Abstract
S-adenosylmethionine (SAM) is a versatile small molecule used in many biological reactions. This review focuses on the mechanistic consideration of SAM-dependent methylation and 3-amino-3-carboxypropylation reactions. Special emphasis is given to methylation and 3-amino-3-carboxypropylation of carbon atoms, for which both nucleophilic mechanisms and radical mechanisms are used, depending on the specific enzymatic reactions. What is the logic behind Nature’s choice of different reaction mechanisms? Here I aim to rationalize the choice of different reaction mechanisms in SAM-dependent alkylation reaction by analyzing a few enzymatic reactions in depth. These reactions include SAM-dependent cyclopropane fatty acid synthesis, DNA cytosine methylation, RNA adenosine C2 and C8 methylation, and 3-amino-3-carboxypropylation involved in diphthamide biosynthesis and wybutosine biosynthesis.
Introduction on SAM-dependent alkylation reactions: methylation and 3-amino-3-carboxypropylation
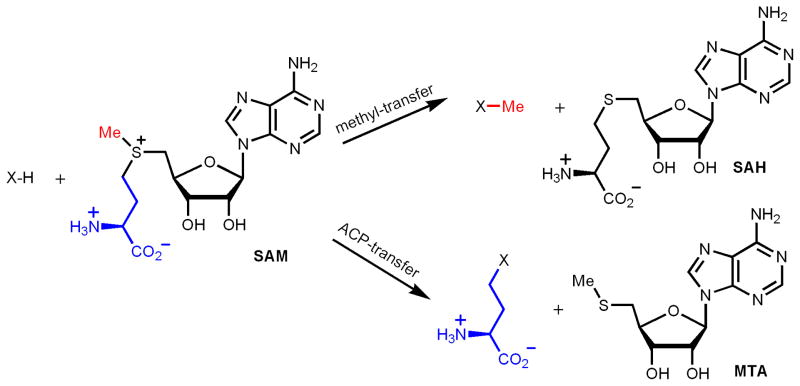
S-adenosylmethionine (SAM) is a versatile molecule used in many biological reactions. It is a commonly used methyl donor in numerous biologically important methylation reactions, including DNA methylation, RNA methylation, and protein methylation. The 3-amino-3-carboxypropyl (ACP) group of SAM can also be transferred to different acceptor molecules, such as RNA and proteins. In addition to serving as a methyl donor and an ACP donor, SAM is also involved in several other types of group transfer reactions. This has been summarized nicely in a review paper by Fontecave, Atta and Mulliez[1]. Here, I will limit the discussion to the methyl-transfer and ACP-transfer reactions of SAM (Figure 1), focusing on the mechanistic considerations for such enzymatic transformations.
Figure 1.
SAM-dependent methyl-transfer and ACP-transfer reactions.
The positively charged sulfonium ion in SAM makes the three carbon atoms that are bonded to the sulfur atom prone to attack by nucleophiles. Indeed, many of the methyl-transfer and ACP-transfer reactions involve nucleophiles attacking the methyl and ACP carbons, respectively. However, it has recently been demonstrated that several methyl-transfer and ACP-transfer reactions use more complicated radical mechanisms. One example is the methylation of C2 and C8 of an adenine ring in ribosomal RNA in bacteria[2–4]. The other example is the ACP-transfer to a histidine residue in translation elongation factor 2 (EF2) in a step required for diphthamide biosynthesis[5, 6]. These new mechanistic findings raised an interesting question: In SAM-dependent methyl-transfer or ACP-transfer reactions, when should a simple nucleophilic mechanism be used, and when should a more sophisticated radical mechanism be used? What is the logic behind Nature’s choice of chemistry in these SAM-dependent alkylation reactions?
SAM-dependent N- or O-alkylation: nucleophilic mechanism
When the alkyl acceptor is a heteroatom (most commonly N, O, and S), the methyl or ACP-transfer reactions occur via simple nucleophilic mechanism. This is because these heteroatoms are nucleophilic due to the presence of lone pair electrons. Methylation of heteroatoms is very abundant in biological systems [7]. One of the examples is protein lysine methylation, which is known to occur via this simple nucleophilic mechanism. Structures of protein lysine methyltransferases showed that the binding of SAM and the protein substrate on the enzyme is consistent with an SN2 type of mechanism [8–10].
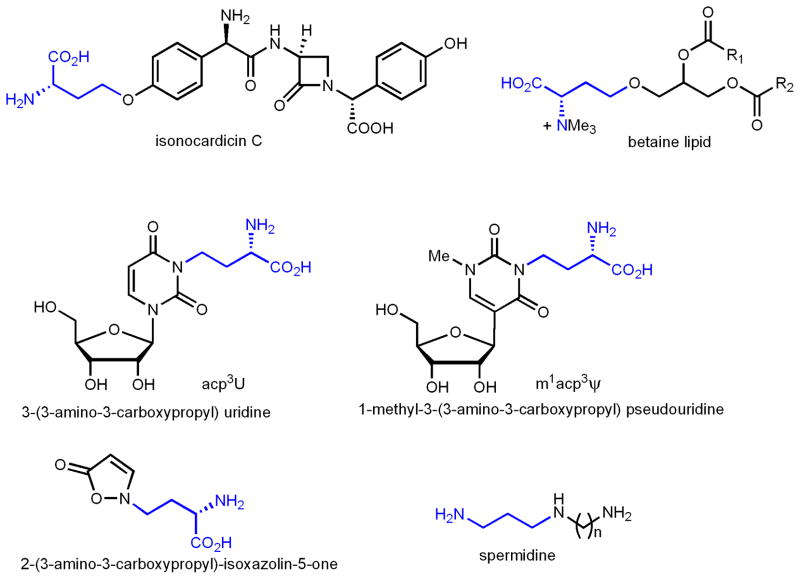
SAM-dependent ACP-transfer reactions are relatively rare compared with methyl-transfer reactions. ACP-transfer to both O and N has been known (Figure 2). Examples of O-ACP transfer include the biosynthesis of a bacterial betaine lipid diacylglyceryl-N, N, N-trimethylhomoserine[11], and the biosynthesis of isonocardicin, a beta-lactam type antibiotics[12]. Examples of N-ACP transfer are mainly from RNA modifications, such as 3-(3- amino-3-carboxypropyl) uridine [13–16] or acp3U and 1-methyl-3-(3-amino-3-carboxypropyl) pseudouridine or m1acp3 Ψ̃ [17, 18]. An ACP-transfer reaction was also demonstrated in the biosynthesis of 2-(3-amino-3-carboxypropyl)-isoxazolin-5-one, a neurotoxic amino acid from Lathyrus odoratus[19]. A reaction that is closely related to the ACP-transfer reaction is found in polyamine biosynthesis, in which SAM is decarboxylated to decarboxy-SAM and then the 3-aminopropyl group is transferred[20]. For many of the N- and O-ACP transfer reactions, the enzymes responsible for these reactions are not known yet. Even for the few reactions that the enzymes are known[11, 12], no detailed structural and mechanistic studies are available. However, preliminary evidence suggests that they also use simple nucleophilic mechanisms, presumably similar to polyamine synthases[20]. For example, radical SAM enzymes [21–23] are typically oxygen-sensitive because the [4Fe-4S] cluster and the radical formed are prone to oxidation. Enzymes that catalyze these ACP-transfer reactions, however, are not oxygen sensitive [11, 12], suggesting that they may use simple nucleophilic mechanism. In addition, these enzymes bear weak sequence homology to known methyltransferases that use simple nucleophilic mechanisms [11, 12].
Figure 2.
Examples of ACP-transfer to O- or N-heteroatoms. In spermidine biosynthesis, SAM is first decarboxylated and then a 3-aminopropyl group is transferred.
SAM-dependent C-alkylation: multiple mechanisms
In SAM-dependent alkylation reactions, when the methyl or ACP acceptors are carbon atoms, the enzymatic reaction mechanisms are more complicated and depend on the electronic properties of the acceptor molecules. A few specific examples will be examined below to illustrate this point and rationalize Nature’s choice of different reaction mechanisms.
Examples of C-methylation
SAM-dependent cyclopropane ring formation
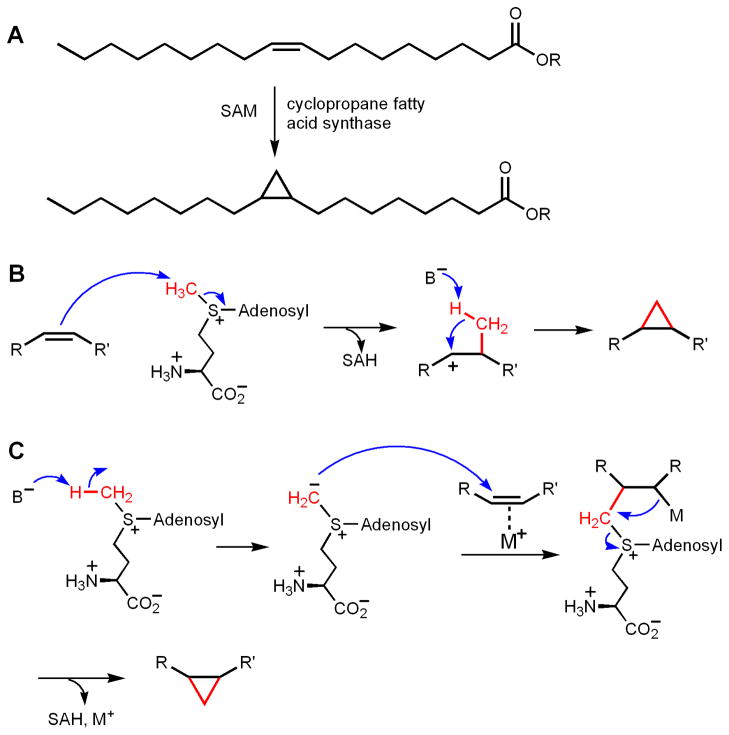
One interesting example of C-methylation is the formation of cyclopropane on unsaturated fatty acids in bacteria (Figure 3A). Although the group that is eventually transferred is a methylene group, mechanistically these reactions are similar to methylation reactions. Two enzymatic systems have been studied, the E. coli cyclopropane fatty acid (CFA) synthase and mycolic acid cyclopropane synthases from Mycobacterium tuberculosis [24–27]. Both types of enzymes make cyclopropane rings using SAM and an alkene functional group in fatty acids. At least two mechanisms have been proposed (Figure 3, B and C). In the first mechanism, the alkene acts as the nucleophile and attacks the methyl group of SAM, forming a carbocation intermediate. The methyl group is then deprotonated and combines with the carbocation to form the cyclopropane ring (Figure 3B). This mechanism was proposed based on the observation that three highly homologous methyltransferases from M. tuberculosis catalyze different reactions: one catalyzes the formation of a cyclopropane ring, one catalyzes the formation of a methylated olefin, and the other catalyzes the formation of a methylated hydroxy compound [28]. The result of these different reaction products from the three homologous enzymes could be explained by the formation of similar carbocation intermediates [28]. In the second mechanism, the methyl group of SAM is first deprotonated to form a sulfonium ylide. The ylide then attacks the double bond to form the cyclopropane ring, possibly facilitated by certain metal ions [25, 29, 30] (Figure 3C). The major experimental support for this mechanism was from model organic reactions showing that copper could catalyze the cyclopropanation of olefins by sulfur ylides [25, 29, 30]. No direct experimental support has been reported for this mechanism. The formation of sulfonium ylide typically requires strong bases, such as NaH. It was estimated that the pKa of trimethyl sulfonium is about 29 [31]. If the methyl group in SAM has a similar pKa, it would be difficult to deprotonate it under physiological condition. Several lines of experimental evidence support the mechanism shown in Figure 3B is operating. One piece of evidence is that in the crystal structure of the M. tuberculosis mycolic acid cyclopropane synthase, McaA1, a positively charged lipid molecule was bound the enzyme, together with SAH [24]. The molecule is thought to mimic the carbocation intermediate in the reaction pathway. Furthermore, the structures of the three mycolic acid cyclopropane synthases all have a seven-stranded α/β fold [24], similar to other methyltransferases [7]. Second, vinylfluorine and epoxide-containing substrate analogs were found to be inhibitors of E. coli CFA synthase, which is consistent with the formation of a carbocation intermediate [32]. Third, for E. coli CFA synthase, kinetic isotope effects measurements and kinetic studies with SAM, Se-SAM, and Te-SAM are also consistent with the mechanism shown in Figure 3B [25]. Interestingly, in the crystal structure of McaA1, a bicarbonate is bound at the enzyme active site. This bound bicarbonate was proposed to be the general base that deprotonates the methyl group in the carbocation intermediate [24]. The bicarbonate was bound by conserved Tyr, His, and Glu residues. Mutation of the Tyr and His residue in E. coli CFA led to a defective enzyme [26, 27], the activity of which can be rescued by high concentrations of sodium bicarbonate [27]. This evidence further supports the hypothesis that the bound bicarbonate is functionally important, although it is not clear why a bicarbonate would be better suited to act as the general base to deprotonate the carbocation intermediate.
Figure 3.
Methyl-transfer to C-C double bond to form cyclopropane rings. (A) The reaction catalyzed by cyclopropane fatty acid synthase. (B) The proposed cyclopropane ring formation mechanism that uses the olefine as the nucleophile. (C) The proposed cyclopropane ring formation mechanism that involves the formation of a sulfonium ylide as the nucleophile.
The cyclopropane synthase examples are important in the sense that they provide a reference point to consider the mechanisms of SAM-dependent C-alkylation reactions. Substrates that are more nucleophilic than or equally nucleophilic as an olefine can be alkylated via simply nucleophilic mechanisms.
DNA cytosine C5 methylation
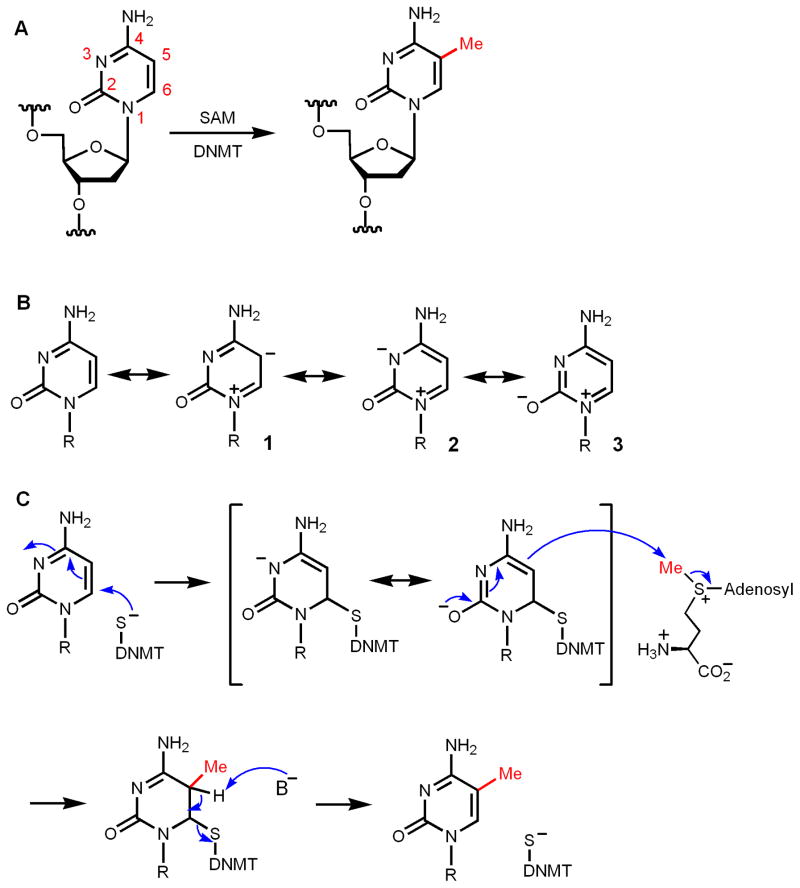
Another interesting SAM-dependent C-methylation reaction is the methylation of the C5 position of cytosine in DNA (Figure 4A). DNA methylation is an important epigenetic modification and has been the subject of intense investigation [33]. The C5 position of cytosine is not nucleophilic. Although an enamine can be nucleophilic (more nucleophilic than an alkene functional group), the enamine moiety in cytosine is not nucleophilic due to the electron-withdrawing by the carbonyl and N3 (the resonance structures 2 and 3, which have negative charges on N and O, should be more stable than 1, Figure 4B). This point can also be seen from the calculated electrostatic potential map of cytosine [34]. Therefore, C5 of cytosine cannot directly act as a nucleophile. The electron-withdrawal by N3 and the carbonyl, however, makes the C5–C6 double bond electron deficient and prone to attack by nucleophiles (Figure 4C) in a reaction that is similar to a Michael reaction. In DNA methyltransferases (DNMT), this nucleophile is the thiolate from a Cys residue [35–38]. The addition product, which is like a deprotonated enamine, is nucleophilic and reacts with SAM via an SN2-like mechanism to capture the methyl group. The resulting intermediate then eliminates the Cys of DNMT to give the methylated cytosine product (Figure 4C). Therefore, methylation of C5 of cytosine is an example of converting an electron deficient methyl acceptor to a nucleophile for the methyl-transfer reaction by addition of an active site Cys thiolate. RNA uracil and cytosine methylation uses the same mechanism [39–42]. This enzyme-assisted nucleophile formation mechanism is also used in thymidylate synthase [43], but the methyl group comes from methylene-tetrahydrofolate, not SAM, in the form of a methylene transfer reaction.
Figure 4.
Mechanism of DNA cytosine C5 methylation. (A) The DNA methylation reaction catalyzed by DNA methyltransferase (DNMT). (B) Resonance structures of cytosine showing that C5 is not nucleophilic due to the presence of the carbonyl at C2. Resonance structure 3 is more stable than 2, which is more stable than 1. (C) Nucleophilic attack by a Cys residue in DNMT convert cytosine to a nucleophile, which then undergoes methylation in an SN2 type reaction. Elimination of the Cys residue gives the methylated cytosine residue.
Methylation of C2 and C8 of adenine on 23S ribosomal RNA
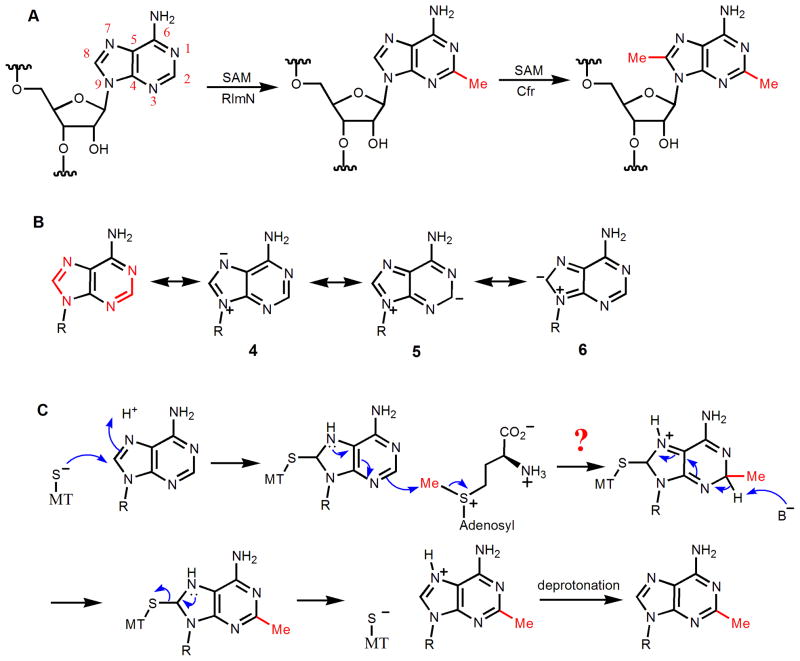
Methylation of C2 and C8 of Ade2503 of bacterial 23S ribosomal RNA are catalyzed by two closely related enzymes, Cfr and RlmN, respectively (Figure 5A) [44, 45]. RlmN is house-keeping methylating enzyme [45], while methylation catalyzed by Cfr is a mechanism that confers resistance to several antibiotics that target the ribosome [46]. The C2 and C8 of adenine exist in two amidine functional groups (highlighted in red in Figure 5B), which indicates that they are more likely to be electrophiles than nucleophiles. Although one can draw resonance structures (5 and 6, Figure 5B) with negative charge on C2 and C8, these resonance structures are not as stable as 4, in which the negative charge is on N7 (Figure 5B). The electrostatic potential map is consistent with this [34]. Since the amidine carbon is electron-deficient, one can imagine an enzyme-assisted nucleophile formation mechanism, similar to the one used in DNA methylation (Figure 4C). However, in this case, a Cys thiolate attacking the C8 would generate an anion that cannot be stabilized by resonating the negative charge to a more electron-deficient functional group (Figure 5C). In contrast, in the case of DNA cytosine methylation, the anion generated can be stabilized by delocalizing the electrons to the more electron-withdrawing carbonyl (Figure 5C).
Figure 5.
Ribosomal RNA methylation catalyzed by RlmN and Cfr. (A) The reactions catalyzed by RlmN and Cfr. RlmN catalyzes the C8 methylation of adenine, while Cfr catalyzes the C2 methylation of adenine. (B) Both C2 and C8 of adenine exist in amidine functional groups, which determine that they are electron deficient. Even though resonance structures (5 and 6) with negative charges on C2 and C8 can be drawn, they are not as stable as the resonance structures (such as 4) that have the negative charge on the more electronegative nitrogen atom. (C) Adenine is also not suitable for enzyme-assisted nucleophile formation mechanisms because the anion generated cannot be stabilized by an electron-withdrawing group, as in the case of cytosine.
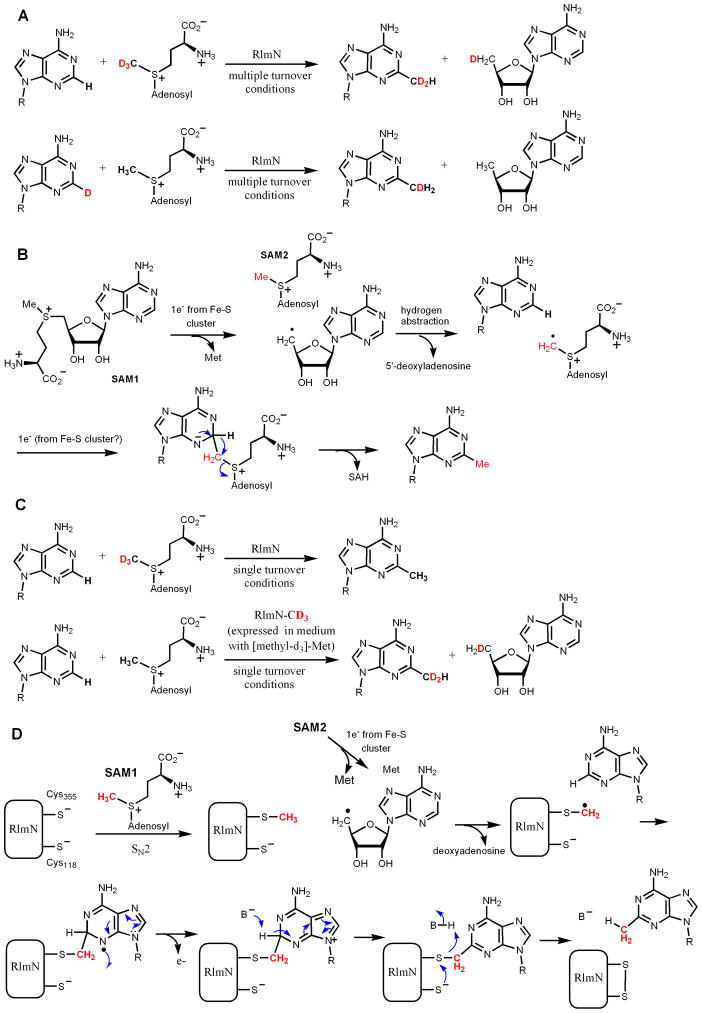
Recently, significant progress has been made regarding the mechanism of RlmN and Cfr. Consistent with the above analysis that nucleophilic mechanisms may be difficult, available data suggest that the methylation of C2 and C8 of adenine in 23S rRNA uses a unique radical mechanism. Both RlmN and Cfr belong to the radical SAM superfamily of enzymes, with a conserved CX3CX2C motif for binding to a [4Fe-4S] cluster [47]. It was demonstrated by Fujimori and coworkers that both enzymes contain Fe-S clusters, require anaerobic conditions and the presence of a reductant for activity, and generate 5′-deoxyadenosine as one of the products [2]. Under multiple turnover conditions, when (methyl-d3)-SAM was used, the methyl group in the RNA product contained two deuterium atoms, while the 5′-deoxyadenosine contained one deuterium (Figure 6A) [3]. When the hydrogen on C2 or C8 is replaced with deuterium, the methyl group in the RNA product contained one deuterium, while 5′-deoxyadenosine product is not labeled with deuterium (Figure 6A) [3]. Based on the data, a mechanism was proposed, in which the 5′-deoxyadenosyl radical abstract a hydrogen atom from the methyl group of another SAM molecule, forming a radical cation, which then added to the adenine ring, leading to the formation of methyl adenosine (Figure 6B).
Figure 6.
Mechanism of RlmN and Cfr-catalyzed rRNA methylation. (A) Isotope labeling experiments by Fujimori and coworkers showed that a “CH2” group was transferred from SAM and the H from C2 of adenine ended up on the methyl group. (B) The mechanism proposed based on the data shown in (A). (C) Isotope labeling experiments by Booker and coworkers showed that under single turnover conditions, the “CH2” group that was transferred was not directly from the labeled SAM, but from a methyl group that was covalently bonded to RlmN/Cfr. (D) The mechanism for RlmN proposed by Booker and coworkers.
On the other hand, Booker and coworkers[4] discovered that in single turnover reaction conditions, when (methyl-d3)-SAM was used, the methyl group in the RNA product contained no deuterium label (Figure 6C), which is different from the results of Fujimori shown in Figure 6A. When RlmN was expressed in medium with (methyl-d3)-methionine, under single turnover conditions, the methyl group in the RNA product contained two deuteriums while the 5′-deoxyadenosine product contained one deuterium (Figure 6C) [4]. Further evidence was presented to show that the immediate methylene donor is a methylated Cys residue (Cys355) of RlmN. The recombinant RlmN protein isolated from E. coli were methylated on Cys355. A mechanism was proposed based on the experimental evidence (Figure 6D) [4]. In this mechanism, Cys355 is first methylated by a SAM molecule. The Fe-S cluster then activate a second molecule of SAM to produce the 5′-deoxyadenosine radical, which then abstract a hydrogen atom from the methyl group on Cys355. The Cys355-CH2 radical is then added to the adenine ring of the RNA substrate to form a nitrogen radical. The radical can lose one electron to yield a positively charged intermediate, which can subsequently be deprotonated to restore the adenine ring. For the release of the methylated product from the covalent enzyme-product complex, Cys118 was proposed to act as the nucleophile and electron donor, leading to the formation of a Cys355-Cys118 disulfide bond. The involvement of Cys118 in the process is supported by the observation that RlmN Cys118A mutant isolated contained RNA but wildtype RlmN did not [4].
The result of the isotope labeling experiments in Fujimori’s study is different from that in Booker’s work (results shown in Figure 6A and 6C, respectively). It is possible that the RlmN protein in Fujimori’s study was not methylated on Cys355. However, the mostly likely reason for the difference is whether the enzymatic reaction was done under single turnover or multiple turnover conditions. Even if RlmN was methylated in Fujimori’s study, under multiple turnover conditions, the isotope labeling results would have been the same as that shown in Figure 6A because under multiple turnover conditions, the methyl group on Cys355 would only be transferred to a very small percentage (equivalent to the amount of methylated enzyme) of the product formed.
Although the two mechanisms differ from each other in certain details and the two enzymes are still under active investigation and debate, they do have several common features. Both mechanisms involved the formation of a 5′-deoxyadenosyl radical and the reaction would ultimately require two SAM molecules for one reaction. Most importantly, both mechanisms suggest that the group that is directly transferred is a methylene group instead of a methyl group. Thus, this type of enzyme should be more accurately termed “methyl synthases” instead of “methyltransferases” [3].
Examples of C-(3-amino-3-carboxypropylation)
There are two known examples of ACP-transfer to carbon atoms. One is in the biosynthesis of diphthamide, a modified histidine residue in archaeal and eukaryotic translation elongation factor 2 (EF2), and the other is the biosynthesis of wybutosine in tRNA. Available evidence suggests that these two ACP-transfer reactions use different mechanisms [5, 6, 48, 49].
ACP-transfer reaction in diphthamide biosynthesis
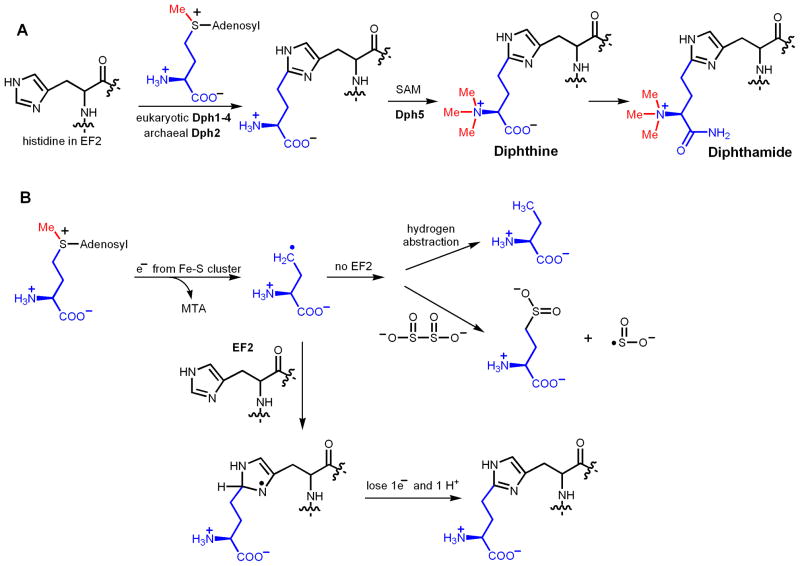
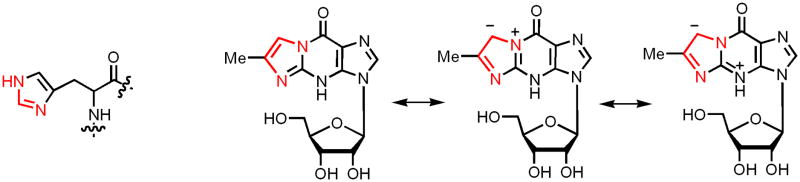
The diphthamide modification was proposed to occur in three steps (Figure 7A), with the first one being the ACP transfer from SAM to the C2 of the imidazole ring.[50] Genetic studies identified four genes, Dph1-Dph4, required for the ACP-transfer step in eukaryotes [51–55]. Dph1 and Dph2 are homologous to each other. Recently, a fifth gene, WD85, was also suggested to be involved in this step [56]. Among Dph1-Dph4, only one protein that is homologous to eukaryotic Dph1 and Dph2 can be found in archaea. Using Pyrococcus horikoshii Dph2 (PhDph2), the ACP-transfer reaction has been reconstituted in vitro [5]. Although the conserved CX3CX2C motif [47] that is found in most radical SAM enzymes is not present in PhDph2, biochemical, structural, and spectroscopic data suggest that PhDph2 contains a [4Fe-4S] cluster [5]. Furthermore, the in vitro reaction requires the presence of a reductant, such as sodium dithionite. Therefore, PhDph2 looks like a radical SAM enzyme. However, in contrast to the known radical SAM enzyme, the formation of 5′-deoxyadenosine was not detected in the reaction. On the other hand, when the substrate protein was not present, the formation of 2-aminobutyrate and homocysteine sulfinic acid was detected, which suggests that the reaction occurs via an ACP radical (Figure 7B) [5]. A possible reaction mechanism has been proposed, in which the ACP radical is added to the imidazole ring and then a hydrogen atom is eliminated to give the desired product [6]. This discovery is interesting because it implies that the enzyme can control which C-S bond in SAM is broken by the electron transfer event from the Fe-S cluster [57]. Thus, diphthamide biosynthesis requires a new radical SAM enzyme that generates an ACP radical.
Figure 7.
The ACP-transfer reaction mechanism in diphthamide biosynthesis. (A) The proposed biosynthesis pathway of diphthamide. The first step is an ACP-transfer reaction. (B) Proposed mechanism for the ACP-transfer reaction. The formation of the ACP radical is supported by the detection of 2-aminobutyrate and homocysteine sulfinic acid when no EF2 was present in the reaction.
An interesting question is why Nature needs to come out with a different radical SAM enzyme to carry out the chemistry of diphthamide biosynthesis? The C2 of the imidazole ring is not nucleophilic since it resembles an amidine carbon, similar to the C2 and C8 of adenine ring. Therefore, a simple nucleophilic mechanism is unlikely to operate here. If using a traditional radical SAM enzyme to generate a 5′-deoxyadenosine radical, the reaction presumably would need to proceed by abstracting the hydrogen on C2 of the imidazole ring to form an imidazole radical, which would then capture the ACP group from a SAM molecule. The problem with such a mechanism is that the bond energy of an sp2 C-H bond is very high [58]. Energetically, it is not feasible for the 5′-deoxyadenosine radical to abstract a hydrogen atom from an sp2 carbon atom. In contrast, when an ACP radical is generated, it can add to the imidazole ring, avoiding the energetically unfavorable hydrogen abstraction from an sp2 carbon.
ACP-transfer reaction in wybutosine biosynthesis
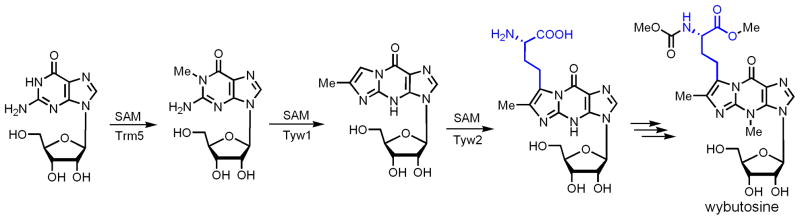
Wybutosine is a modified nucleoside at the 3′-position adjacent to the anticodon of eukaryotic phenylalanine tRNA [59]. The proposed biosynthesis pathway is shown in Figure 8 [48]. A traditional radical SAM enzyme, Tyw1, is required for the formation of the imidazole ring, although detailed reaction mechanism is not known yet [60]. The ACP-transfer step is catalyzed by Tyw2 [48], which has similarity to methyltransferases that catalyze nucleophilic methyl-transfer reactions [48, 49]. The crystal structure of Tyw2 from archaeal species showed that the ACP group, instead of methyl group, is directed to the RNA binding site [49]. This is constant with a SN2 type of mechanism. Mostly likely, the substituted imidazole ring in the substrate acts as the nucleophile to attack SAM, leading to the ACP transfer.
Figure 8.
The proposed biosynthesis pathway of wybutosine. Tyw1 is a radical SAM enzyme catalyzing the formation of the third ring in the modified base. Tyw2 is the enzyme that catalyzes the ACP-transfer reaction.
Why can the ACP transfer reaction in wybutosine simply use a nucleophilic mechanism? This is especially interesting considering that in both ACP transfer reactions in diphthamide biosynthesis and wybutosine biosynthesis, the ACP acceptors are imidazole rings. The logic behind Nature’s choice of chemistry here can be rationalized based on the intrinsic nucleophilicity of the carbon atom in the imidazole ring (Figure 9). As described above, the C2 of the imidazole ring in diphthamide biosynthesis is electron deficient since it is in an amidine functional group. In contrast, the ACP-accepting carbon atom of the imidazole ring in wybutosine biosynthesis is electron rich because it can be considered as an enamine (Figure 9). Resonance electron donation by one of the nitrogen atoms in the ring makes the C-C double bond more electron rich than an isolated C-C double bond, such as that found in the CFA synthase substrate.
Figure 9.
The ACP-accepting carbon atoms in diphthamide biosynthesis and wybutosine biosynthesis have different electronic properties, which determine the choice of different enzymatic reaction mechanisms. In diphthamide biosynthesis, the carbon atom exists in an amidine functional group and thus is electron deficient. In wybutosine biosynthesis, the carbon atom exists in an enamine functional group, which is electron rich.
Summary
From the several examples discussed above, some general principles can be devised to rationalize, and perhaps more importantly, to predict the reaction mechanisms used in different SAM-dependent enzymatic reactions. In general, the electronic properties of the substrates, especially the atoms that will be alkylated, should be analyzed carefully. If the methyl (or methylene in the case of RlmN/Cfr) or ACP acceptor is electron rich, a simple nucleophilic mechanism would be used. These include methyl or ACP transfer to heteroatoms, such as O, N, and S, and electron rich C atoms, such as those in an olefin (in cyclopropane fatty acid biosynthesis) or an enamine (in wybutosine biosynthesis). When the methyl/methylene or ACP acceptor is sufficiently electron deficient, such as C5 of cytosine, then a Cys thiolate can attack the electron deficient substrate and convert it to a good nucleophile to carry out nucleophilic methyl-transfer. When the methyl/methylene or ACP acceptor is neither electron deficient nor electron rich, a nucleophilic mechanism would be difficult and radical mechanisms are required. Interestingly, all known examples (RlmN/Cfr-catalyzed methylation and PhDph2-catalyzed ACP transfer) in this category involve the amidine part of a heterocyclic compound.
The above summary and arguments are based on the assumption that if the electronic properties of the alkyl acceptor allows the use of simple nucleophilic mechanisms, Nature would choose to use nucleophilic mechanisms. Only when the chemistry is too difficult to achieve via nucleophilic mechanisms, will Nature choose to use more complicated radical mechanisms. Based on known reactions catalyzed by radical SAM enzymes [21–23], this assumption seems to be correct. However, it is hard to predict whether there are to-be-discovered enzymatic reactions that do not fit this assumption. Therefore, when using the above summary and argument to predict the unknown enzymatic reaction mechanism, it should be kept in mind that the assumption may not be correct in rare cases,
Highlights.
Enzymatic reaction mechanisms for SAM-dependent alkylation reactions are discussed.
Focus on alkylation of carbon atoms, for which both polar and radical mechanisms are used.
The choice of polar or radical mechanism is rationalized
The rationalization could help to predict mechanisms of new SAM-dependent alkylation reactions
Acknowledgments
Work from H.L.’s laboratory in this area is supported by NIH/NIGMS R01GM088276.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fontecave M, Atta M, Mulliez E. S-adenosylmethionine: nothing goes to waste. Trends In Biochemical Sciences. 2004;29:243–249. doi: 10.1016/j.tibs.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 2.Yan F, LaMarre JM, Röhrich R, Wiesner J, Jomaa H, Mankin AS, Fujimori DG. RlmN and Cfr are radical SAM enzymes involved in methylation of ribosomal RNA. J Am Chem Soc. 2010;132:3953–3964. doi: 10.1021/ja910850y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yan F, Fujimori DG. RNA methylation by radical SAM enzymes RlmN and Cfr proceeds via methylene transfer and hydride shift. Proc Natl Acad Sci USA. 2011;108:3930–3934. doi: 10.1073/pnas.1017781108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grove TL, Benner JS, Radle MI, Ahlum JH, Landgraf BJ, Krebs C, Booker SJ. A radically different mechanism for S-adenosylmethionine-dependent methyltransferases. Science. 2011 doi: 10.1126/science.1200877. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y, Zhu X, Torelli AT, Lee M, Dzikovski B, Koralewski RM, Wang E, Freed J, Krebs C, Ealick SE, Lin H. Diphthamide biosynthesis requires an organic radical generated by an iron-sulphur enzyme. Nature. 2010;465:891–896. doi: 10.1038/nature09138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu X, Dzikovski B, Su X, Torelli AT, Zhang Y, Ealick SE, Freed JH, Lin H. Mechanistic understanding of Pyrococcus horikoshii Dph2, a [4Fe-4S] enzyme required for diphthamide biosynthesis. Mol BioSystems. 2011;7:74–81. doi: 10.1039/c0mb00076k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schubert HL, Blumenthal RM, Cheng X. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem Sci. 2003;28:329–335. doi: 10.1016/S0968-0004(03)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao B, Jing C, Wilson JR, Walker PA, Vasisht N, Kelly G, Howell S, Taylor IA, Blackburn GM, Gamblin SJ. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature. 2003;421:652–656. doi: 10.1038/nature01378. [DOI] [PubMed] [Google Scholar]
- 9.Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, Ivanov GS, McKinney K, Tempst P, Prives C, Gamblin SJ, Barlev NA, Reinberg D. Regulation of p53 activity through lysine methylation. Nature. 2004;432:353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- 10.Trievel RC, Beach BM, Dirk LMA, Houtz RL, Hurley JH. Structure and catalytic mechanism of a SET domain protein methyltransferase. Cell. 2002;111:91–103. doi: 10.1016/s0092-8674(02)01000-0. [DOI] [PubMed] [Google Scholar]
- 11.Riekhof WR, Andre C, Benning C. Two enzymes, BtaA and BtaB, are sufficient for betaine lipid biosynthesis in bacteria. Arch Biochem Biophys. 2005;441:96–105. doi: 10.1016/j.abb.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 12.Reeve AM, Breazeale SD, Townsend CA. Purification, characterization, and cloning of an S-adenosylmethionine-dependent 3-amino-3-carboxypropyltransferase in nocardicin biosynthesis. J Biol Chem. 1998;273:30695–30703. doi: 10.1074/jbc.273.46.30695. [DOI] [PubMed] [Google Scholar]
- 13.Ohashi Z, Maeda M, McCloskey JA, Nishimura S. 3-(3-Amino-3-carboxypropyl)uridine. Novel modified nucleoside isolated from Escherichia coli phenylalanine transfer ribonucleic acid. Biochemistry. 1974;13:2620–2625. doi: 10.1021/bi00709a023. [DOI] [PubMed] [Google Scholar]
- 14.Kowalak JA, Bruenger E, Crain PF, McCloskey JA. Identities and phylogenetic comparisons of posttranscriptional modifications in 16 S ribosomal RNA from Haloferax volcanii. J Biol Chem. 2000;275:24484–24489. doi: 10.1074/jbc.M002153200. [DOI] [PubMed] [Google Scholar]
- 15.Friedman S, Li HJ, Nakanishi K, Van Lear G. 3-(3-Amino-3-carboxypropyl)uridine. Structure of the nucleoside in Escherichia coli transfer ribonucleic acid that reacts with phenoxyacetoxysuccinimide. Biochemistry. 1974;13:2932–2937. doi: 10.1021/bi00711a024. [DOI] [PubMed] [Google Scholar]
- 16.Nishimura S, Taya Y, Kuchino Y, Ohashi Z. Enzymatic synthesis of 3-(3-amino-3-carboxypropyl) uridine in phenylalanine transfer RNA: Transfer of the 3-amino-3-carboxypropyl group from S-adenosylmethionine. Biochem Biophys Res Commun. 1974;57:702–708. doi: 10.1016/0006-291x(74)90603-2. [DOI] [PubMed] [Google Scholar]
- 17.Saponara AG, Enger MD. The isolation from ribonucleic acid of substituted uridines containing alpha-aminobutyrate moieties derived from methionine. Biochim Biophys Acta. 1974;349:61–77. doi: 10.1016/0005-2787(74)90009-4. [DOI] [PubMed] [Google Scholar]
- 18.Maden BEH, Forbes J, de Jonge P, Klootwijk J. Presence of a hypermodified nucleotide in hela cell 18 S and Saccharomyces carlsbergensis 17 S ribosomal RNAs. Febs Letters. 1975;59:60–63. doi: 10.1016/0014-5793(75)80341-3. [DOI] [PubMed] [Google Scholar]
- 19.Ikegami F, Sakai R, Ishikawa T, Kuo YH, Lambein F, Murakoshi I. Biosynthesis in vitro of 2-(3-amino-3-carboxypropyl)-isoxazolin-5-one, the neurotoxic amino acid in Lathyrus odoratus. Biol Pharm Bull. 1993;16:732–734. doi: 10.1248/bpb.16.732. [DOI] [PubMed] [Google Scholar]
- 20.Ikeguchi Y, Bewley MC, Pegg AE. Aminopropyltransferases: function, structure and genetics. J Biochem. 2006;139:1–9. doi: 10.1093/jb/mvj019. [DOI] [PubMed] [Google Scholar]
- 21.Frey PA, Hegeman AD, Ruzicka FJ. The radical SAM superfamily. Crit Rev Biochem Mol Biol. 2008;43:63 –88. doi: 10.1080/10409230701829169. [DOI] [PubMed] [Google Scholar]
- 22.Duschene KS, Veneziano SE, Silver SC, Broderick JB. Control of radical chemistry in the AdoMet radical enzymes. Curr Opin Chem Biol. 2009;13:74–83. doi: 10.1016/j.cbpa.2009.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vey JL, Drennan CL. Structural insights into radical generation by the radical SAM superfamily. Chem Rev. 2011;111:2487–2506. doi: 10.1021/cr9002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang C-c, Smith CV, Glickman MS, Jacobs WR, Sacchettini JC. Crystal structures of mycolic acid cyclopropane synthases from Mycobacterium tuberculosis. J Biol Chem. 2002;277:11559–11569. doi: 10.1074/jbc.M111698200. [DOI] [PubMed] [Google Scholar]
- 25.Iwig DF, Grippe AT, McIntyre TA, Booker SJ. Isotope and elemental effects indicate a rate-limiting methyl transfer as the initial step in the reaction catalyzed by Escherichia coli cyclopropane fatty acid synthase. Biochemistry. 2004;43:13510–13524. doi: 10.1021/bi048692h. [DOI] [PubMed] [Google Scholar]
- 26.Iwig DF, Uchida A, Stromberg JA, Booker SJ. The activity of Escherichia coli cyclopropane fatty acid synthase depends on the presence of bicarbonate. J Am Chem Soc. 2005;127:11612–11613. doi: 10.1021/ja053899z. [DOI] [PubMed] [Google Scholar]
- 27.Courtois F, Ploux O. Escherichia coli cyclopropane fatty acid synthase: Is a bound bicarbonate ion the active-site base? Biochemistry. 2005;44:13583–13590. doi: 10.1021/bi051159x. [DOI] [PubMed] [Google Scholar]
- 28.Yuan Y, Barry CE. A common mechanism for the biosynthesis of methoxy and cyclopropyl mycolic acids in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 1996;93:12828–12833. doi: 10.1073/pnas.93.23.12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pohl S, Law JH, Ryhage R. The path of hydrogen in the formation of cyclopropane fatty acids. Biochimica Et Biophysica Acta. 1963;70:583–585. doi: 10.1016/0006-3002(63)90794-7. [DOI] [PubMed] [Google Scholar]
- 30.Cohen T, Herman G, Chapman TM, Kuhn D. Laboratory model for the biosynthesis of cyclopropane rings. Copper-catalyzed cyclopropanation of olefins by sulfur ylides. J Am Chem Soc. 1974;96:5627–5628. [Google Scholar]
- 31.Rios A, O’Donoghue AC, Amyes TL, Richard JP. Formation and stability of organic zwitterions -The carbon acid pKas of the trimethylsulfonium and tetramethylphosphonium cations in water. Can J Chem. 2005;83:1536–1542. [Google Scholar]
- 32.Molitor EJ, Paschal BM, Liu H-w. Cyclopropane fatty acid synthase from Escherichia coli: Enzyme purification and inhibition by vinylfluorine and epoxide-containing substrate analogues. ChemBioChem. 2003;4:1352–1356. doi: 10.1002/cbic.200300767. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 34.Bonaccorsi R, Pullman A, Scrocco E, Tomasi J. The molecular electrostatic potentials for the nucleic acid bases: Adenine, thymine, and cytosine. Theoretical Chemistry Accounts: Theory. Computation, and Modeling (Theoretica Chimica Acta) 1972;24:51–60. [Google Scholar]
- 35.Wu JC, Santi DV. Kinetic and catalytic mechanism of HhaI methyltransferase. J Biol Chem. 1987;262:4778–4786. [PubMed] [Google Scholar]
- 36.Klimasauskas S, Kumar S, Roberts RJ, Cheng X. Hhal methyltransferase flips its target base out of the DNA helix. Cell. 1994;76:357–369. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- 37.Chen L, MacMillan AM, Verdine GL. Mutational separation of DNA binding from catalysis in a DNA cytosine methyltransferase. J Am Chem Soc. 1993;115:5318–5319. [Google Scholar]
- 38.Chen L, MacMillan AM, Chang W, Ezaz-Nikpay K, Lane WS, Verdine GL. Direct identification of the active-site nucleophile in a DNA (cytosine-5)-methyltransferase. Biochemistry. 1991;30:11018–11025. doi: 10.1021/bi00110a002. [DOI] [PubMed] [Google Scholar]
- 39.Kealey JT, Gu X, Santi DV. Enzymatic mechanism of tRNA (m5U54)methyltransferase. Biochimie. 1994;76:1133–1142. doi: 10.1016/0300-9084(94)90042-6. [DOI] [PubMed] [Google Scholar]
- 40.Alian A, Lee TT, Griner SL, Stroud RM, Finer-Moore J. Structure of a TrmA-RNA complex: A consensus RNA fold contributes to substrate selectivity and catalysis in m5U methyltransferases. Proc Natl Acad Sci USA. 2008;105:6876–6881. doi: 10.1073/pnas.0802247105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Y, Santi DV. m5C RNA and m5C DNA methyl transferases use different cysteine residues as catalysts. Proc Natl Acad Sci USA. 2000;97:8263–8265. doi: 10.1073/pnas.97.15.8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Foster PG, Nunes CR, Greene P, Moustakas D, Stroud RM. The first structure of an RNA m5C methyltransferase, Fmu, provides insight into catalytic mechanism and specific binding of RNA substrate. Structure. 2003;11:1609–1620. doi: 10.1016/j.str.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 43.Carreras CW, Santi DV. The catalytic mechanism and structure of thymidylate synthase. Annu Rev Biochem. 1995;64:721–762. doi: 10.1146/annurev.bi.64.070195.003445. [DOI] [PubMed] [Google Scholar]
- 44.Giessing AMB, Jensen S, Sr, Rasmussen A, Hansen LH, Gondela A, Long K, Vester B, Kirpekar F. Identification of 8-methyladenosine as the modification catalyzed by the radical SAM methyltransferase Cfr that confers antibiotic resistance in bacteria. RNA. 2009;15:327–336. doi: 10.1261/rna.1371409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toh S-M, Xiong L, Bae T, Mankin AS. The methyltransferase YfgB/RlmN is responsible for modification of adenosine 2503 in 23S rRNA. RNA. 2008;14:98–106. doi: 10.1261/rna.814408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kehrenberg C, Schwarz S, Jacobsen L, Hansen LH, Vester B. A new mechanism for chloramphenicol, florfenicol and clindamycin resistance: methylation of 23S ribosomal RNA at A2503. Mol Microbiol. 2005;57:1064–1073. doi: 10.1111/j.1365-2958.2005.04754.x. [DOI] [PubMed] [Google Scholar]
- 47.Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucl Acids Res. 2001;29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Noma A, Kirino Y, Ikeuchi Y, Suzuki T. Biosynthesis of wybutosine, a hyper-modified nucleoside in eukaryotic phenylalanine tRNA. EMBO J. 2006;25:2142–2154. doi: 10.1038/sj.emboj.7601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Umitsu M, Nishimasu H, Noma A, Suzuki T, Ishitani R, Nureki O. Structural basis of AdoMet-dependent aminocarboxypropyl transfer reaction catalyzed by tRNA-wybutosine synthesizing enzyme, TYW2. Proc Natl Acad Sci USA. 2009;106:15616–15621. doi: 10.1073/pnas.0905270106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dunlop PC, Bodley JW. Biosynthetic labeling of diphthamide in Saccharomyces cerevisiae. J Biol Chem. 1983;258:4754–4758. [PubMed] [Google Scholar]
- 51.Chen JY, Bodley JW, Livingston DM. Diphtheria toxin-resistant mutants of Saccharomyces cerevisiae. Mol Cell Biol. 1985;5:3357–3360. doi: 10.1128/mcb.5.12.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mattheakis L, Shen W, Collier R. DPH5, a methyltransferase gene required for diphthamide biosynthesis in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:4026–4037. doi: 10.1128/mcb.12.9.4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mattheakis LC, Sor F, Collier RJ. Diphthamide synthesis in Saccharomyces cerevisiae: structure of the DPH2 gene. Gene. 1993;132:149. doi: 10.1016/0378-1119(93)90528-b. [DOI] [PubMed] [Google Scholar]
- 54.Moehring TJ, Danley DE, Moehring JM. In vitro biosynthesis of diphthamide, studied with mutant Chinese hamster ovary cells resistant to diphtheria toxin. Mol Cell Biol. 1984;4:642–650. doi: 10.1128/mcb.4.4.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu S, Milne GT, Kuremsky JG, Fink GR, Leppla SH. Identification of the proteins required for biosynthesis of diphthamide, the target of bacterial ADP-ribosylating toxins on translation elongation factor 2. Mol Cell Biol. 2004;24:9487–9497. doi: 10.1128/MCB.24.21.9487-9497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carette JE, Guimaraes CP, Varadarajan M, Park AS, Wuethrich I, Godarova A, Kotecki M, Cochran BH, Spooner E, Ploegh HL, Brummelkamp TR. Haploid genetic screens in human cells identify host factors used by pathogens. Science. 2009;326:1231–1235. doi: 10.1126/science.1178955. [DOI] [PubMed] [Google Scholar]
- 57.Broderick JB. Biochemistry: A radically different enzyme. Nature. 2010;465:877–878. doi: 10.1038/465877a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Energetics of Organic Free Radicals. Vol. 4 Chapman and Hall; London: 1996. [Google Scholar]
- 59.Blobstein SH, Grunberger D, Weinstein IB, Nakanishi K. Isolation and structure determination of the fluorescent base from bovine liver phenylalanine transfer ribonucleic acid. Biochemistry. 1973;12:188–193. doi: 10.1021/bi00726a002. [DOI] [PubMed] [Google Scholar]
- 60.Suzuki Y, Noma A, Suzuki T, Senda M, Senda T, Ishitani R, Nureki O. Crystal structure of the radical SAM enzyme catalyzing tricyclic modified base formation in tRNA. J Mol Biol. 2007;372:1204–1214. doi: 10.1016/j.jmb.2007.07.024. [DOI] [PubMed] [Google Scholar]