by Race
Among the 391 cases with PhIP DNA adduct assessment, men reported consuming an average of 7.5 different beverages (of the 15 types of beverages queried) with seven types of beverages consumed the mode and median response. Overall, cases in the study averaged 7.4 beverage servings per day. Table 1 shows how reporting of consumption of specific beverages varied by race. African Americans and whites did not differ in the percent of men consuming milk, tea, tomato juice, citrus juice or liquor, but a significantly lower percentage of African-American men reported consuming coffee (71.2% vs. 88.2%, p<0.001), beer (34.1% vs. 56.6%, p<0.001), diet pop (27.1% vs. 47.1%, p<0.001), and wine (21.8 vs. 44.3%, p<0.001) compared with white men. A higher percentage of African-American cases reported consuming other juices (77.6% vs. 53.8%, p<0.001), fruit juices (48.1% vs. 9.5%, p<0.001), regular pop (73.5% vs. 52.9%, p<0.001) and meal replacement (9.4% vs. 3.6%, p=0.02).
Table 1.
Percent Beverage consumption in Prostate Cancer Cases by Race
| Beverage | African American (n=170) | White (n=221) | P Value |
|---|---|---|---|
| Milk | 69.2 | 67.4 | 0.704 |
| Coffee | 71.2 | 88.2 | <.001 |
| Tea | 52.4 | 51.6 | 0.880 |
| Tomato juice | 37.6 | 40.7 | 0.537 |
| Citrus juice | 84.7 | 81.9 | 0.463 |
| Other juice | 77.6 | 53.8 | <.001 |
| Fruit drink | 48.8 | 9.5 | <.001 |
| Meal replacement | 9.4 | 3.6 | 0.018 |
| Diet pop | 27.1 | 47.1 | <.001 |
| Regular pop | 73.5 | 52.9 | <.001 |
| Water | 93.5 | 95.9 | 0.280 |
| Beer | 34.1 | 56.6 | <.001 |
| Any wine | 21.8 | 44.3 | <.001 |
| Red wine | 18.2 | 37.1 | <.001 |
| White wine | 15.9 | 30.3 | <.001 |
| Liquor | 31.2 | 38.5 | 0.135 |
Beverage Consumption and PhIP-DNA Adduct Levels in Prostate
Mean PhIP-DNA adduct levels by type of beverage consumed are reported in table 2. Means are reported separately for tumor and adjacent non-tumor tissue and were adjusted for race, age, grilled meat consumption, Gleason score and tumor volume, which are the five factors we found previously to be associated with PhIP-DNA adduct levels in prostate (8, 23). In prostate tumor cells, significantly lower mean PhIP-DNA adduct levels were found among cases that reported consuming red wine (p=0.006) and any wine (p=0.04). Cases who consumed white wine or beer also had lower PhIP-DNA adduct levels in tumor, but the p value for these differences were not significant (p=0.09 for both). In non-tumor cells, the same trends were observed between consumers and non-consumers. In prostate non-tumor cells, red wine consumption showed the strongest inverse relationship with PhIP-DNA adduct levels. After correcting for multiple comparisons within the two cell types, the red wine inverse association in prostate non-tumor cells was the only association that remained statistically significant at the 0.05 alpha level (Pcorrected = 0.032). We found no differences in adjusted PhIP-DNA adduct levels between whites and African-Americans (0.154 ± 0.015 vs. 0.156 ± 0.015; p=0.6), and race stratified comparisons showed similar beverage consumption associations compared with the analysis of the full sample.
Table 2.
Mean adjusted PhIP-DNA adduct level in prostate nontumor and tumor cells of prostate cancer cases across beverage consumption categories*
| Beverage | NonTumor | Tumor | ||||||
|---|---|---|---|---|---|---|---|---|
| nC | nNC | Consumers | Nonconsumers | P value | Consumers | Nonconsumers | P value | |
| Milk | 266 | 124 | 0.154±0.015 | 0.157±0.015 | 0.58 | 0.097±0.009 | 0.097±0.009 | 0.83 |
| Coffee | 316 | 75 | 0.154±0.015 | 0.160±0.015 | 0.36 | 0.097±0.009 | 0.098±0.009 | 0.92 |
| Tea | 203 | 188 | 0.156±0.015 | 0.154±0.015 | 0.62 | 0.098±0.009 | 0.097±0.009 | 0.66 |
| Tomato juice | 154 | 237 | 0.158±0.015 | 0.154±0.015 | 0.34 | 0.098±0.009 | 0.097±0.009 | 0.59 |
| Citrus juice | 325 | 66 | 0.155±0.015 | 0.158±0.015 | 0.52 | 0.097±0.009 | 0.100±0.009 | 0.39 |
| Other juice | 251 | 140 | 0.155±0.015 | 0.155±0.015 | 0.97 | 0.096±0.009 | 0.100±0.009 | 0.16 |
| Fruit drink | 104 | 287 | 0.161±0.015 | 0.153±0.015 | 0.15 | 0.098±0.009 | 0.097±0.009 | 0.85 |
| Meal replacement | 24 | 367 | 0.159±0.017 | 0.155±0.015 | 0.70 | 0.100±0.010 | 0.097±0.009 | 0.67 |
| Diet pop | 150 | 241 | 0.158±0.015 | 0.154±0.015 | 0.33 | 0.098±0.009 | 0.097±0.009 | 0.78 |
| Regular pop | 242 | 149 | 0.157±0.015 | 0.152±0.015 | 0.34 | 0.098±0.009 | 0.095±0.009 | 0.36 |
| Water | 370 | 20 | 0.156±0.015 | 0.149±0.017 | 0.51 | 0.098±0.009 | 0.087±0.011 | 0.13 |
| Beer | 183 | 208 | 0.151±0.015 | 0.159±0.015 | 0.06 | 0.095±0.009 | 0.100±0.009 | 0.09 |
| Any wine | 135 | 256 | 0.150±0.015 | 0.158±0.015 | 0.11 | 0.093±0.009 | 0.100±0.009 | 0.04 |
| Red wine | 113 | 278 | 0.145±0.015 | 0.160±0.015 | 0.002 | 0.091±0.009 | 0.100±0.009 | 0.006 |
| White wine | 94 | 297 | 0.152±0.015 | 0.157±0.015 | 0.35 | 0.093±0.009 | 0.099±0.009 | 0.09 |
| Liquor | 138 | 253 | 0.155±0.015 | 0.156±0.015 | 0.83 | 0.097±0.009 | 0.098±0.009 | 0.84 |
Adduct level measured as mean absorbance adjusted for batch effects, grilled meat consumption, age, race, grade and tumor volume ± standard error
Red wine and grilled red meat exposure associations with PhIP-DNA adduct levels by race
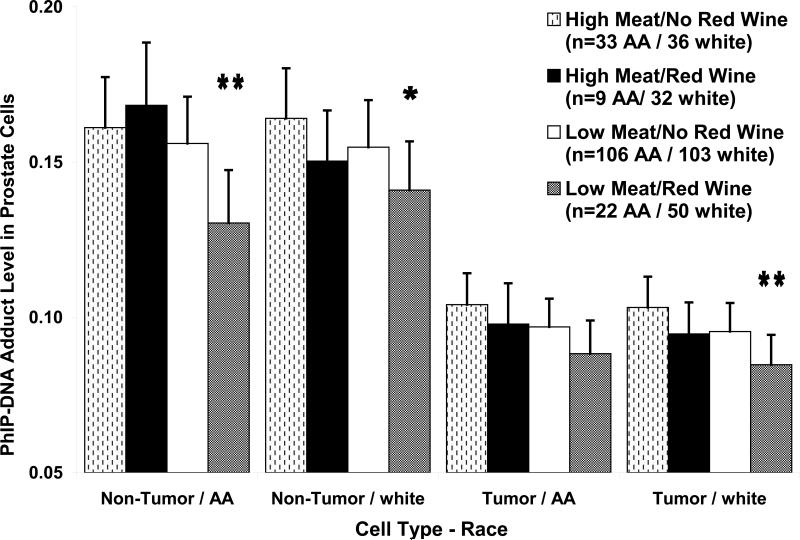
We next conducted analyses stratified by red wine and grilled red meat consumption and race to discern their joint effects on PhIP-DNA adduct levels in prostate (figure 1). In non-tumor cells, the African-American and white cases with high grilled red meat consumption that did not drink red wine had 23% (p=0.009) and 16% (p=0.01) higher PhIP-DNA adduct levels compared with their respective race matched counterparts with low grilled meat consumption who drank red wine. In African-American cases, the inverse association between red wine exposure and PhIP-DNA adduct levels in non-tumor cells was only observed in cases with low grilled red meat exposure, whereas in whites red wine consumption was inversely associated with PhIP-DNA adducts in both the high and low grilled red meat exposure groups. In tumor cells, the African-American and white cases with high grilled red meat consumption that did not drink red wine had 18% (p=0.06) and 22% (p=0.005) higher PhIP-DNA adduct levels compared with low grilled meat consumers who drank red wine.
Figure 1.
Mixed model estimated PhIP-DNA adduct level means and standard errors stratified by cell type (non-tumor/tumor), race (white/black), grilled meat intake (high/low) and red wine consumption (yes/no). Model adjusted for tumor grade, tumor volume and batch effects. Listed p values (* p<0.05; ** p<0.01) are for comparisons of each meat/red wine consuption group with the high grilled meat intake/no red wine consumption group within each cell type/race stratum.
To determine how genetic factors related to the metabolism of PhIP might affect the relationship between PhIP-DNA adducts and red wine consumption, we tested several race- and tissue type-specific multivariable models that included covariates for genetic factors potentially associated with PhIP-DNA adducts (table 3). The UGT1A10 and SULT1A1 PCA factors were derived from race-specific principal components models that tested for the number of principal variance components that would account for 80 percent or more of the variation within the UGT1A10 and SULT1A1 tagSNPs genotyped. For whites, 80.4% of UGT1A10 common variation was described by six principal components (denoted PCA1...PCA6). For African Americans, 81.2% of UGT1A10 common variation was described by 12 principal components (denoted PCA1...PCA12). For SULT1A1 variation, 93.0% of the common variation was described by the first two of three principal components in whites, while in African Americans the first three of four principal components described 90.1% of SULT1A1 common variation. These genetic factors included two race-specific UGT1A10 PCA factors that were most strongly associated with PhIP-DNA adduct levels and one SULT1A1 PCA factor that showed an association with PhIP-DNA adduct levels in African-American cases.
Table 3.
Race-specific Associations of Red Wine Consumption and Genetic Factors with PhIP-DNA Adduct Levels in Prostate
| Model 1a | Model 2b | |||
|---|---|---|---|---|
| β ± SE | p value | β ± SE | p value | |
| African-American Cases | ||||
| Non-Tumor Cells | (n=170) | (n=122) | ||
| Red Wine Consumption | -0.015 ± 0.009 | 0.09 | -0.016 ± 0.010 | 0.12 |
| SULT1A1 PCA3 | 0.014 ± 0.004 | 0.0003 | ||
| UGT1A1 PCA4 | -0.011 ± 0.004 | 0.003 | ||
| UGT1A1 PCA6 | 0.006 ± 0.004 | 0.13 | ||
| African Ancestry | 0.114 ± 0.038 | 0.004 | ||
| Tumor Cells | ||||
| Red Wine Consumption | -0.006 ± 0.006 | 0.32 | -0.005 ± 0.007 | 0.52 |
| SULT1A1 PCA3 | 0.010 ± 0.003 | 0.0004 | ||
| UGT1A1 PCA4 | -0.007 ± 0.003 | 0.01 | ||
| UGT1A1 PCA6 | -0.0004 ± 0.003 | 0.89 | ||
| African Ancestry | 0.035 ± 0.028 | 0.22 | ||
| White Cases | ||||
| Non-Tumor Cells | (n=221) | (n=180) | ||
| Red Wine Consumption | -0.013 ± 0.006 | 0.03 | -0.016 ± 0.006 | 0.01 |
| UGT1A1 PCA3 | -0.004 ± 0.003 | 0.22 | ||
| UGT1A1 PCA5 | -0.005 ± 0.003 | 0.11 | ||
| Tumor Cells | ||||
| Red Wine Consumption | -0.010 ± 0.004 | 0.02 | -0.013 ± 0.004 | 0.005 |
| UGT1A1 PCA3 | -0.002 ± 0.002 | 0.47 | ||
| UGT1A1 PCA5 | -0.002 ± 0.002 | 0.43 | ||
adjusted for age, tumor volume, gleason grade and red meat consumption
adjusted for covariates in a and genetic factors listed
In whites, a significant negative association with red wine consumption was observed in both non-tumor and tumor cells that increased by approximately 30% after adjusting for the two UGT1A10 principal components most strongly associated with PhIP-DNA adduct levels. In a multivariable model of PhIP-DNA adduct levels in tumor cells, the association of red wine consumption was significant at the p=0.01 level. In African-American cases, before adjusting for genetic factors the beta coefficients for red wine consumption in non-tumor cells were of similar magnitude and direction as was observed in whites, although of lower significance likely owing to the smaller sample size. However, unlike in white cases, in African-American cases adjustment for genetic factors left the effect estimates for red wine consumption unchanged in both non-tumor and tumor cells. The additional covariate of African ancestry showed a statistically significant (p=0.004) association with PhIP-DNA adduct in non-tumor cells. Specific genetic factors were also more prominently associated with PhIP-DNA adduct levels in African-American cases, most notably the third principal component for SULT1A1 common variation and the fourth principal component for UGT1A10 common variation.
Discussion
PhIP-DNA adducts are a measurable biologic effective dose of dietary heterocyclic amine exposure (32). Having previously demonstrated an association between consumption of grilled red meat and PhIP-DNA adduct levels in human prostate (23), in the present study we tested for associations between consumption of 15 different beverages and PhIP-DNA adduct levels in prostate. Red wine consumption had the strongest association with PhIP-DNA adduct levels being inversely related to adduct levels in both prostate tumor and non-tumor cells.
Recently, red wine and its most biologically active component resveratrol, a polyphenolic phytoestrogen found in high concentrations in the skins of grapes used to make red wine (33), has drawn attention as a potential chemopreventive agent for prostate cancer (34). Epidemiologic support for the chemopreventive effects of red wine on prostate cancer comes mainly from a case-control study that showed an inverse dose response with average glasses of red wine consumed per week and risk of prostate cancer (35), however, cohort studies have failed to replicate this finding (36-38). Resveratrol has generated considerable research interest due to its potential wide range of biological effects that include antioxidant, chemopreventive, cardioprotective, neuroprotective, anti-inflammatory and anti-viral activities (39, 40). Much of the research into the potential biologic mechanisms that underlie resveratrol's chemopreventive effects has focused on its antioxidant capacity, but one proposed mechanism that may have relevance in relation in prostate cancer is resveratrol's potency in decreasing the mutagenic potential of food-derived heterocyclic amines (HA) such as -2 amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) and 2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline (MeIQx) at micromolar concentrations (41). One possible explanation for the inhibitory effects of resveratrol on PhIP mutagenicity is that the activation of PhIP to its reactive DNA binding species and conjugation of resveratrol, resulting in its reduced bioavailability, both occur through the same sulfation enzymatic reaction (42, 43). In primary cultures of human mammary epithelial cells, administration of resveratrol inhibited sulfotransferase activity twice as effectively as it inhibited acetyltransferase activity and reduced PhIP-DNA adduct levels to about 50 percent of that observed in untreated control cells (11). Furthermore, the most common functional genetic variant of the SULT1A1 gene, the codon 213 His/Arg substitution, affects the enzymatic activity of this key sulfotransferase protein toward both PhIP (13) and resveratrol (16).
In humans, PhIP detoxification occurs primarily in the liver by glucuronidation of PhIP and N-OH-PhIP by UDP-glucuronosyltransferase with increased levels in urine of the main metabolite, N-OH-PhIP-N2-glucuronide, corresponding to lower levels of DNA adducts found in the colon of individuals exposed to PhIP (44). The UDP-glucuronosyltransferase (UGT) superfamily of enzymes are coded by the UGT1 locus on chromosome 2q37 that consists of a set of common exons in the 3' region designated 2, 3, 4, and 5 and an exon 1 unique to each gene in the UGT family (45). UGT enzymes catalyze the glucuronidation of a variety of endogenous compounds and are integrally involved in the detoxification of many carcinogens, the clearance of drugs and the metabolism of a variety of endogenous compounds (46). Among the UGT enzymes, UGT1A10 has exhibited significantly higher glucuronidation rates against PhIP and NOH-PhIP than any other UGT family member (18).
Sulfation plays a key role in the activation of N-hydroxy derivatives of carcinogenic heterocylica amines, such as PhIP (47). A role for SULT1A1 in PhIP-induced prostate carcinogenesis is biologically plausible since the SULT1A1 transcript has been detected in human prostate (48). In studies using functional recombinant proteins from cDNA inserted in Salmonella typhimurium strain, N-hydroxy-PhIP was activated specifically by human SULT1A1, but not by NAT1 or NAT2 (49). In humans that ingested a radioactively labeled PhIP compound, SULT1A1 activity is positively correlated with PhIP-DNA adduct formation in colon and a much stronger predictor of DNA adduct levels than NAT2 activity (44), which would be the alternative activation pathway of N2-Hydroxy-PhIP. Similar results were found in PhIP exposure experiments conducted with prostate cytosols prepared from benign prostate tissue collected from men undergoing transuretheral resections of the prostate in that SULT1A1 protein levels, but not NAT2 protein levels, were correlated with DNA adduct levels (50). SULT1A1-catalyzed sulfation of PhIP results in an unstable sulfoxy metabolite of N-hydroxy-PhIP that readily binds to DNA, or a 4'-SO4-PhIP metabolite that is excreted in the urine (51). While SULT1A1 can then both potentially activate and detoxify PhIP, making it difficult to discern the overall role of SULT1A1 in PhIP-induced carcinogenesis, clearly SULT1A1 activity is influenced by resveratrol and plays a role in DNA adduct formation.
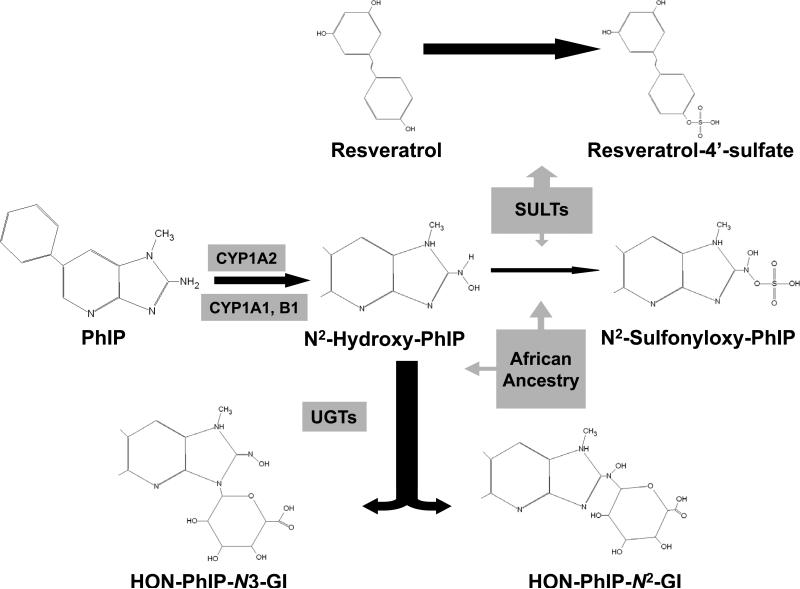
We found that the level of African ancestry was strongly associated with PhIP-DNA adduct levels in non-tumor prostate cells. Nowell et al. found a more pronounced association with increasing SUL1A1 activity and prostate cancer risk in African Americans compared with whites (52). Common genetic variation is known to be associated with gene expression levels (53) and recent work suggests that differences in common genetic variations among African-American and European-American may lead to group-specific alterations in cancer-related pathways that control host response, inflammation, and tumor angiogenesis (54). In light of these findings, we hypothesize that our “African ancestry” variable may be a surrogate for differential gene expression that could influence how PhIP is detoxified by UGTs as well as the effects that resveratrol and SULT1A1 have on N2-hydroxy-PhIP activation (figure 2).
Figure 2.
2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP), is the major genotoxic heterocyclic amine generated from cooking meats at high temperatures. PhIP is metabolized by CYP forms to its active intermediate, N-Hydroxy-PhIP, which can then be detoxified through conjugation by various UDP-glucuronosyltransferase (UGT) forms to PhIP-N2-glucuronide and PhIP-N3-glucuronide. Alternatively, N-Hydroxy-PhIP can undergo sulfation to form N2-Sulfonyloxy-PhIP, which can bind DNA to form carcinogen adducts. Reseveratrol is also a substrate for sulfotransferases, and can competitively inhibit the activation of PhIP via this pathway. We also hypothesize that other genetic factors related to African ancestry interact with the glucuronidation and sulfation pathways of N-Hydroxy-PhIP metabolism. Note that we depict only one branch of the PhIP metabolic pathway in this figure and further note that SULT1A1 can also react with N-hydroxy-PhIP to form 4'-SO4-PhIP that is excreted in urine.
In summary, we have found red wine consumption to be inversely associated with PhIPDNA adduct levels in prostate tumor and non-tumor cells of men with prostate cancer. Since PhIP is known to be a potential prostate carcinogen, red wine may hold renewed promise as a chemopreventive agent for prostate cancer assuming: 1) PhIP exposure that results in DNA damage through adduct formation significantly increases prostate risk; 2) the active agent in red wine, resveratrol, through competitive inhibition with SULT1A1 (or some other yet to be defined mechanism) can significantly reduce the carcinogenic effects of PhIP exposure in human prostate. The first point has yet to be proven, but is an area of active investigation for our group. Previous work in controlled experimental systems (11) and our results suggest the second point is plausible, but given the cross-sectional nature of our results, further studies where the dose and timing of resveratrol consumption with respect to PhIP exposures can be controlled are needed to validate how resveratrol acts on PhIP metabolism in humans. In conclusion, our results suggest that the effects of PhIP exposure can be mitigated by red wine consumption, but before any chemoprevention efforts are directed toward this pathway, the influence of inherited genetic factors that vary by race on PhIP metabolism needs to be better understood.
Acknowledgments
This work was supported by the National Institute of Environmental Health Sciences R01 ES11126 grant awarded to BAR and Department of Defense grant PC050775 awarded to CHB
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Rodriguez C, McCullough ML, Mondul AM, Jacobs EJ, Chao A, Patel AV, et al. Meat consumption among Black and White men and risk of prostate cancer in the Cancer Prevention Study II Nutrition Cohort. Cancer Epidemiol Biomarkers Prev. 2006;15:211–6. doi: 10.1158/1055-9965.EPI-05-0614. [DOI] [PubMed] [Google Scholar]
- 3.Nakai Y, Nelson WG, De Marzo AM. The Dietary Charred Meat Carcinogen 2-Amino-1-Methyl-6-Phenylimidazo[4,5-b]Pyridine Acts as Both a Tumor Initiator and Promoter in the Rat Ventral Prostate. Cancer Res. 2007;67:1378–84. doi: 10.1158/0008-5472.CAN-06-1336. [DOI] [PubMed] [Google Scholar]
- 4.Shirai T, Cui L, Takahashi S, Futakuchi M, Asamoto M, Kato K, et al. Carcinogenicity of 2-amino-1-methyl-6-phenylimidazo [4,5-b]pyridine (PhIP) in the rat prostate and induction of invasive carcinomas by subsequent treatment with testosterone propionate. Cancer Lett. 1999;143:217–21. doi: 10.1016/s0304-3835(99)00128-7. [DOI] [PubMed] [Google Scholar]
- 5.Keating GA, Bogen KT. Estimates of heterocyclic amine intake in the US population. J Chromatogr B Analyt Technol Biomed Life Sci. 2004;802:127–33. doi: 10.1016/j.jchromb.2003.10.047. [DOI] [PubMed] [Google Scholar]
- 6.Kidd LC, Stillwell WG, Yu MC, Wishnok JS, Skipper PL, Ross RK, et al. Urinary excretion of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) in White, African-American, and Asian-American men in Los Angeles County. Cancer Epidemiol Biomarkers Prev. 1999;8:439–45. [PubMed] [Google Scholar]
- 7.Bogen KT, Keating GA, Chan JM, Paine LJ, Simms EL, Nelson DO, et al. Highly elevated PSA and dietary PhIP intake in a prospective clinic-based study among African Americans. Prostate Cancer Prostatic Dis. 2007;10:261–9. doi: 10.1038/sj.pcan.4500941. [DOI] [PubMed] [Google Scholar]
- 8.Tang D, Liu JJ, Bock CH, Neslund-Dudas C, Rundle A, Savera AT, et al. Racial differences in clinical and pathological associations with PhIP-DNA adducts in prostate. Int J Cancer. 2007;121:1319–24. doi: 10.1002/ijc.22806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nozawa H, Nakao W, Takata J, rimoto-Kobayashi S, Kondo K. Inhibition of PhIP-induced mammary carcinogenesis in female rats by ingestion of freeze-dried beer. Cancer Lett. 2006;235:121–9. doi: 10.1016/j.canlet.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 10.Turesky RJ, Richoz J, Constable A, Curtis KD, Dingley KH, Turteltaub KW. The effects of coffee on enzymes involved in metabolism of the dietary carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine in rats. Chem Biol Interact. 2003;145:251–65. doi: 10.1016/s0009-2797(03)00022-x. [DOI] [PubMed] [Google Scholar]
- 11.Dubuisson JG, Dyess DL, Gaubatz JW. Resveratrol modulates human mammary epithelial cell O-acetyltransferase, sulfotransferase, and kinase activation of the heterocyclic amine carcinogen N-hydroxy-PhIP. Cancer Lett. 2002;182:27–32. doi: 10.1016/s0304-3835(02)00061-7. [DOI] [PubMed] [Google Scholar]
- 12.Dawson DA. Beyond black, white and Hispanic: race, ethnic origin and drinking patterns in the United States. J Subst Abuse. 1998;10:321–39. doi: 10.1016/s0899-3289(99)00009-7. [DOI] [PubMed] [Google Scholar]
- 13.Nowell S, Ambrosone CB, Ozawa S, MacLeod SL, Mrackova G, Williams S, et al. Relationship of phenol sulfotransferase activity (SULT1A1) genotype to sulfotransferase phenotype in platelet cytosol. Pharmacogenetics. 2000;10:789–97. doi: 10.1097/00008571-200012000-00004. [DOI] [PubMed] [Google Scholar]
- 14.Ozawa S, Tang YM, Yamazoe Y, Kato R, Lang NP, Kadlubar FF. Genetic polymorphisms in human liver phenol sulfotransferases involved in the bioactivation of N-hydroxy derivatives of carcinogenic arylamines and heterocyclic amines. Chem Biol Interact. 1998;109:237–48. doi: 10.1016/s0009-2797(97)00135-x. [DOI] [PubMed] [Google Scholar]
- 15.Dalhoff K, Buus JK, Enghusen PH. Cancer and molecular biomarkers of phase 2. Methods Enzymol. 2005;400:618–27. doi: 10.1016/S0076-6879(05)00035-2. [DOI] [PubMed] [Google Scholar]
- 16.Ung D, Nagar S. Variable sulfation of dietary polyphenols by recombinant human sulfotransferase (SULT) 1A1 genetic variants and SULT1E1. Drug Metab Dispos. 2007;35:740–6. doi: 10.1124/dmd.106.013987. [DOI] [PubMed] [Google Scholar]
- 17.Girard H, Thibaudeau J, Court MH, Fortier LC, Villeneuve L, Caron P, et al. UGT1A1 polymorphisms are important determinants of dietary carcinogen detoxification in the liver. Hepatology. 2005;42:448–57. doi: 10.1002/hep.20770. [DOI] [PubMed] [Google Scholar]
- 18.Dellinger RW, Chen G, Blevins-Primeau AS, Krzeminski J, Amin S, Lazarus P. Glucuronidation of PhIP and N-OH-PhIP by UDP-glucuronosyltransferase 1A10. Carcinogenesis. 2007;28:2412–8. doi: 10.1093/carcin/bgm164. [DOI] [PubMed] [Google Scholar]
- 19.Rybicki BA, Neslund-Dudas C, Nock NL, Schultz LR, Eklund L, Rosbolt J, et al. Prostate cancer risk from occupational exposure to polycyclic aromatic hydrocarbons interacting with the GSTP1 Ile105Val polymorphism. Cancer Detect Prev. 2006;30:412–22. doi: 10.1016/j.cdp.2006.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patterson RE, Kristal AR, Tinker LF, Carter RA, Bolton MP, Agurs-Collins T. Measurement characteristics of the Women's Health Initiative food frequency questionnaire. Ann Epidemiol. 1999;9:178–87. doi: 10.1016/s1047-2797(98)00055-6. [DOI] [PubMed] [Google Scholar]
- 21.Thompson IM, Tangen C, Goodman P. The Prostate Cancer Prevention Trial: design, status, and promise. World J Urol. 2003;21:28–30. doi: 10.1007/s00345-002-0315-y. [DOI] [PubMed] [Google Scholar]
- 22.Sinha R. An epidemiologic approach to studying heterocyclic amines. Mutat Res. 2002;506-507:197–204. doi: 10.1016/s0027-5107(02)00166-5. [DOI] [PubMed] [Google Scholar]
- 23.Tang D, Liu JJ, Rundle A, Neslund-Dudas C, Savera AT, Bock CH, et al. Grilled meat consumption and PhIP-DNA adducts in prostate carcinogenesis. Cancer Epidemiol Biomarkers Prev. 2007;16:803–8. doi: 10.1158/1055-9965.EPI-06-0973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takahashi S, Tamano S, Hirose M, Kimoto N, Ikeda Y, Sakakibara M, et al. Immunohistochemical demonstration of carcinogen-DNA adducts in tissues of rats given 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP): detection in paraffin-embedded sections and tissue distribution. Cancer Res. 1998;58:4307–13. [PubMed] [Google Scholar]
- 25.Shirai T, Sano M, Tamano S, Takahashi S, Hirose M, Futakuchi M, et al. The prostate: a target for carcinogenicity of 2-amino-1-methyl-6- phenylimidazo[4,5-b]pyridine (PhIP) derived from cooked foods. Cancer Res. 1997;57:195–8. [PubMed] [Google Scholar]
- 26.Xu Z, Kaplan NL, Taylor JA. TAGster: efficient selection of LD tag SNPs in single or multiple populations. Bioinformatics. 2007;23:3254–5. doi: 10.1093/bioinformatics/btm426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng CT, Chen JC, Yeh KT, Wang YF, Hou MF, Lee TP, et al. The relationship among the polymorphisms of SULT1A1, 1A2 and different types of cancers in Taiwanese. Int J Mol Med. 2003;11:85–9. [PubMed] [Google Scholar]
- 28.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9:811–8. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 29.SAS Institute Inc. SAS/STAT Users's Guide, Version 9. SAS Institute Inc.; Cary, NC: 2002. [Google Scholar]
- 30.Gauderman WJ, Murcray C, Gilliland F, Conti DV. Testing association between disease and multiple SNPs in a candidate gene. Genet Epidemiol. 2007;31:383–95. doi: 10.1002/gepi.20219. [DOI] [PubMed] [Google Scholar]
- 31.Hoggart CJ, Shriver MD, Kittles RA, Clayton DG, McKeigue PM. Design and analysis of admixture mapping studies. Am J Hum Genet. 2004;74:965–78. doi: 10.1086/420855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin D, Kaderlik KR, Turesky RJ, Miller DW, Lay JO, Jr., Kadlubar FF. Identification of N-(Deoxyguanosin-8-yl)-2-amino-1-methyl-6-phenylimidazo [4,5-b]pyridine as the major adduct formed by the food-borne carcinogen, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine, with DNA. Chem Res Toxicol. 1992;5:691–7. doi: 10.1021/tx00029a016. [DOI] [PubMed] [Google Scholar]
- 33.Kopp P. Resveratrol, a phytoestrogen found in red wine. A possible explanation for the conundrum of the ‘French paradox’? Eur J Endocrinol. 1998;138:619–20. doi: 10.1530/eje.0.1380619. [DOI] [PubMed] [Google Scholar]
- 34.Walsh PC. Alcohol consumption and risk of prostate cancer in middle-aged men. J Urol. 2005;173:1170. [PubMed] [Google Scholar]
- 35.Schoonen WM, Salinas CA, Kiemeney LA, Stanford JL. Alcohol consumption and risk of prostate cancer in middle-aged men. Int J Cancer. 2005;113:133–40. doi: 10.1002/ijc.20528. [DOI] [PubMed] [Google Scholar]
- 36.Sutcliffe S, Giovannucci E, Leitzmann MF, Rimm EB, Stampfer MJ, Willett WC, et al. A prospective cohort study of red wine consumption and risk of prostate cancer. Int J Cancer. 2007;120:1529–35. doi: 10.1002/ijc.22498. [DOI] [PubMed] [Google Scholar]
- 37.Velicer CM, Kristal A, White E. Alcohol use and the risk of prostate cancer: results from the VITAL cohort study. Nutr Cancer. 2006;56:50–6. doi: 10.1207/s15327914nc5601_7. [DOI] [PubMed] [Google Scholar]
- 38.Schuurman AG, Goldbohm RA, van den Brandt PA. A prospective cohort study on consumption of alcoholic beverages in relation to prostate cancer incidence (The Netherlands). Cancer Causes Control. 1999;10:597–605. doi: 10.1023/a:1008925103542. [DOI] [PubMed] [Google Scholar]
- 39.Bhat KPL, Kosmeder JW, Pezzuto JM. Biological effects of resveratrol. Antioxid Redox Signal. 2001;3:1041–64. doi: 10.1089/152308601317203567. [DOI] [PubMed] [Google Scholar]
- 40.Baur JA, Sinclair DA. Therapeutic potential of resveratrol: the in vivo evidence. Nat Rev Drug Discov. 2006;5:493–506. doi: 10.1038/nrd2060. [DOI] [PubMed] [Google Scholar]
- 41.Boyce A, Doehmer J, Gooderham NJ. Phytoalexin resveratrol attenuates the mutagenicity of the heterocyclic amines 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine and 2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline. J Chromatogr B Analyt Technol Biomed Life Sci. 2004;802:217–23. doi: 10.1016/j.jchromb.2003.10.057. [DOI] [PubMed] [Google Scholar]
- 42.Buonarati MH, Turteltaub KW, Shen NH, Felton JS. Role of sulfation and acetylation in the activation of 2-hydroxyamino-1-methyl-6-phenylimidazo[4,5-b]pyridine to intermediates which bind DNA. Mutat Res. 1990;245:185–90. doi: 10.1016/0165-7992(90)90048-o. [DOI] [PubMed] [Google Scholar]
- 43.Walle T, Hsieh F, DeLegge MH, Oatis JE, Jr., Walle UK. High absorption but very low bioavailability of oral resveratrol in humans. Drug Metab Dispos. 2004;32:1377–82. doi: 10.1124/dmd.104.000885. [DOI] [PubMed] [Google Scholar]
- 44.Malfatti MA, Dingley KH, Nowell-Kadlubar S, Ubick EA, Mulakken N, Nelson D, et al. The urinary metabolite profile of the dietary carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine is predictive of colon DNA adducts after a low-dose exposure in humans. Cancer Res. 2006;66:10541–7. doi: 10.1158/0008-5472.CAN-06-1573. [DOI] [PubMed] [Google Scholar]
- 45.Owens IS, Basu NK, Banerjee R. UDP-glucuronosyltransferases: gene structures of UGT1 and UGT2 families. Methods Enzymol. 2005;400:1–22. doi: 10.1016/S0076-6879(05)00001-7. [DOI] [PubMed] [Google Scholar]
- 46.Nagar S, Remmel RP. Uridine diphosphoglucuronosyltransferase pharmacogenetics and cancer. Oncogene. 2006;25:1659–72. doi: 10.1038/sj.onc.1209375. [DOI] [PubMed] [Google Scholar]
- 47.Ozawa S, Chou HC, Kadlubar FF, Nagata K, Yamazoe Y, Kato R. Activation of 2-hydroxyamino-1-methyl-6-phenylimidazo[4,5-b] pyridine by cDNA-expressed human and rat arylsulfotransferases. Jpn J Cancer Res. 1994;85:1220–8. doi: 10.1111/j.1349-7006.1994.tb02933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dooley TP, Haldeman-Cahill R, Joiner J, Wilborn TW. Expression profiling of human sulfotransferase and sulfatase gene superfamilies in epithelial tissues and cultured cells. Biochem Biophys Res Commun. 2000;277:236–45. doi: 10.1006/bbrc.2000.3643. [DOI] [PubMed] [Google Scholar]
- 49.Muckel E, Frandsen H, Glatt HR. Heterologous expression of human N-acetyltransferases 1 and 2 and sulfotransferase 1A1 in Salmonella typhimurium for mutagenicity testing of heterocyclic amines. Food Chem Toxicol. 2002;40:1063–8. doi: 10.1016/s0278-6915(02)00032-7. [DOI] [PubMed] [Google Scholar]
- 50.Al-Buheissi SZ, Patel HR, Meinl W, Hewer A, Bryan RL, Glatt H, et al. N-Acetyltransferase and sulfotransferase activity in human prostate: potential for carcinogen activation. Pharmacogenet Genomics. 2006;16:391–9. doi: 10.1097/01.fpc.0000204998.22301.09. [DOI] [PubMed] [Google Scholar]
- 51.Chou HC, Lang NP, Kadlubar FF. Metabolic activation of N-hydroxy arylamines and N-hydroxy heterocyclic amines by human sulfotransferase(s). Cancer Res. 1995;55:525–9. [PubMed] [Google Scholar]
- 52.Nowell S, Ratnasinghe DL, Ambrosone CB, Williams S, Teague-Ross T, Trimble L, et al. Association of SULT1A1 phenotype and genotype with prostate cancer risk in African-Americans and Caucasians. Cancer Epidemiol Biomarkers Prev. 2004;13:270–6. doi: 10.1158/1055-9965.epi-03-0047. [DOI] [PubMed] [Google Scholar]
- 53.Zhang W, Duan S, Kistner EO, Bleibel WK, Huang RS, Clark TA, et al. Evaluation of genetic variation contributing to differences in gene expression between populations. Am J Hum Genet. 2008;82:631–40. doi: 10.1016/j.ajhg.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martin DN, Boersma BJ, Yi M, Reimers M, Howe TM, Yfantis HG, et al. Differences in the tumor microenvironment between African-American and European-American breast cancer patients. PLoS ONE. 2009;4:e4531. doi: 10.1371/journal.pone.0004531. [DOI] [PMC free article] [PubMed] [Google Scholar]