Abstract
Tyrosyl radicals (Y•s) are prevalent in biological catalysis and are formed under physiological conditions by the coupled loss of both a proton and an electron. Fluorotyrosines (FnYs, n=1–4) are promising tools for studying the mechanism of Y• formation and reactivity, as their pKas and peak potentials span four units and 300 mV, respectively, between pH 6–10. In this manuscript, we present the directed evolution of aminoacyl-tRNA synthetases (aaRS) for 2,3,5-trifluorotyrosine (2,3,5-F3Y) and demonstrate their ability to charge an orthogonal tRNA with a series of FnYs, while maintaining high specificity over Y. An evolved aaRS is then used to site-specifically incorporate FnYs into the two subunits (α2 and β2) of E. coli class Ia ribonucleotide reductase (RNR), an enzyme that employs stable and transient Y•s to mediate long-range, reversible radical hopping during catalysis. Each of four conserved Ys in RNR is replaced with FnY(s) and the resulting proteins isolated in good yields. FnYs incorporated at position 122 of β2, the site of a stable Y• in the wt RNR, generate long-lived FnY•s that are characterized by EPR spectroscopy. Furthermore, we demonstrate that the radical pathway in the mutant Y122(2,3,5)F3Y-β2 is energetically and/or conformationally modulated such that the enzyme retains its activity, but that a new on-pathway Y• can accumulate. The distinct EPR properties of the 2,3,5-F3Y• facilitate spectral subtractions that make detection and identification of new Y•s straightforward.
Examples of amino acid radicals participating in charge transfer reactions are prevalent in nature.1 Of these, tyrosyl radicals (Y•s) have been shown to mediate a number of key metabolic transformations, including O2 evolution, fatty acid oxidation, peroxide disproportionation, and prostaglandin synthesis.2 The thermodynamics of Y oxidation require that proton transfer (PT) accompanies electron transfer (ET) at physiological pH, a process that may occur by either a stepwise or coupled (PCET) mechanism. To investigate the mechanism of Y• formation and Y•-mediated catalysis in metalloenzymes, a method by which the native Y• may be subtly perturbed is necessary. A series of N-acetyl-fluorotyrosinamides (N-Ac-FnY-NH2s) have been characterized in solution and found to have phenolic pKas from 5.6–8.4 and peak potentials ranging from 705–968 mV.3,4 This series of Y analogs provides a means of systematically modulating the chemical properties governing both the PT and ET events.
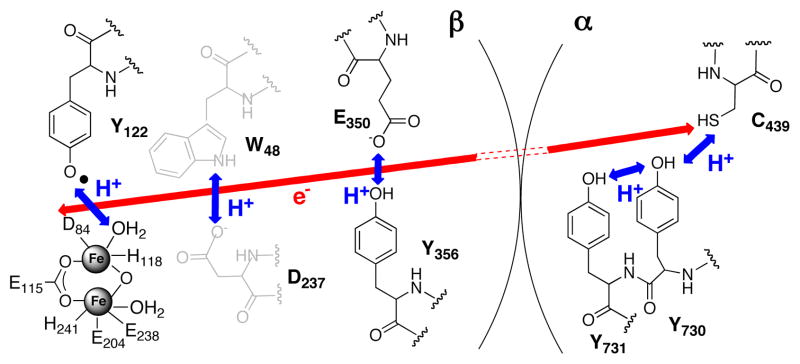
Ribonucleotide reductase (RNR) catalyzes the formation of all four 2′-deoxynucleotides from the corresponding nucleoside diphosphates (NDPs) by a mechanism involving protein and nucleotide radicals.1 In the class Ia RNRs, a stable difer-ric-Y• cofactor in the β2 subunit initiates catalysis by transiently oxidizing a cysteine at the active site in the α2 subunit. The mechanism of radical initiation has been best studied in the E. coli RNR, and is believed to involve an unprecedented mechanism of reversible long-range (>35 Å) PCET along a pathway of absolutely conserved redox active amino acids: Y122• ↔ [W48?] ↔ Y356 in β2 to Y731↔ Y730↔ C439 in α2 (Scheme 1).5,6 This hypothesis has been tested by the site-specific incorporation of more than a half-dozen unnatural amino acids (UAAs) into positions along the pathway, using both expressed protein ligation (EPL)7 and in vivo nonsense suppression8,9 techniques. Studies in which a series of FnYs were incorporated at position 356 of β2 by EPL were particularly informative, as pH rate profiles of the resulting proteins indicated that this residue was redox active, that proton and electron movement is orthogonal at this position (Scheme 1) and that an ordered hydrogen bonding network involving the Y356 phenol is not necessary for catalysis.10,11
Scheme 1.
Working mechanism for long-range PCET pathway in E. coli class Ia RNR
The current hypothesis holds that protons (blue arrows) move orthogonally to the electron (red arrow) in β2 and co-linearly with the electron in α2. The mechanism across the α/β interface is unknown. There is no direct evidence that W48 and its putative H+ acceptor, D237, participate in long-range PCET during turnover.
FnYs incorporated in place of other conserved Ys along the pathway would be comparably informative. However, the EPL technology used to generate Y356FnY-β2s can not be readily extended to positions in the protein interior. While other strategies for incorporation of FnYs exist, namely global incorporation4,12 or site-specific insertion of photocaged FnYs,13 these techniques are applicable to mono- and di-FnYs only and are minimally useful in RNR, as the former lacks specificity and the latter has complications arising from photolysis.14
Herein, a robust method for the in vivo site-specific incorporation of a series of FnYs is reported. This method requires evolution of an orthogonal aaRS-tRNA pair and has been successfully used to incorporate over 70 UAAs into E. coli, S. cervevisiae, and mammalian proteins in response to a nonsense (or “blank”) codon.15,16 Originally, we sought to evolve an aaRS specific for 2,3,5-trifluorotyrosine (2,3,5-F3Y, 1), as previous data suggested that its redox potential is sufficiently increased relative to Y at appropriate pHs such that its incorporation at position 356 of β2 changes the rate-determining step in RNR from a conformational change17 to PCET.11 Three fluorine substituents together with the low phenolic pKa (6.4) were expected to allow evolution of an RS selective for the phenolate of 1 over the Y phenol.
To selectively incorporate 1 into proteins, an orthogonal Methanococcus jannaschii aaRS-tRNA pair that suppresses the amber stop codon (TAG) was evolved to uniquely encode this amino acid by a double sieve selection process based on cellular viability.16 A library of TyrRS mutants was generated by randomizing eight positions near the active site, based on the known structure of Tyr-bound MjTyrRS,18 including those residues closest to the aromatic ring (Y32, L65, and H70) and those involved in H-bonding to the Y phenol (Y32 and D158). The library was subjected to three positive and negative selection cycles, after which individual candidates were isolated and assessed for viability in the presence and absence of 1 at different chloramphenicol (Cm) concentrations. The selection resulted in two primary clones, E3 (Y32L, L65G, H70N, F108F, Q109Q, D158S, I159Y, L162H) and E11 (Y32H, L65Y, H70G, F108Y, Q109A, D158N, I159I, L162R), that conferred Cm resistance (>150 μg/mL) only in the presence of 1 (Figure S1 and Table S1 of Supporting Information). Given the mutations and the structure,18 we tried to understand the basis for recognition of 1 by the aaRSs (Figure S2 of SI). We had predicted that the negative charge on the phenolate of 1 would be the basis for discrimination against Y, and anticipated the introduction of positively charged side chains in the mutants. E3 contains a L162H mutation, but the protonation state of this residue is unknown. E11 contains a single basic side chain (L162R); however, a crystal structure of the p-acetylphenylalanine RS containing the same mutation revealed that the R side chain was directed away from the active site.19 Y32 and/or D158 are mutated in both 2,3,5-F3Y-RSs to residues that could conceivably behave as H-bond donors to the phenolate of 1. The substitution of Gly at either position 65 (E3) or 70 (E11) may enlarge the substrate binding pocket. Thus, there exists minimal evidence indicating that the observed discrimination occurs on the basis of the phenolate. Instead, it appears that the fluorine substitutents provide the key recognition elements. The introduction and retention of many polar side chains provide opportunities for the three F atoms to participate in multipolar interactions of the nature C-F…H-N/O or C-F…C=O.20 Given the sensitivity of these interactions to distance and geometry, it is impossible to speculate on the specific atoms involved without a high resolution structure. Crystallization attempts are underway to better under-stand the basis of F-protein interactions in the 2,3,5-F3Y-RSs. The nature of these interactions is of general interest, given the large number of fluorinated pharmaceuticals.20
The aaRS hits were cloned into the previously described pEVOL system21 for enhanced expression of proteins containing 1. We confirmed the selective incorporation of 1 into proteins by aaRSs E3 and E11 using pET-GFPY151TAG, a plasmid that encodes a C-terminally His6 tagged GFP.15 Full-length, fluorescent protein is observed only when the TAG codon is suppressed by an amino-acylated tRNA. E3 afforded 10 mg/L of purified GFP, while E11 yielded 4 mg/L. Both aaRSs incorporated 1 into GFP with an observed mass of 27759 Da (expected: 27758 Da, Figure S3 of SI). In the absence of 1, low levels of phenylalanine incorporation could be detected for E3; however, this background incorporation is not detected in the presence of 1 (a property observed with several previously evolved aaRSs).22a No background incorporation was detected for E11 in the absence of 1.
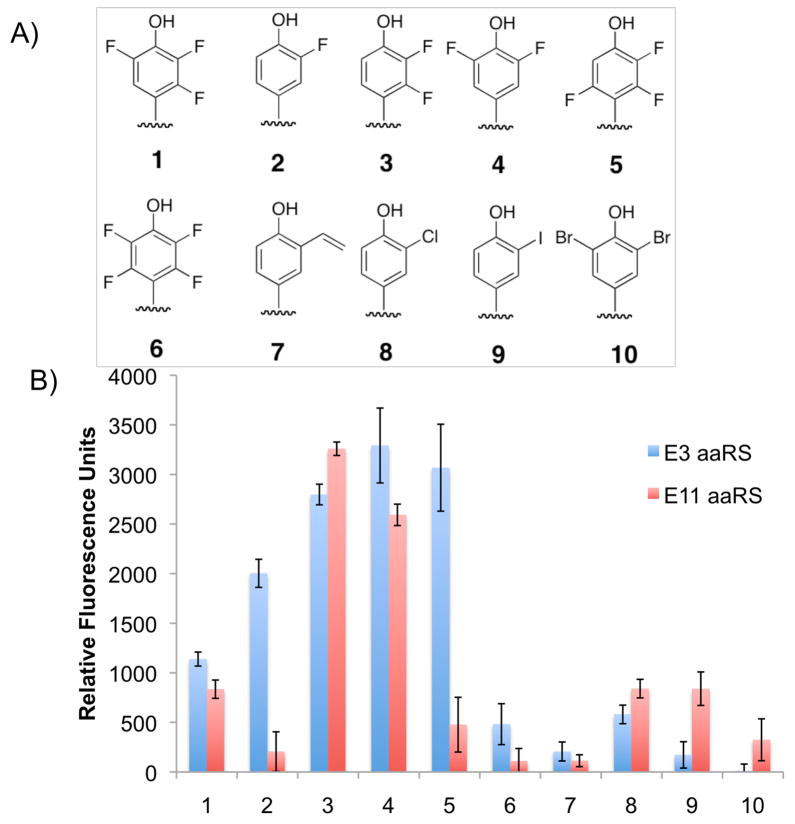
Recent experiments have shown that some aaRSs exhibit a degree of polyspecificity.22 This feature arises from the lack of other UAAs in the selection medium to eliminate synthetases that can recognize them. To determine if these 2,3,5-F3Y aaRSs are polyspecific, they were screened against a diverse library of UAAs using a GFP fluorescence assay.22a Of the ~80 UAAs in the library, both aaRSs were capable of aminoacylating 3-fluorotyrosine (2) and 3-chlorotyrosine (8) (Figure S4 of SI). The results of this first screen indicated the aaRSs exhibit some polyspecificity for other halogenated Ys, prompting a second GFP screen on Ys with a narrower range of substitutions (including 3–7, 9, 10). The evolved aaRSs were capable of incorporating all members of the FnY series (3-FY (2), 2,3-F2Y (3), 3,5-F2Y (4), 2,3,6-F3Y (5), and 2,3,5,6-F4Y (6)), as well as several other halogenated analogs (Figure 1). The incorporation of each UAA was confirmed by LC/MS analysis of the mutant GFPs (Figure S5 of SI). Interestingly, several of the analogs (2–5) were incorporated into GFP at higher levels than 1, thus our evolved aaRSs are henceforth called “FnY-RSs.” LC/MS analysis of E. coli lysate indicated that the reduced cellular uptake is one reason for the low incorporation of 6 relative to the other FnYs.
Figure 1.
Polyspecificity of FnY-RSs. A) Y analogs that screened positive for GFP incorporation. B) GFP fluorescence assay of the two evolved aaRSs (blue = E3; red = E11) based on suppression of GFPY151TAG in the presence of 1 mM UAA.
The FnY-RS E3 was then used to incorporate FnYs into positions 730 and 731 of the RNR α2 subunit (Scheme 1). These residues have recently been shown to be sites of transient Y• formation during radical propagation by substitution of 3-aminotyrosine (NH2Y)8 and 3-nitrotyrosine (NO2Y)9 at these positions using similar technology. Cells were transformed with pEVOL-FnY-RS-E3 and pET-nrdAY730TAG, encoding for α with an N-His6 tag and a stop codon at the position of Y730,23 and grown in the presence of 1. SDS PAGE analysis of whole cells after induction revealed a ~1:1 ratio of full-length and truncated proteins (Figure S6 of SI). The desired protein, Y730(2,3,5)F3Y-α2, was isolated by Ni-NTA chromatography to give 10 mg of protein/g cell paste (~50 mg/L culture). An analogous protocol was used to incorporate 3, 4 and 5 into position 730 of α2. The Y730FnY-α2s were purified in yields of 2.5–3 mg/g (Table S2 of SI). Consistent with the GFP data (Figure 1), the yield of protein containing 6 was reduced 10-fold relative to other FnYs. The 730-optimized protocol was then used to incorporate 1 and 4 into position 731 of α2 with comparable isolated protein yields. Finally, the expression protocol was used with minor modification to incorporate 1 at position 356 of β2 and the protein purified (2 mg/g).
Incorporation of FnYs into position 122 of β2, the site of the stable Y• (t1/2 ~ 4 days, 4 °C) in the wt, was of mechanistic interest. The reduction of Y122• occurs concomitant with oxidation of C439 in α2 (Scheme 1), yet the intermediates of radical propagation in the wt enzyme are kinetically masked by rate-limiting conformational changes.17 Thus, changes to Y122• or any of the pathway residues are undetectable by rapid biophysical techniques. Furthermore, even in those mutant RNRs in which radicals can be trapped and/or conformational gating can be partially lifted, observation and identification of new radicals by X-band EPR is complicated by their extensive spectral overlap. Recently, site-specific insertion of NO2Y at position 122 of β2 was described. Reaction of apo-Y122NO2Y-β2 with Fe2+ and O2 afforded assembly of a diferric-NO2Y122• cluster.24 Use of the potent oxidant NO2Y122• as a radical initiator decoupled ET from the rate-limiting conformational change and allowed the first detection of a transient Y• on the pathway.24,25 However, the high redox potential of NO2Y relative to Y (Δ ~200 mV, pH 7) introduced complications to the system, namely the short half-life of NO2Y122• (t1/2 ~ 40 s, 25 °C) and the mutant’s inability to perform multiple turnovers.
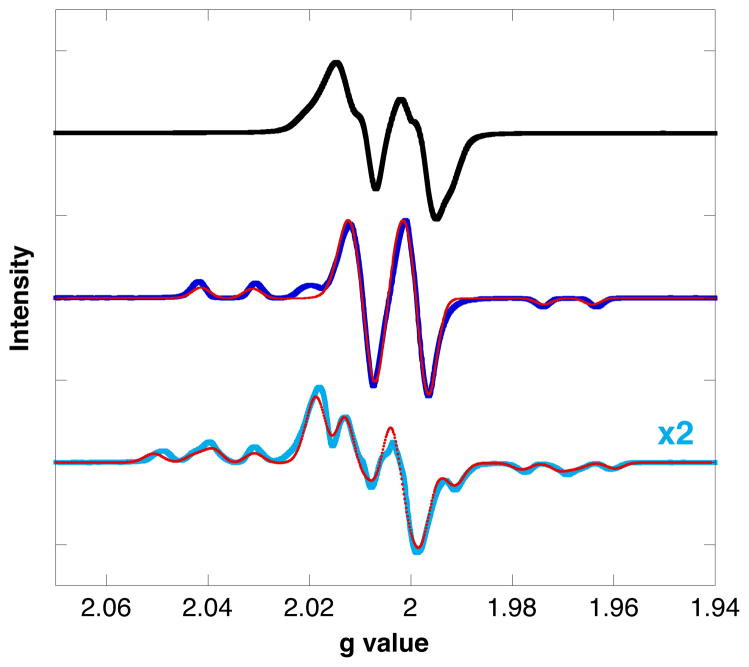
Given these results, we sought to modulate the properties of position 122 more subtly using FnYs. Given the known perturbation of the phenolic pKa at this position (>2.5 units),9 UAAs 1 and 4 should be protonated at physiological pH and are predicted to be <10 mV harder and ~50 mV easier to oxidize than Y•, respectively. Using pEVOL-FnY-RS-E3 and pBAD-nrdBY122TAG, encoding untagged β2 with a stop codon at position 122,24 the proteins Y122(2,3,5)F3Y-β2 and Y122(3,5)F2Y-β2 were expressed and isolated by anion-exchange chromatography in yields ≥30 mg/g. The two mutants were characterized by UV-VIS and EPR spectroscopy to determine whether they contained any FnY•. The as-isolated wt subunit contains 1.2 Y•/β2, with a sharp, diagnostic absorbance at 411 nm and a doublet X-band EPR spectrum dominated by coupling to one of the β-methylene protons (Figure 2, black). As-isolated Y122(3,5)F2Y-β2 and Y122(2,3,5)F3Y-α2 had λmaxs at 396 and 404 nm, respectively, consistent with the values determined for 3,5-F2Y• and 2,3,5-F3Y• generated by photooxidation of benzophenone-FnY-OMe dipeptides.4
Figure 2.
X-band EPR spectra of E. coli wt Y122• (black), 3,5-F2Y122• (dark blue), and 2,3,5-F3Y• (cyan). Spectra were recorded at 77 K and normalized for radical concentration. Simulations (red) were performed in EasySpin.26
X-band EPR analysis of the purified proteins at 77 K indicated that the as-isolated 3,5-F2Y and 2,3,5-F3Y mutants contained 0.2 and 0.02 FnY122•/β2, suggesting that the FnY122•s are inherently less stable that the native Y122•. Higher radical content was obtained for F3Y122 mutants (0.4–0.9 F3Y122•/β2) by in vitro reconstitution of the cofactor.27 Both the 3,5-F2Y122• and the 2,3,5-F3Y122• were stable for more than 30 min at 4 °C. The EPR spectra of the putative 3,5-F2Y• and 2,3,5-F3Y• at position 122 of β2 (Figure 2, blue and cyan) are quite distinct from the EPR spectra of FnY•s generated on the corresponding free amino acids by UV-photolysis.4 The spectra of the latter are relatively featureless, with line broadening arising from a large distribution of dihedral angles for the β-methylene protons. In contrast, structural constraints imposed by the protein give rise to FnY122• spectra with well-defined couplings to the fluorine nuclei (150–180 MHz) and one of the β-methylene protons (36–48 MHz). Spectral simulations (Figure 2, red, and Table S3) were conducted using the parameters for 3-FY122• as an initial reference.4 High-field EPR/ENDOR analysis of the FnY•s is ongoing (Myers, Minnihan, Stubbe, and Britt).
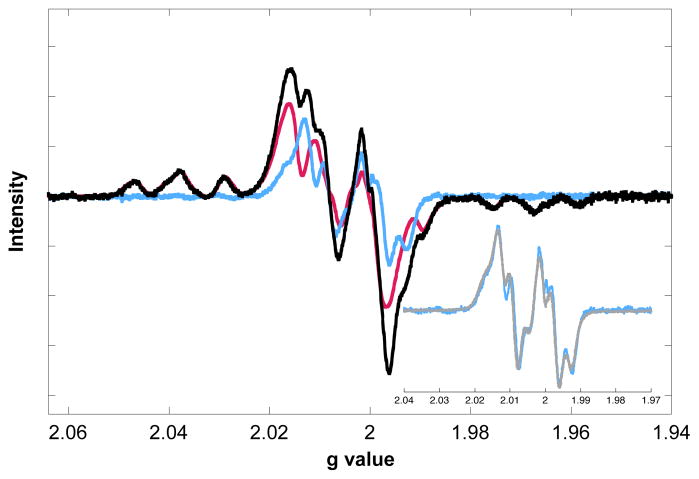
Nucleotide reductase assays of Y122(2,3,5)F3Y-β2 revealed the mutant has 30% the wt β2 activity (scaled for radical). This result suggests that introduction of 2,3,5-F3Y at position 122 slightly perturbs radical propagation energetics and/or the conformational changes that gate it, but not to the extent that it prevents multiple catalytic turnovers. The reactivity of this mutant was further investigated by EPR spectroscopy. Y122(2,3,5)F3Y-β2 was mixed on ice with wt-α2, CDP (substrate) and ATP (allosteric effector) under single-turnover conditions in an EPR tube and quenched in a dry ice/acetone bath at 20 sec. The spectrum of the reaction mixture (Figure 3, black) is a composite of two species: 2,3,5-F3Y122• (pink) and a new radical (blue). Subtraction of the former from the composite was conducted using the fluorine hyperfine couplings, which occur in unobstructed regions of the spectrum. Calculating the double integral intensities of the two species indicates that the new radical constitutes 25% of the total spin. The new radical is assigned as Y356• on the basis of additional EPR experiments in which the pathway was blocked at specific positions by insertion of a redox-inert F. Indeed, when Y122(2,3,5)F3Y-β2 was reacted with Y731F-α2, a very similar new radical was formed in identical yield (Figure S7 of SI). A reaction with the double mutant Y122(2,3,5)F3Y/Y356F-β2 and wt-α2 gave no evidence of a new radical. The assignment is further substantiated by its strong similarity to the radical observed when NO2Y122• is used as a radical initiator (Figure 3, inset).24 Extensive evidence suggests that the new radical in the NO2Y mutant is primarily located at Y356.25
Figure 3.
EPR spectrum of the reaction of Y122(2,3,5)F3Y-β2, wt-α2, CDP, and ATP quenched at 20 s. Subtraction of the 2,3,5-F3Y122• contribution (pink) from the reaction spectrum (black) gives a new radical (blue). Inset: Overlay of the new radical with Y356• formed by Y122NO2Y-β2 (gray).
While it is necessary to determine the kinetic competence of the new radical, observation of a putative Y356• is promising in two regards. First, the ability to observe an on-pathway Y• (while maintaining significant catalytic activity) will have important mechanistic implications. Investigations are under-way to determine what properties of F3Y allow observation of this new radical. We anticipate deriving additional insight into the long-range PCET mechanism by modulating the driving force and phenolic pKa at positions on the pathway in a step-wise fashion. Similarly, FnYs may be utilized for mechanistic studies of ET and/or PT in model proteins and native enzymes.
Additionally, this result demonstrates the utility of FnY•s as spectroscopic handles. The width of the FnY• X-band EPR spectra and the presence of distinct fluorine hyperfine couplings, which give rise to features in the low and high field spectral regions, allow accurate subtraction of FnY•s from other g ~2 radicals (e.g. Y•, [β-2H2]Y•, NH2Y•, NO2Y•).8,24 Thus, FnY•s afford the first opportunity to identify low concentrations of radical species that would otherwise go undetected in complicated reaction mixtures.25 FnY•s may prove similarly useful in high-field EPR and ENDOR studies. For proteins that do not use radicals, FnYs may be incorporated as probes of structure and/or local environment for 19F-NMR studies.28 Thus, application of FnYs as spectroscopic probes may be extended to a broad biophysical research community.
Supplementary Material
Acknowledgments
This work was supported by NIH grant GM29595 (J.S.) and Grant DE-FG03-00ER46051 from the Division of Materials Sciences, Department of Energy (P.G.S.). D.D.Y. acknowledges a NIH Ruth L. Kirchstein Postdoctoral Fellowship F32CA144213. We thank the Gao Lab (Boston College) for providing Fmoc-F4Y.
Footnotes
Author Contributions
The manuscript was written through contributions of all authors.
ASSOCIATED CONTENT
Supporting Information. Methods; FnY-RS selection; structure of MjTyrRS active site; GFP polyspecificity screen; LC-MS of FnY-GFPs; expression/purification of FnY-RNRs; EPR reaction spectra for Y122(2,3,5)F3Y-β2 with blocked mutants. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Stubbe J, van der Donk WA. Chem Rev. 1998;98:705. doi: 10.1021/cr980059c. [DOI] [PubMed] [Google Scholar]
- 2.a) Barry BA, el-Deeb MK, Sandusky PO, Babcock GT. J Biol Chem. 1990;265:20139. [PubMed] [Google Scholar]; b) Gupta A, Mukherjee A, Matsui K, Roth JP. J Am Chem Soc. 2008;130:11274. doi: 10.1021/ja8042273. [DOI] [PubMed] [Google Scholar]; c) Zhao X, Suarez J, Khajo A, Yu S, Metlitsky L, Magliozzo RS. J Am Chem Soc. 2010;132:8268. doi: 10.1021/ja103311e. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Tsai AL, Kulmacz RJ. Arch Biochem Biophys. 2010;493:103. doi: 10.1016/j.abb.2009.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim K, Cole PA. J Am Chem Soc. 1998;120:6851. [Google Scholar]
- 4.Seyedsayamdost MR, Reece SY, Nocera DG, Stubbe J. J Am Chem Soc. 2006;128:1569. doi: 10.1021/ja055926r. [DOI] [PubMed] [Google Scholar]
- 5.Uhlin U, Eklund H. Nature. 1994;370:533. doi: 10.1038/370533a0. [DOI] [PubMed] [Google Scholar]
- 6.Stubbe J, Nocera DG, Yee CS, Chang MCY. Chem Rev. 2003;103:2167. doi: 10.1021/cr020421u. [DOI] [PubMed] [Google Scholar]
- 7.a) Yee CS, Seyedsayamdost MR, Chang MCY, Nocera DG, Stubbe J. Biochemistry. 2003;42:14541. doi: 10.1021/bi0352365. [DOI] [PubMed] [Google Scholar]; b) Chang MCY, Yee CS, Nocera DG, Stubbe J. J Am Chem Soc. 2004;126:16702. doi: 10.1021/ja044124d. [DOI] [PubMed] [Google Scholar]; c) Seyedsayamdost MR, Stubbe J. J Am Chem Soc. 2006;128:2522. doi: 10.1021/ja057776q. [DOI] [PubMed] [Google Scholar]
- 8.Seyedsayamdost MR, Xie J, Chan CT, Schultz PG, Stubbe J. J Am Chem Soc. 2007;129:15060. doi: 10.1021/ja076043y. [DOI] [PubMed] [Google Scholar]
- 9.Yokoyama K, Uhlin U, Stubbe J. J Am Chem Soc. 2010;132:8385. doi: 10.1021/ja101097p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yee CS, Chang MCY, Ge J, Nocera DG, Stubbe J. J Am Chem Soc. 2003;125:10506. doi: 10.1021/ja036242r. [DOI] [PubMed] [Google Scholar]
- 11.Seyedsayamdost MR, Yee CS, Reece SY, Nocera DG, Stubbe J. J Am Chem Soc. 2006;128:1562. doi: 10.1021/ja055927j. [DOI] [PubMed] [Google Scholar]
- 12.Rappaport F, Boussac A, Force DA, Peloquin J, Brynda M, Sugiura M, Un S, Britt RD, Diner BA. J Am Chem Soc. 2009;131:4425. doi: 10.1021/ja808604h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilkins BJ, Marionni S, Young DD, Liu J, Wang Y, Di Salvo ML, Deiters A, Cropp TA. Biochemistry. 2010;49:1557. doi: 10.1021/bi100013s. [DOI] [PubMed] [Google Scholar]
- 14.Incorporation of o-nitrobenzyl-FnYs was previously attempted at position 730 of α2. The location of this residue in the protein interior resulted in inefficient decaging. Additionally, recovery of protein activity after photolysis was poor, likely the result of side reactions with neighboring redox-active amino acids.
- 15.Young TS, Schultz PG. J Biol Chem. 2010;285:11039. doi: 10.1074/jbc.R109.091306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.a) Xie J, Schultz PG. Methods. 2005;36:227. doi: 10.1016/j.ymeth.2005.04.010. [DOI] [PubMed] [Google Scholar]; b) Liu CC, Schultz PG. Annu Rev Biochem. 2010;79:413. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 17.Ge J, Yu G, Ator MA, Stubbe J. Biochemistry. 2003;42:10071. doi: 10.1021/bi034374r. [DOI] [PubMed] [Google Scholar]
- 18.Kobayashi T, Nureki O, Ishitani R, Yaremchuk A, Tukalo M, Cusack S, Sakamoto K, Yokoyama S. Nat Struct Biol. 2003;10:425. doi: 10.1038/nsb934. [DOI] [PubMed] [Google Scholar]
- 19.Turner JM, Graziano J, Spraggon G, Schultz PG. J Am Chem Soc. 2005;127:14976. doi: 10.1021/ja0549042. [DOI] [PubMed] [Google Scholar]
- 20.a) Muller K, Faeh C, Diederich F. Science. 2007;317:1881. doi: 10.1126/science.1131943. [DOI] [PubMed] [Google Scholar]; b) Zhou P, Zou J, Tian F, Shang Z. J Chem Inf Model. 2009;49:2344. doi: 10.1021/ci9002393. [DOI] [PubMed] [Google Scholar]
- 21.Young TS, Ahmad I, Yin JA, Schultz PG. J Mol Biol. 2010;395:361. doi: 10.1016/j.jmb.2009.10.030. [DOI] [PubMed] [Google Scholar]
- 22.a) Young DD, Young TS, Jahnz M, Ahmad I, Spraggon G, Schultz PG. Biochemistry. 2011;50:1894. doi: 10.1021/bi101929e. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Miyake-Stoner SJ, Refakis CA, Hammill JT, Lusic H, Hazen JL, Deiters A, Mehl RA. Biochemistry. 2010;49:1667. doi: 10.1021/bi901947r. [DOI] [PubMed] [Google Scholar]
- 23.Minnihan EC, Seyedsayamdost MR, Uhlin U, Stubbe J. J Am Chem Soc. 2011;133:9430. doi: 10.1021/ja201640n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yokoyama K, Uhlin U, Stubbe J. J Am Chem Soc. 2010;132:15368–79. doi: 10.1021/ja1069344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yokoyama K, Smith AA, Corzilius B, Griffin RG, Stubbe J. Submitted. [Google Scholar]
- 26.Stoll S, Schweiger A. J Magn Reson. 2006;178:42. doi: 10.1016/j.jmr.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 27.Bollinger JM, Jr, Tong WH, Ravi N, Huynh BH, Edmondson DE, Stubbe J. Methods Enzymol. 1995;258:278. doi: 10.1016/0076-6879(95)58052-2. [DOI] [PubMed] [Google Scholar]
- 28.Li C, Wang GF, Wang Y, Creager-Allen R, Lutz EA, Scronce H, Slade KM, Ruf RA, Mehl RA, Pielak GJ. J Am Chem Soc. 2010;132:321. doi: 10.1021/ja907966n. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.