Abstract
IL-4 expression is known to be activated in CD4 T cells when they are differentiated to Th2 but not Th1 cells. However, CD4 T cells selected by MH class II-expressing thymocytes, named thymocyte-selected CD4 T cells (T-CD4 T cells), express IL-4 under both Th1 and Th2 conditions. In this study, we investigated molecular mechanisms by which IL-4 gene expression is regulated in T-CD4 T cells. We found that T-CD4 T cells express IL-4 soon after selection in the thymus. Deficiency of DNase I hypersensitive (HS) sites HS5a and HS5 at the 3′-enhancer region in the IL-4 gene decreased IL-4 production, but T-CD4 T cells were able to make IL-4 under the Th1-inducing condition. Consistent with this, IL-4 was expressed in Th1 differentiated T-CD4 T cells in the absence of recombination signal binding protein-J that interacts with HS5. When HS5 was examined separately from other endogenous regulatory elements using a reporter system, CD4 T cells that are selected by thymic epithelial cells cannot transcribe the IL-4 reporter gene with HS5 alone. However, HS5 was able to induce the expression of the IL-4 reporter gene in T-CD4 T cells. Interestingly, the Th1 differentiating signal led to deacetylation at HS5 of the IL-4 endogenous gene, whereas the Th2-inducing environment had no effect. Therefore, in T-CD4 T cells, HS5 plays an essential role during the induction phase of IL-4 expression, but the maintenance of IL-4 expression in Th1 cells requires additional regulatory elements.
On Ag stimulation, naive CD4 T cells can differentiate into Th1, Th2, or Th17 effecter cells, which rapidly produce IFN-γ, IL-4, or IL-17, respectively (1-4). The hallmark cytokine of Th1 cells is IFN-γ, which is instrumental for cell-mediated immunity. Th2 cells produce IL-4, IL-5, and IL-13 that are involved in controlling immune responses against extracellular parasites (5). In addition, IL-4 and IL-5 are strongly implicated in atopic and allergic disease because of their role in regulating IgE-mediated immune responses via mast cells and eosinophils. IL-17, together with other cytokines and chemokines released by activated Th17 cells, plays an important role in inflammatory autoimmune diseases (6-11). Thus, proper regulation of Th differentiation is critical for controlling both cellular and humoral immune responses, and for maintaining immune homeostasis.
The nonoverlapping cytokine expression patterns in Th1 and Th2 cells are controlled by inheritable states of transcriptional activation and repression established during the differentiation process. For example, programmed chromatin modifications in the Il13-Il4 locus correlate well with the transcriptional competence of Th2 cytokine genes in a lineage-specific manner. Chromatin modifications control the accessibility of transcriptional activators and repressors in discrete regions of the locus that have been identified as DNase I hypersensitive (HS) sites (5).
Clusters of HS sites have been characterized at the Il13-Il4 locus on the basis of the lineage specificity and activation dependence. HSS1, HSS2, HS0, HS1, HS2, HS3, HS5, and HS5a are Th2 specific (12-14). All sites are constitutive except that HS5a formation is activation dependent (12-14). HSS3 and HS4 are also constitutive and commonly observed in naive, Th1, and Th2 cells (5, 14). Comparative cross-species analyses of genomic sequences revealed considerable conservations of noncoding sequences, and the HS sites in the Il13-Il4 locus often correlate with the conserved regions. Conserved noncoding sequences 1 and 2 correspond to HSS1 and HSS2 and to HS5, respectively (5, 14, 15). CD4 T cells from mice lacking conserved noncoding sequence 1 or mice with disrupted HS5 and the 3′ enhancer marked by HS5a have a reduction in their ability to secrete Th2 cytokines (16-18). However, Th2 cytokine production is not abolished completely in either of the mutant mice (18, 19), suggesting that the activity of either element alone cannot explain the lineage-specific transcriptional competency of Th2 cytokine genes. Therefore, genetic deletion experiments have not been sufficient to define the functional role of those elements in lineage-specific gene expression.
To gain further insights into the role of the cis-acting activity of HS5a and HS5 on the lineage specificity at the single-cell level, a transgenic (Tg) mouse model was developed using GFP as a reporter (17). A study of this GFP Tg model showed that, unlike conventional CD4 T cells, HS5 alone was sufficient to activate IL-4 expression in the memory phenotype (MP) CD4 T cells, as well as NKT cells (17). Depleting MP CD4 T cells resulted in a reduction of Th2 cytokine production on secondary stimulation, suggesting its importance in IL-4 production by MP CD4 T cells (17). HS5 contains multiple putative binding sites for recombination signal binding protein-J (RBP-J), a critical modulator for Notch signaling (17). Ligand binding to Notch receptor leads to a series of proteolytic processing events (20, 21), and the released intracellular domain translocates to the nucleus and acts as a transcriptional activator through an association with a DNA binding protein, RBP-J (22). RBP-J conditional knockout mice showed the reduction in IgG1 and IgE, which seems to be caused mainly by an abrogation of the initial IL-4 production by MP CD4 T cells (17).
We and others have demonstrated that CD4 T cells can be selected by MHC class II-expressing thymocytes (23, 24), and that thymocyte-selected CD4 (T-CD4 hereon) T cells exhibit the unusual effecter function (25). Human thymocytes express MHC class II, and thymocyte-mediated CD4 T cell selection provides a mechanism for several documented observations that could not be otherwise explained (26-34). A recent report showed the presence of T-CD4 T cells in humans, further strengthening the relevance of studying T-CD4 T cells (35). T-CD4 T cells also participate in protecting allergen-induced allergic airway inflammation and EAE (25, 36) suggesting an immune regulatory role of T-CD4 T cells. Unlike CD4 T cells that are developed by thymic epithelial cells (TECs; epithelial cell-selected CD4 T cell [E-CD4 T cells hereon]), T-CD4 T cells produce Th1 and Th2 cytokines shortly after activation in vivo and in vitro, which is similar to NKT cells (25). Furthermore, T-CD4 T cells produce Th2 cytokines even after being differentiated into the Th1 cell lineage. Surprisingly, IL-4 expression in T-CD4 T cells does not require Stat6, which is also dispensable for NKT but not E-CD4 T cells (25, 37). T-CD4 T cells deficient in IL-4 production can produce IL-5 and IL-13, eliminating IL-4 as a required Th2-inducing cytokine (38). Thus, IL-4 expression in T-CD4 cells seems to be regulated by a distinct mechanism.
To have a better understanding of IL-4 regulation in T-CD4 T cells, we focused on HS5a and HS5 to determine whether this enhancer region is essential for IL-4 gene expression in T-CD4 T cells. In this study, we report that the deletion of these 3′ enhancers did not abrogate IL-4 expression in T-CD4 T cells even under the Th1-inducing condition. Furthermore, HS5 plays an important role to induce IL-4 reporter expression in T-CD4 T cells. However, IL-4 reporter expression mediated by HS5 was abolished by the Th1- but not Th2-inducing signal, revealing the differential role of HS5 for the induction versus maintenance of IL-4 expression.
Materials and Methods
Mice
Mice carrying the human type III CIITA transgene (Tg) were described previously (38). Tg mice were bred to carry both the CD45.1 and CD45.2 congenic markers, and non-Tg littermates were used as wild-type (WT) controls. GFP reporter Tg system (Δ5a and Δ5a/5) and RBP-J conditional knockouts were previously described (17). Δ5a mice were crossed with CIITATg mice to generate Δ5a/Tg. Mice lacking DNase hypersensitive site 5/5a−/− were reported previously (18). 4get mice described previously (39) were purchased from The Jackson Laboratory (Bar Harbor, ME). CD45.1+ C57BL/6.SJL (B6) mice and the MHC class II Aβ-deficient mice on the C57BL/6.SJL background (Abb−/−) carrying the CD45.1 congenic marker were purchased from Taconic (Germantown, NY). All of the earlier mentioned mice were on C57BL/6.SJL (B6) background. All mice were housed in the animal facility at The University of Michigan Medical School under specific pathogen-free conditions and used at 6–12 wk of age. All animal experiments were performed under protocols approved by the institution.
Bone marrow chimeric mice
For bone marrow (BM) transfer experiments, the recipient B6/SJL or Abb−/− mice were lethally irradiated with 950 rad 24 h before receiving BM transfers. Total BM cells were harvested from the femurs and tibias of donor mice (2–3 mo of age) and depleted of mature T cells, B cells, and MHC class II+ lymphocytes by using a mixture of Abs containing anti-CD4 (RL172), anti-CD8 (TIB105, TIB210), anti-CD19 (1D3), and anti-MHC class II (M5/114), followed by complement-mediated lyses. BM cells from two different types of donor mice were mixed at a ratio of 1:1, and each recipient mouse received 5 × 106 cells in 400 μl of 1× PBS via tail vein injection. All BM chimeras were reconstituted for at least 8–12 wk before analysis of T cell development and function.
Flow cytometry
All Abs used for flow cytometry were purchased from BD Pharmingen (San Diego, CA). Cells were preincubated with the anti-FcγR mAb 2.4G2 to block nonspecific Ab binding before they were stained with the following FITC-, PE-, PerCP-, CyChrome-, APC-, or biotin-conjugated Abs: CD4 (L3T4), CD45.1 (A20), CD45.2 (104), anti–IL-4 (11B11), and anti–IFN-γ (XMG1.2). Fluorochrome-conjugated streptavidin was used to visualize staining by biotinylated primary Abs. Events were acquired on a FACS-Canto (Becton Dickinson) flow cytometer, and the data were analyzed with the FlowJo software.
CD4 T cell preparation and differentiation
CD4 T cells were purified from single-cell suspension of splenocytes from chimeric mice with anti-mouse CD4 microbeads (Miltenyi Biotec, Auburn, CA). CD4 T cells (1 × 106/ml) were stimulated with 5 μg/ml plate-bound anti-CD3ε (145-2C11), 1 μg/ml anti-CD28 (37.51), and 50 units IL-2 (Roche, Indianapolis, IN) for 5–7 d to induce Th differentiation. For Th1 differentiation, 3.5 ng/ml IL-12 and 10 μg/ml anti–IL-4 (11B11) were added. Th2 cultures were supplemented with 10 ng/ml IL-4 and 10 μg/ml anti–IFN-γ (R4-6A2).
Intracellular cytokine staining
CD4+ T cells differentiated in Th1 and Th2 for 5–7 d were restimulated with 50 ng/ml phorbol myristyl acetate and 1.5 μM ionomycin (Calbiochem, San Diego, CA) for 5 h. Monensin (Sigma, St. Louis, MO) at 3 μM was added during the last 3 h of stimulation. Activated Th1 and Th2 cells were stained with fluorochrome-conjugated anti-CD45.1 and anti-CD45.2 Abs. Cells were then fixed in 2–4% paraformaldehyde for 10 min at room temperature and permeabilized with 0.2% saponin (Sigma), followed by staining with anti–IL-4 (11B11) and anti–IFN-γ (XMG1.2) for flow cytometry.
Chromatin immunoprecipitation analysis
Chromatin immunoprecipitation (ChIP) analysis was performed according to the ChIP assay protocol (Upstate Biotechnology) as described previously (25). In brief, 2–3 × 106 CD4 T cells from WT and Tg mice were fixed in 1% formaldehyde for 10 min at room temperature, washed, lysed, and sonicated with three pulses to generate chromatin fragments of 200–500 bp in length. Anti-acetylated histone H3 Ab (Upstate Biotechnology) was added (3 μl per immunoprecipitation) to the diluted lysates and incubated overnight. Protein A-Sepharose CL-4B beads (GE Healthcare) were added for 1 h. After washes, the immunocomplexes were eluted, the cross-links were reversed, and DNA was purified by phenol/chloroform extraction and resuspended in 50 μl Tris-EDTA buffer. The primer pairs used for CD3ε were 5′-CATTTCCAAGTGACGTGG-3′ and 5′-AACACACTGGCTGCATGC-3′; for HS5, 5′-TCACATGAGCCTTTGCAAGACA-3′ and 5′-GGCTGCGGATGACTGATCAG-3′; and for IL-4p, 5′-GGTAAAGCCTCATTCCATGGTCCTG-3′ and 5′-TCTGGGCCAATCAGCACCTCTCTT-3′.
RNA analysis and quantitative real-time PCR
Total RNA from CD4 T cells was extracted using TRIzol-mediated lysis according to the manufacturer’s recommendations (Invitrogen) and reverse transcribed using the SuperScript First-Strand cDNA Synthesis System (Invitrogen). Quantitative real-time PCR was performed with SYBR Green PCR Master Mix (Applied Biosystems). All PCR reactions were done in triplicate, and the data were analyzed by the comparative threshold cycle (ΔCt) method and normalized to GAPDH. The primer pairs used for GAPDH were 5′-CCAGGTTGTCTCCTGCGACT-3′ and 5′-ATACCAGGAAATGAGCTTGACAAAGT3′; for IL-4, 5′-ACAGGAGAAGGGACGCCAT-3′ and 5′-GAAGCCCTACAGACGAGCTCA-3′; and for IFN-γ, 5′-TCAAGTGGCATAGATGTGGAAGAA-3′ and 5′-TGGCTCTGCAGGATTTTCATG-3′.
Statistical analysis
The Student two-tailed t test was used to calculate statistical significance. A p value <0.05 was considered statistically significant (*p < 0.05; **p < 0.01).
Results
Important but not essential role of the 3′ enhancer to express the IL-4 gene
To determine the role of HS5a and HS5 for IL-4 gene expression in T-CD4 T cells, we transferred BM cells from mice lacking both HS5a and HS5 (HS5a/5−/−) together with BM prepared from WT or CIITA Tg mice to WT or Aβ−/− mice resulting in [HS5a/5−/−+WT→B6] and [HS5a/5−/−+Tg→Aβ−/−] mice, respectively. In [HS5a/5−/−+WT→B6] mice, thymocytes originated from both HS5a/5−/− and WT BM cells are selected by host TECs; thus, all CD4 T cells are E-CD4 T cells. However, the same HS5a/5−/− cells in [HS5a/5−/−+Tg→Aβ−/−] mice cannot be selected by TECs because of the deficiency of Aβ expression in the host mice. Therefore, HS5a/5−/− cells undergo positive selection mediated by CIITA-expressing and thus MHC class II+ thymocytes generating T-CD4 T cells. We have demonstrated that this selection pathway is efficiently operated in this type of chimera (24). To identify the cells originated from the three parties, we used a congenic marker CD45. CD4 T cells from the chimeras were differentiated under the Th1- and Th2-inducing conditions to measure IFN-γ and IL-4 production.
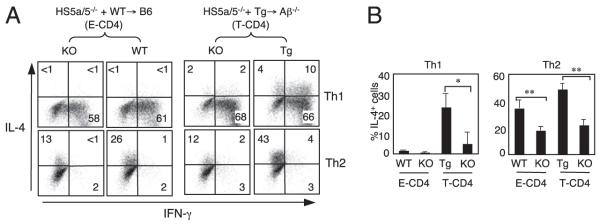
In agreement with the published studies (18), CD4 T cells from spleens of [HS5a/5−/−+WT→B6] mice showed reduced IL-4 expression as compared with their respective controls when differentiated under the Th2-inducing condition (Fig. 1A, left group, Fig. 1B). When the same cells were differentiated to Th1 cells, they did not produce an appreciable level of IL-4 with or without HS5a/5a. However, in [HS5a/5−/−+Tg→Aβ−/−] mice, HS5a/5−/− originated CD4 T cells produced IL-4 under the Th1 condition, although the number of IL-4+ cells was less in HS5a/5−/− cells than that of Tg partner cells (Fig. 1A, 1B). Therefore, HS5a/5 seems to play an important role for IL-4 expression in both E- and T-CD4 T cells.
FIGURE 1.
The HS5 and HS5a region in the IL-4 locus is important but not essential to express IL-4. A and B, BM from HS5a/5−/− mice (knockout [KO]) (CD45.2) were mixed with those from WT or CIITA Tg mice (CD45.1/2) and cotransferred into B6 or Aβ−/− recipients (CD45.1). Eight to 12 wk after BM transplantations, splenic CD4 T cells from BM chimeras were differentiated into Th1 or Th2 cells and analyzed IL-4 and IFN-γ expression by intracellular cytokine staining. The cells were also stained with an anti-CD45.1 and anti-CD45.2 Ab to distinguish the two donors and the recipient cells. Graphs in B show the mean ± SD of four mice. *p < 0.05, **p < 0.01.
IL-4 gene expression in T-CD4 T cells on positive selection in the thymus
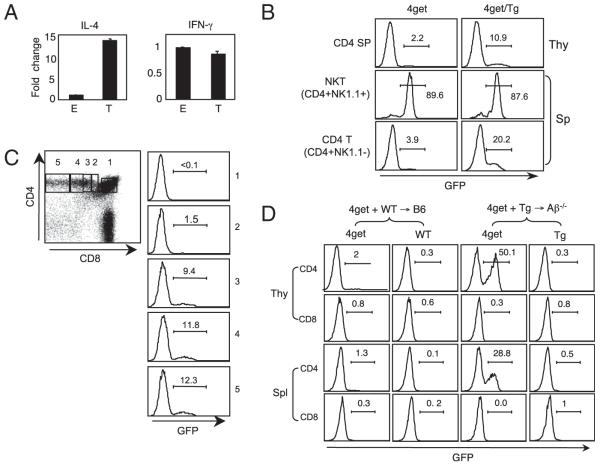
The different requirement of regulatory elements to express IL-4 in T-CD4 cells is likely due to the thymic selection pathway. To investigate IL-4 gene expression during T-CD4 T cell development, we compared mRNA levels of freshly sorted CD4 single positive (SP) from WT→B6 and Tg→Aβ−/− chimeras that generate E- and T-CD4 T cells, respectively. Consistent with the reported observations (25), the amount of IL-4 transcripts in freshly isolated T-CD4 SP cells was far greater than E-CD4 SP cells, whereas the level of IFN-γ mRNA was comparable between the two (Fig. 2A). Next, we examined IL-4 gene expression at a single-cell level using an IL-4 reporter system known as 4get (39). 4get reporter mice were crossed with CIITA Tg mice (4get/Tg) in which CD4 cells are developed by thymocytes, as well as TECs, whereas CD4 cells in 4get mice are selected exclusively by TECs (24). Because T-CD4 T cells express the IL-4 gene, GFP+ cells are expected to be detected in 4get/Tg but not in 4get mice. Indeed, 4get/Tg mice showed GFP+ cell population in the thymus and the spleen (Fig. 2B). Invariant NKT (iNKT) cells express the IL-4 gene constitutively; thus, iNKT cells in 4get mice are GFP+ (40). Consistent with this report, iNKT cells in both mice were GFP+ (Fig. 2B). We then examined GFP+ cells during CD4 T cell maturation measured by the gradual loss of CD8 from double-positive thymocytes. Fig. 2C illustrates the inverse correlation between the percentage of GFP+ cells and the level of CD8. CD8 SP cells did not express a detectable level of GFP (data not shown).
FIGURE 2.
Expression of IL-4 during T-CD4 T cell development. A, IL-4 expression is enhanced in T-CD4 thymocytes. E- and T-CD4 T cells were electronically sorted from the thymus of [WT→B6] and [CIITATg→Aβ−/−] chimeric mice and subjected to RNA preparation immediately. IL-4 and IFN-γ mRNA were quantified by quantitative RT-PCR, and the results were expressed as ratios relative to the housekeeping gene GAPDH. B, GFP expression of 4get and 4get mice expressing CIITA transgene (4get/Tg). Thymocytes and splenocytes from 4get and 4get/Tg mice were freshly isolated and used to assess GFP expression. Numbers indicate the percentage of cells expressing GFP. Representative profiles are shown. C, GFP expression during CD4 T cell development in the 4get/Tg thymus. Total thymocytes were used to measure GFP expression. Numbers in the dot plot correspond to each histogram. D, Thymic selection dictates IL-4 expression measured by GFP. Chimeric mice [4get+WT→B6] and [4get+Tg→Ab−/−] were generated to obtain E- and T-CD4 T cells, respectively. Eight to 12 wk after BM transplantations, total thymocytes and splenocytes were prepared and examined for GFP expression without manipulation in vitro. The numbers shown are the percentage of GFP+ cells. A–C, Data are representative of more than three independent experiments; (D) data are representative of two independent experiments.
The frequency of GFP+ CD4 T cells in 4get/Tg mice is typically at a range of 10–15%, suggesting that TEC-mediated development of CD4 T cells is dominant when both selection pathways are present. If this were the case, it is anticipated to get increased GFP+ cells if only thymocyte-mediated selection is available. To test this possibility, we constructed [4get+WT→B6] and [4get+Tg→Aβ−/−] chimeras to obtain 4get cells selected by TECs and thymocytes, respectively. After 8–12 wk of reconstitution, we isolated thymocytes and splenocytes, and analyzed the GFP expression level in these chimeric mice. As expected, there was a substantial increase in GFP+ cells in [4get+Tg→Aβ−/−] (Fig. 2D, right group). In contrast, 4get cells developed by TECs expressed GFP poorly (Fig. 2D, left group).
HS5 is required to express the IL-4 gene in T-CD4 T cells
Although the 4get system allows us to examine IL-4 expression in cells without manipulation, we cannot study the regulatory elements with the 4get mice. To investigate the role of the 3′-enhancer region, we used another GFP reporter Tg system that has been characterized previously (17). This reporter transgene contains the 5′ enhancer and the IL-4 promoter followed by GFP as a common element (17) (Fig. 3A). To this construct the 3′-enhancer region containing HS5 but not HS5a was added, resulting in Δ5a Tg mice. We compared these mice with a related Tg line carrying the construct Δ5a/5, in which an additional 1413-bp region containing HS5 has been deleted (17) (Fig. 3A).
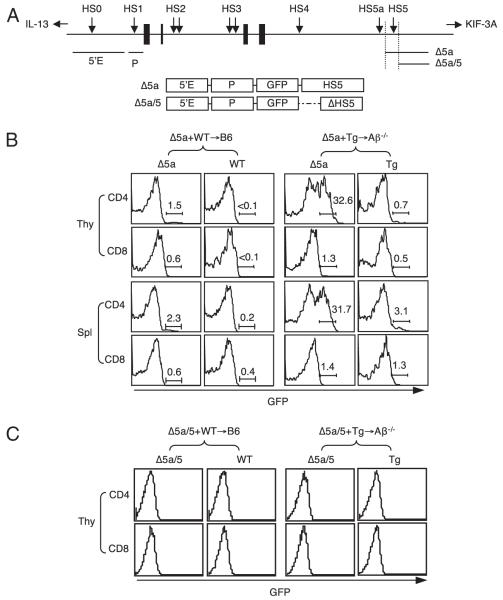
FIGURE 3.
HS5 together with the 5′ enhancer and the promoter can induce IL-4 expression in T-CD4 T cells. A, Schematic diagram of the IL-4 locus and the transgene constructs used in this study. Black boxes denote IL-4 exons; arrows denote HS sites; horizontal bars represent fragments used for Tg constructs. B, HS5-mediated expression of GFP in T-CD4 T cells. BM from Δ5a mice (CD45.2) shown in A were mixed with WT or Tg BM (CD45.1/2) and cotransferred to either B6 or Aβ−/− hosts (CD45.1). Chimeric mice were reconstituted for 8–12 wk. Freshly isolated total thymocytes and splenocytes were used to assess GFP expression by flow cytometry. The numbers shown are the percentage of GFP+ cells. C, Lack of GFP expression in the absence of HS5. Mixed BM chimeras were constructed as in A except BM cells from Δ5a/5 reporter mice shown in A were used. GFP expression of CD4 and CD8 T cells in the thymus is shown.
Using the reporter mice, we constructed two groups of chimeric mice, [Δ5a+WT→WT] and [Δ5a+Tg→Aβ−/−], and examined GFP expression of freshly isolated HS5 cells from the thymus and the spleen. GFP positivity would indicate the expression of the transgene directed by HS5. In E-CD4 T cells developed in [Δ5a+WT→WT] mice, only a small fraction of GFP+ cells was detectable in both thymuses and spleens (Fig. 3B, left group), which likely corresponds to MP cells shown previously (17). However, when the same Δ5a cells were developed by thymocytes in [Δ5a+Tg→Aβ−/−] chimeras, 30–40% GFP+ CD4 T cells was present in the thymus (Fig. 3B, right group). A similar proportion of GFP+ CD4 T cells was present in the spleen. In addition, GFP expression was restricted to CD4 T cells, as evidenced by very low GFP+ CD8 T cells in the [Δ5a+Tg→Aβ−/−] mice (Fig. 3B). These data suggest that the region containing HS5 plays an important role to express the IL-4 gene in T-CD4 T cells.
We next asked whether HS5 is required for IL-4 expression. To test this, we constructed BM chimeras with cells from Δ5a/5 mice that do not have HS5. Neither [Δ5a/5+WT→WT] nor [Δ5a/5+ Tg→Aβ−/−] mice generated CD4 T cells that express GFP (Fig. 3C). Therefore, HS5 is indispensable to express the IL-4 gene in T-CD4 T cells from a minimal transgene.
RBP-J–deficient T-CD4 T cells can express IL-4 even under the Th1 differentiation condition
The data shown earlier demonstrate the significance of HS5 for IL-4 expression in T-CD4 T cells. The main transcription factor recognizing HS5 is RBP-J (17), and the deficiency of RBP-J selectively impairs Th2 differentiation in E-CD4 T cells (41, 42). Therefore, given the fact that HS5 is critical to express IL-4 in T-CD4 T cells, RBP-J could be essential as well. To test this, we transferred BM cells from RBP-J−/− mice together with either WT or Tg BM cells to WT or Aβ−/− hosts generating [RBPJ−/−+WT→B6] and [RBPJ−/−+Tg→Aβ−/−] chimeras, respectively. Under Th2-inducing conditions, RBPJ−/− CD4 T cells from both groups of mice showed reduced IL-4 expression in a similar fashion as described previously (Fig. 4A, 4B) (41, 42). On Th1 differentiation, RBPJ−/− T-CD4 T cells from [RBPJ−/− +Tg→ Aβ−/−] mice were able to make IL-4. Although RBPJ−/− cells appeared to produce a lower amount of IL-4 than the partner cells, the difference was not statistically significant, suggesting a marginal role of RBP-J for IL-4 expression in T-CD4 T cells.
FIGURE 4.
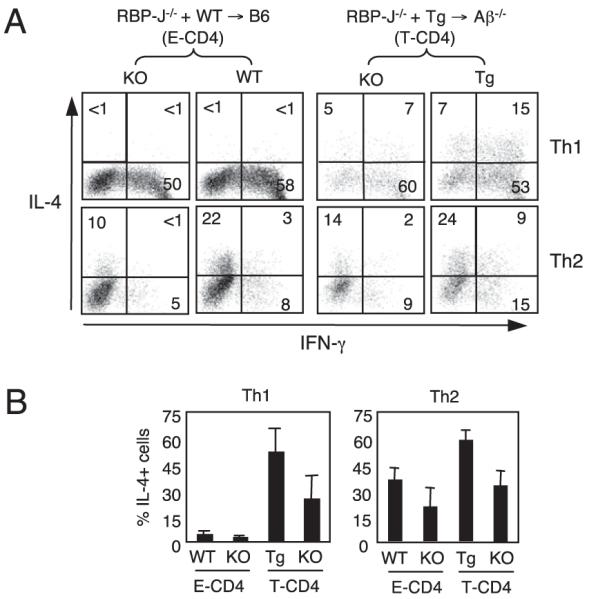
Marginal role of RBP-J in T-CD4 T cells to produce IL-4. A, IL-4 expression in T-CD4 T cells in the absence of RBP-J. BM from RBP-J−/− mice (CD45.2) were mixed with those from WT or Tg mice (CD45.1/2) and cotransferred into B6 or Aβ−/− recipients (CD45.1). Splenic CD4 T cells from BM chimera shown were differentiated as in Fig. 1A and analyzed for IL-4 and IFN-γ expression. Mice were analyzed 8–10 wk after BM transplantation. The percentage of positive cells in each quadrant is shown. B, Data shown as mean ± SD from three to four mice in each group.
HS5 is sufficient for maintenance of IL-4 in Th2 but not in Th1 cells
We showed in Fig. 3B and 3C that freshly isolated CD4 SP thymocytes from [Δ5a+Tg→Aβ−/−] mice were able to express GFP. This suggests that HS5 is a key regulatory element to induce IL-4 gene expression during T-CD4 T cell development. We also demonstrated that T-CD4 T cells produce IL-4 even under the Th1 condition (Figs. 1, 4). Therefore, we asked whether HS5 has the same regulatory function to maintain IL-4 expression when CD4 T cells differentiate to Th1 and Th2 cells. To test this, splenic CD4 T cells were prepared from [Δ5a+WT→WT] (E-CD4) and [Δ5a+Tg→Aβ−/−] (T-CD4) mice and cultured under Th1 or Th2 skewing conditions. We first examined Th1 and Th2 cytokine production by measuring IL-4 together with IFN-γ. As expected, comparison between Th2 cultures of WT and Tg BM originated cells showed comparable percentages of IL-4+ cells (Fig. 5A, bottom panels). In addition, Th2 differentiated HS5 cells from both groups of mice also contained an equivalent IL-4+ population. However, Th1 cells from [Δ5a+Tg→Aβ−/−] mice that generate T-CD4 T cells produced IL-4 under the Th1 condition (Fig. 5A). Together, cells exhibited the proper Th1 and Th2 cell phenotype.
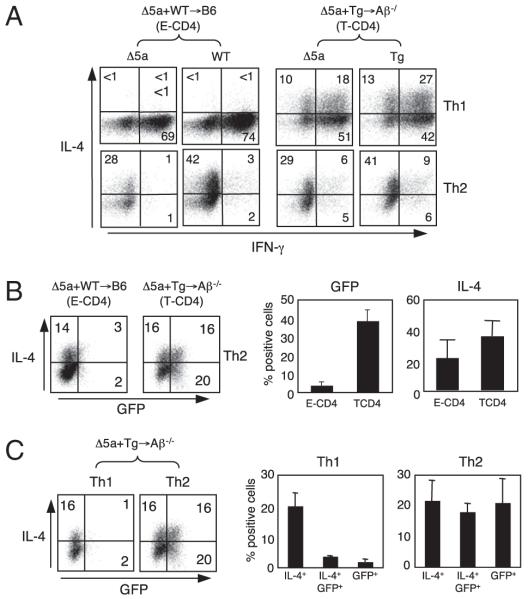
FIGURE 5.
Differential role of HS5 for IL-4 expression in T-CD4 T cells. A, IL-4 production by T-CD4 T cells on Th1 and Th2 differentiation. Splenic CD4 T cells from BM chimera shown were differentiated as in Fig. 1A and analyzed for IL-4 and IFN-γ expression. B, Th2 differentiated cells from the same chimeras in A were analyzed to compare the expression of IL-4 and GFP. Cells from Δ5a Tg mice from each chimera are shown. Numbers indicate percentage of cells in each quadrant. Bar graphs are mean ± SD of three to four mice in each group. C, Δ5a originated CD4 T cells from [Δ5a+Tg→Aβ−/−] mice were differentiated to Th1 and Th2, and analyzed for IL-4 and GFP expression. Bar graphs (right panels) show each subpopulation in the dot plot.
Next, using the same sets of chimeras, we assessed GFP (transgene) and IL-4 (endogenous gene) expression of Th2 differentiated cells that were originated from Δ5a BM. Partner cells were not analyzed because they do not express the transgene. Fig. 5B shows that GFP expression was much greater in T-CD4 T cells compared with that of E-CD4 T cells on Th2 differentiation. In addition, the percentage of GFP+ population was comparable with the percentage of freshly isolated cells shown in Fig. 3. However, the percentages of IL-4–expressing cells between E- and T-CD4 T cells were comparable (Fig. 5B). This suggests that HS5 alone cannot direct GFP expression even under the Th2 culture condition if CD4 T cells were selected by TECs. In contrast, when CD4 T cells are developed by thymocytes, HS5 can turn on the IL-4 gene immediately after selection and maintains during Th2 differentiation.
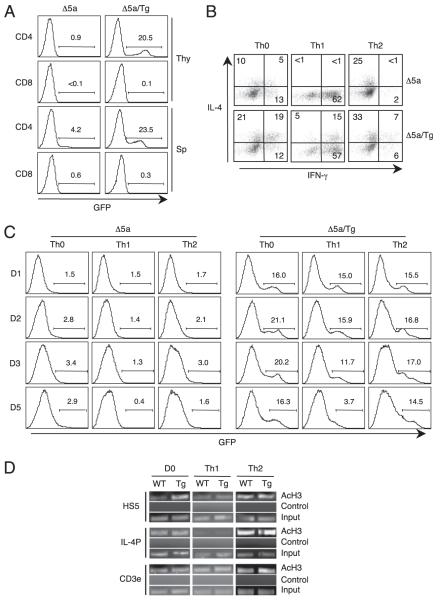
To further investigate the role of HS5 in T-CD4 T cells generated in [Δ5a+Tg→Aβ−/−] mice, we compared GFP expression of Δ5a cells after differentiated to Th1 or Th2 cells. The Th1 culture had few GFP+ cells, whereas Th2 differentiated Δ5a T-CD4 T cells had a substantial population expressing GFP (Fig. 5C, left panel). Furthermore, the Th2 culture showed the comparable representation of three subpopulations: IL-4+, IL-4+GFP+, and GFP+ cells (Fig. 5C, compare middle and right panels). These data suggest that new GFP transcripts have been accumulated in the Th2 but not the Th1 culture. Although the Th1 and Th2 cultures were started from the cell population containing the same proportion of GFP-expressing cells (Fig. 3B), a majority of cells lost GFP expression when cells were differentiated to Th1 cells.
Gradual loss of HS5-mediated IL-4 expression by the Th1-inducing signal
To gain insights toward the regulation of IL-4 expression under the Th1 condition in T-CD4 T cells, we generated mice expressing both the Δ5a GFP reporter and CIITA transgene (Δ5a/Tg). In Δ5a/Tg mice, CD4 T cells can undergo either TEC- or thymocyte-mediated selection. As expected, Δ5a/Tg mice showed the GFP+ CD4 T cell population in the thymus and spleen (Fig. 6A). When CD4 T cells from these mice were differentiated under non-skewing (Th0), Th1, or Th2 conditions, CD4 T cells from Δ5a/Tg mice expressed the expected pattern of IL-4 and IFN-γ (Fig. 6B). We also examined GFP expression during the course of differentiation. We found that the percentages of GFP+ cells were relatively constant in cells cultured under Th0 and Th2 conditions over 5 d. However, when cells were differentiated to Th1 cells, the GFP-expressing cell population was gradually decreased (Fig. 6C).
FIGURE 6.
Loss of HS5-mediated IL-4 expression by the Th1-inducing signal. A, Total thymocytes (Thy) or splenocytes (Sp) from HS5 mice (Δ5a) shown in Fig. 3A or HS5 crossed with CIITATg mice (Δ5a/Tg) were compared for GFP expression. B, Splenic CD4 T cells were differentiated under non-skewing (Th0), Th1, or Th2 conditions from Δ5a and Δ5a/Tg mice as shown in Fig. 3A and were analyzed for IL-4 and IFN-γ. C, Splenic CD4 T cells from Δ5a mice or Δ5a/Tg mice were differentiated as in B and analyzed for GFP expression at the indicated time points. Numbers indicate the percentage of GFP+ cells. D, Differential acetylation of histone H3 at HS5. WT and CIITA Tg mice were used to isolate splenic CD4 T cells. Freshly isolated (D0) or cells cultured for 5 d under the Th1 and Th2 differentiation were subjected to ChIP assays as in the Materials and Methods followed by PCR. The same precipitated DNA was used for PCR to assess the level of acetylation at HS5, IL-4 promoter (IL-4P), and CD3ε. No Ab group was used as a negative control. CD3ε and input DNA was used as an internal control. Data shown are representative from three independent experiments.
Our data showed that HS5 is essential to induce IL-4 expression but not sufficient to maintain the expression under the Th1 condition. This prompted us to ask whether HS5 is remodeled during T-CD4 T cell development and whether the chromatin configuration is changed by the Th1 differentiation signal. To address this, we performed ChIP assays to measure the level of histone acetylation. Splenic CD4 T cells from WT and CIITA Tg mice were prepared and used immediately or differentiated to Th1 and Th2 cells for 5 d. The ChIP products from these cells would show the degree of histone acetylation at the endogenous IL-4 gene. We found that HS5 was highly acetylated in Tg cells before differentiation (D0), but Th1 differentiated cells no longer sustained the same level of histone acetylation (Fig. 6D). The IL-4 promoter did not show significant differences in histone acetylation between WT and Tg cells.
Discussion
It is accepted that IL-4 expression in E-CD4 T cells is mainly restricted to cells that have undergone Th2 differentiation programming. In contrast, iNKT cells and MP CD4 T cells express IL-4 constitutively (17). As we have shown in this article and also reported previously (25), T-CD4 T cells express the IL-4 gene in a unique fashion, which is shared by iNKT cells. T-CD4 T cells and iNKT cells have preformed IL-4 mRNA and do not require Stat6 to express IL-4. These common properties between T-CD4 T cells and iNKT cells seem to be stemmed from the fact that the two types of cells are selected by thymocytes (24, 43). The IL-4 gene is turned on in T-CD4 T cells immediately after selection, suggesting that signaling delivered during thymocyte–thymocyte interactions changes the intracellular environments leading to remodeling of the IL-4 locus. This is clearly demonstrated by the activation of GFP expression in immature CD4 T cells and subsequent accumulation of GFP+ cells as they matured (Fig. 2). Once the IL-4 locus is remodeled, the expression seems to be maintained throughout the life span of CD4 T cells. iNKT cells show the similar pattern.
What makes T-CD4 T cells to express the IL-4 gene on selection in the thymus? One explanation could be the difference in TCR signaling between E- and T-CD4 T cells. Our studies have demonstrated that T-CD4 T cell development prefers stronger TCR signaling than E-CD4 T cells (Y. Qiao and C.-H. Chang, unpublished data). Therefore, it is possible that T-CD4 T cells expressing TCR that can deliver strong signaling would receive an adequate cue to remodel the IL-4 locus. Similarly, iNKT cells are also known to have strong TCR signaling, and they express IL-4 constitutively. In contrast, a strong signal eliminates E-CD4 T cells by negative selection, and thus the surviving E-CD4 T cells would express TCR that can deliver the moderate strength of signals. It is tempting to speculate that MP CD4 T cells express TCR that may fall into the high end of the signaling range, allowing them to express IL-4. Interestingly, only 30–40% of T-CD4 T cells express IL-4 (Fig. 3B, right group). Again, it is possible that these two populations differ in their TCR repertoire.
The other possibility is the signal from the signaling lymphocytic activation molecule/SLAM-associated protein (SLAM/SAP) pathway. Unlike E-CD4 T cells, both T-CD4 and iNKT cells do not develop in the absence of SLAM/SAP signaling (44-47). Therefore, the SLAM/SAP signaling pathway could be responsible for the activation of the IL-4 locus. Indeed, we showed that T-CD4 T cells deficient in SAP showed fewer IL-4+ cells in the periphery (44). It is likely that signaling from both TCR and the SLAM/SAP pathway participates in remodeling the IL-4 locus.
Another unique feature of T-CD4 T cells is the expression of promyelocytic leukemia zinc finger (PLZF) (35). PLZF is a transcription factor necessary for iNKT cell development (48). In addition, PLZF expression correlates with IL-4 production in iNKT cells (48). A recent report demonstrated that a subset of human CD4 T cells expresses PLZF and also expresses IL-4, supporting the correlation between PLZF and IL-4 expression (35). However, although PLZF deficiency reduced IL-4 production in iNKT cells (48), overexpressing PLZF in CD4 T cells did not activate IL-4 expression as in T-CD4 T cells (49). Therefore, PLZF may be necessary but not sufficient to activate the IL-4 gene. Perhaps PLZF together with signaling delivered by thymocyte–thymocyte interaction is required to remodel the IL-4 locus. Whatever the mechanisms, it is warranted to investigate the role of PLZF for constitutive expression of IL-4 in T-CD4 T cells.
HS sites serve as a regulatory site by accommodating protein binding and can function positively, as well as negatively. The 3′-enhancer region tested in this study is necessary for IL-4 expression in E-CD4 T cells but plays a less critical role in T-CD4 T cells. In the absence of both HS5a and HS5, IL-4 was produced even in Th1 differentiated T-CD4 T cells, although the amount of IL-4 was reduced. This suggests that IL-4 gene transcriptional potential is mediated, in part, by HS5, but other elements also contribute. In line with this observation, the impact caused by the deficiency of RBP-J that binds to HS5 was moderate. Therefore, the optimum expression of the IL-4 gene in T-CD4 T cells seems to share some of the regulatory elements found in E-CD4 T cells.
The IL-4 mini-transgene system revealed the differential role and the potential of the 3′-enhancer region. Particularly, HS5 shows a unique function during the induction versus maintenance phase in T-CD4 T cells. Under the Th2 differentiation condition, HS5 function does not appear to be different because the percentages of GFP+ cells in Th2 differentiated cells are comparable with that of freshly isolated cells. In contrast, when T-CD4 T cells were differentiated to Th1 cells, transgene expression was lost, suggesting a negative signal by the Th1 skewing condition. It seems that the Th1-inducing signal eliminates a positive regulatory factor or induces the expression of a negative factor, which can lead to turning off IL-4 expression. The loss of histone acetylation of HS5 on Th1 differentiation correlates with the loss of GFP expression. Interestingly, this event seems to be IL-4 independent because IL-4 was made by T-CD4 T cells during Th1 differentiation. Moreover, the endogenous IL-4 gene is expressed under the same Th1-inducing condition supporting the hypothesis that the presence of other cis-acting regulatory elements in addition to HS5 is required to sustain expression. Nevertheless, the data revealed the unique role of HS5 during the induction and maintenance phase of IL-4 gene expression in T-CD4 T cells.
T-CD4 T cells in humans also express IL-4 constitutively (35). If T-CD4 T cells in humans behave similarly as murine T-CD4 T cells, they would express IL-4 under all conditions, modulating immunity. Recent reports showed that IL-4 produced in the thymus induces the generation of innate/memory-like CD8 T cells (50, 51). Therefore, it is highly possible that T-CD4 T cells with the capability to produce IL-4 in the thymus likely regulate CD8 T cell-mediated immunity. We and others also have shown the regulatory role of T-CD4 T cells during airway inflammation and EAE (25, 36). Although it remains to be confirmed whether IL-4 produced by T-CD4 T cells affects these immune diseases directly, it is conceivable that T-CD4 T cells would participate in immune responses as a positive and negative regulator similar to iNK-T cells. Further investigation to gain a better understanding of IL-4 gene expression in T-CD4 T cells bears high significance.
Abbreviations used in this article
- BM
bone marrow
- ChIP
chromatin immunoprecipitation
- E-CD4 T cell
epithelial cell-selected CD4 T cell
- HS
DNase I hypersensitive
- iNKT
invariant NKT
- MP
memory phenotype
- PLZF
promyelocytic leukemia zinc finger
- RBP-J
recombination signal binding protein-J
- SLAM/SAP
signaling lymphocytic activation molecule/SLAM-associated protein
- SP
single positive
- T-CD4 T cell
thymocyte-selected CD4 T cell
- TEC
thymic epithelial cell
- Tg
transgenic
Footnotes
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Mosmann TR, Cherwinski H, Bond MW, Giedlin MA, Coffman RL. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J. Immunol. 1986;136:2348–2357. [PubMed] [Google Scholar]
- 2.Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, Murphy KM, Weaver CT. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat. Immunol. 2005;6:1123–1132. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- 3.Park H, Li Z, Yang XO, Chang SH, Nurieva R, Wang YH, Wang Y, Hood L, Zhu Z, Tian Q, Dong C. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat. Immunol. 2005;6:1133–1141. doi: 10.1038/ni1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weaver CT, Harrington LE, Mangan PR, Gavrieli M, Murphy KM. Th17: an effector CD4 T cell lineage with regulatory T cell ties. Immunity. 2006;24:677–688. doi: 10.1016/j.immuni.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 5.Agarwal S, Rao A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity. 1998;9:765–775. doi: 10.1016/s1074-7613(00)80642-1. [DOI] [PubMed] [Google Scholar]
- 6.Langrish CL, Chen Y, Blumenschein WM, Mattson J, Basham B, Sedgwick JD, McClanahan T, Kastelein RA, Cua DJ. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J. Exp. Med. 2005;201:233–240. doi: 10.1084/jem.20041257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cua DJ, Sherlock J, Chen Y, Murphy CA, Joyce B, Seymour B, Lucian L, To W, Kwan S, Churakova T, et al. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature. 2003;421:744–748. doi: 10.1038/nature01355. [DOI] [PubMed] [Google Scholar]
- 8.Yen D, Cheung J, Scheerens H, Poulet F, McClanahan T, McKenzie B, Kleinschek MA, Owyang A, Mattson J, Blumenschein W, et al. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J. Clin. Invest. 2006;116:1310–1316. doi: 10.1172/JCI21404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iwakura Y, Ishigame H. The IL-23/IL-17 axis in inflammation. J. Clin. Invest. 2006;116:1218–1222. doi: 10.1172/JCI28508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harrington LE, Mangan PR, Weaver CT. Expanding the effector CD4 T-cell repertoire: the Th17 lineage. Curr. Opin. Immunol. 2006;18:349–356. doi: 10.1016/j.coi.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 11.McKenzie BS, Kastelein RA, Cua DJ. Understanding the IL-23-IL-17 immune pathway. Trends Immunol. 2006;27:17–23. doi: 10.1016/j.it.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 12.Agarwal S, Avni O, Rao A. Cell-type-restricted binding of the transcription factor NFAT to a distal IL-4 enhancer in vivo. Immunity. 2000;12:643–652. doi: 10.1016/s1074-7613(00)80215-0. [DOI] [PubMed] [Google Scholar]
- 13.Guo L, Hu-Li J, Paul WE. Probabilistic regulation of IL-4 production in Th2 cells: accessibility at the Il4 locus. Immunity. 2004;20:193–203. doi: 10.1016/s1074-7613(04)00025-1. [DOI] [PubMed] [Google Scholar]
- 14.Takemoto N, Koyano-Nakagawa N, Yokota T, Arai N, Miyatake S, Arai K. Th2-specific DNase I-hypersensitive sites in the murine IL-13 and IL-4 intergenic region. Int. Immunol. 1998;10:1981–1985. doi: 10.1093/intimm/10.12.1981. [DOI] [PubMed] [Google Scholar]
- 15.Loots GG, Locksley RM, Blankespoor CM, Wang ZE, Miller W, Rubin EM, Frazer KA. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science. 2000;288:136–140. doi: 10.1126/science.288.5463.136. [DOI] [PubMed] [Google Scholar]
- 16.Grogan JL, Wang ZE, Stanley S, Harmon B, Loots GG, Rubin EM, Locksley RM. Basal chromatin modification at the IL-4 gene in helper T cells. J. Immunol. 2003;171:6672–6679. doi: 10.4049/jimmunol.171.12.6672. [DOI] [PubMed] [Google Scholar]
- 17.Tanaka S, Tsukada J, Suzuki W, Hayashi K, Tanigaki K, Tsuji M, Inoue H, Honjo T, Kubo M. The interleukin-4 enhancer CNS-2 is regulated by Notch signals and controls initial expression in NKT cells and memory-type CD4 T cells. Immunity. 2006;24:689–701. doi: 10.1016/j.immuni.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 18.Solymar DC, Agarwal S, Bassing CH, Alt FW, Rao A. A 3′ enhancer in the IL-4 gene regulates cytokine production by Th2 cells and mast cells. Immunity. 2002;17:41–50. doi: 10.1016/s1074-7613(02)00334-5. [DOI] [PubMed] [Google Scholar]
- 19.Mohrs M, Blankespoor CM, Wang ZE, Loots GG, Afzal V, Hadeiba H, Shinkai K, Rubin EM, Locksley RM. Deletion of a coordinate regulator of type 2 cytokine expression in mice. Nat. Immunol. 2001;2:842–847. doi: 10.1038/ni0901-842. [DOI] [PubMed] [Google Scholar]
- 20.Maillard I, Adler SH, Pear WS. Notch and the immune system. Immunity. 2003;19:781–791. doi: 10.1016/s1074-7613(03)00325-x. [DOI] [PubMed] [Google Scholar]
- 21.Schroeter EH, Kisslinger JA, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 22.Honjo T. The shortest path from the surface to the nucleus: RBP-J kappa/Su(H) transcription factor. Genes Cells. 1996;1:1–9. doi: 10.1046/j.1365-2443.1996.10010.x. [DOI] [PubMed] [Google Scholar]
- 23.Choi EY, Jung KC, Park HJ, Chung DH, Song JS, Yang SD, Simpson E, Park SH. Thymocyte-thymocyte interaction for efficient positive selection and maturation of CD4 T cells. Immunity. 2005;23:387–396. doi: 10.1016/j.immuni.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 24.Li W, Kim MG, Gourley TS, McCarthy BP, Sant’Angelo DB, Chang CH. An alternate pathway for CD4 T cell development: thymocyte-expressed MHC class II selects a distinct T cell population. Immunity. 2005;23:375–386. doi: 10.1016/j.immuni.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 25.Li W, Sofi MH, Yeh N, Sehra S, McCarthy BP, Patel DR, Brutkiewicz RR, Kaplan MH, Chang CH. Thymic selection pathway regulates the effector function of CD4 T cells. J. Exp. Med. 2007;204:2145–2157. doi: 10.1084/jem.20070321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saito Y, Kametani Y, Hozumi K, Mochida N, Ando K, Ito M, Nomura T, Tokuda Y, Makuuchi H, Tajima T, Habu S. The in vivo development of human T cells from CD34(+) cells in the murine thymic environment. Int. Immunol. 2002;14:1113–1124. doi: 10.1093/intimm/dxf087. [DOI] [PubMed] [Google Scholar]
- 27.Yahata T, Ando K, Nakamura Y, Ueyama Y, Shimamura K, Tamaoki N, Kato S, Hotta T. Functional human T lymphocyte development from cord blood CD34+ cells in nonobese diabetic/Shi-scid, IL-2 receptor gamma null mice. J. Immunol. 2002;169:204–209. doi: 10.4049/jimmunol.169.1.204. [DOI] [PubMed] [Google Scholar]
- 28.Hiramatsu H, Nishikomori R, Heike T, Ito M, Kobayashi K, Katamura K, Nakahata T. Complete reconstitution of human lymphocytes from cord blood CD34+ cells using the NOD/SCID/gammacnull mice model. Blood. 2003;102:873–880. doi: 10.1182/blood-2002-09-2755. [DOI] [PubMed] [Google Scholar]
- 29.Traggiai E, Chicha L, Mazzucchelli L, Bronz L, Piffaretti JC, Lanzavecchia A, Manz MG. Development of a human adaptive immune system in cord blood cell-transplanted mice. Science. 2004;304:104–107. doi: 10.1126/science.1093933. [DOI] [PubMed] [Google Scholar]
- 30.Klein C, Cavazzana-Calvo M, Le Deist F, Jabado N, Benkerrou M, Blanche S, Lisowska-Grospierre B, Griscelli C, Fischer A. Bone marrow transplantation in major histocompatibility complex class II deficiency: a single-center study of 19 patients. Blood. 1995;85:580–587. [PubMed] [Google Scholar]
- 31.Godthelp BC, Van Eggermond MC, Van Tol MJ, Vossen JM, van den Elsen PJ. T cell immune reconstitution after allogeneic bone marrow transplantation in bare lymphocyte syndrome. Hum. Immunol. 2000;61:898–907. doi: 10.1016/s0198-8859(00)00156-7. [DOI] [PubMed] [Google Scholar]
- 32.Markert ML, Sarzotti M, Ozaki DA, Sempowski GD, Rhein ME, Hale LP, Le Deist F, Alexieff MJ, Li J, Hauser ER, et al. Thymus transplantation in complete DiGeorge syndrome: immunologic and safety evaluations in 12 patients. Blood. 2003;102:1121–1130. doi: 10.1182/blood-2002-08-2545. [DOI] [PubMed] [Google Scholar]
- 33.Markert ML, Alexieff MJ, Li J, Sarzotti M, Ozaki DA, Devlin BH, Sedlak DA, Sempowski GD, Hale LP, Rice HE, et al. Postnatal thymus transplantation with immunosuppression as treatment for DiGeorge syndrome. Blood. 2004;104:2574–2581. doi: 10.1182/blood-2003-08-2984. [DOI] [PubMed] [Google Scholar]
- 34.Rice HE, Skinner MA, Mahaffey SM, Oldham KT, Ing RJ, Hale LP, Markert ML. Thymic transplantation for complete DiGeorge syndrome: medical and surgical considerations. J. Pediatr. Surg. 2004;39:1607–1615. doi: 10.1016/j.jpedsurg.2004.07.020. [DOI] [PubMed] [Google Scholar]
- 35.Lee YJ, Jeon YK, Kang BH, Chung DH, Park CG, Shin HY, Jung KC, Park SH. Generation of PLZF+ CD4+ T cells via MHC class II-dependent thymocyte-thymocyte interaction is a physiological process in humans. J. Exp. Med. 2010;207:237–246. doi: 10.1084/jem.20091519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park WS, Bae Y, Chung DH, Choi YL, Kim BK, Sung YC, Choi EY, Park SH, Jung KC. T cell expression of CIITA represses Th1 immunity. Int. Immunol. 2004;16:1355–1364. doi: 10.1093/intimm/dxh132. [DOI] [PubMed] [Google Scholar]
- 37.Kaplan MH, Wurster AL, Smiley ST, Grusby MJ. Stat6-dependent and -independent pathways for IL-4 production. J. Immunol. 1999;163:6536–6540. [PubMed] [Google Scholar]
- 38.Patel DR, Li W, Park JS, Sofi MH, Gourley TS, Hangoc G, Kaplan MH, Chang CH. Constitutive expression of CIITA directs CD4 T cells to produce Th2 cytokines in the thymus. Cell. Immunol. 2005;233:30–40. doi: 10.1016/j.cellimm.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 39.Mohrs M, Shinkai K, Mohrs K, Locksley RM. Analysis of type 2 immunity in vivo with a bicistronic IL-4 reporter. Immunity. 2001;15:303–311. doi: 10.1016/s1074-7613(01)00186-8. [DOI] [PubMed] [Google Scholar]
- 40.Matsuda JL, Gapin L, Baron JL, Sidobre S, Stetson DB, Mohrs M, Locksley RM, Kronenberg M. Mouse V alpha 14i natural killer T cells are resistant to cytokine polarization in vivo. Proc. Natl. Acad. Sci. USA. 2003;100:8395–8400. doi: 10.1073/pnas.1332805100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Amsen D, Blander JM, Lee GR, Tanigaki K, Honjo T, Flavell RA. Instruction of distinct CD4 T helper cell fates by different notch ligands on antigen-presenting cells. Cell. 2004;117:515–526. doi: 10.1016/s0092-8674(04)00451-9. [DOI] [PubMed] [Google Scholar]
- 42.Tanigaki K, Tsuji M, Yamamoto N, Han H, Tsukada J, Inoue H, Kubo M, Honjo T. Regulation of alphabeta/gammadelta T cell lineage commitment and peripheral T cell responses by Notch/RBP-J signaling. Immunity. 2004;20:611–622. doi: 10.1016/s1074-7613(04)00109-8. [DOI] [PubMed] [Google Scholar]
- 43.Bendelac A. Positive selection of mouse NK1+ T cells by CD1-expressing cortical thymocytes. J. Exp. Med. 1995;182:2091–2096. doi: 10.1084/jem.182.6.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li W, Sofi MH, Rietdijk S, Wang N, Terhorst C, Chang CH. The SLAM-associated protein signaling pathway is required for development of CD4+ T cells selected by homotypic thymocyte interaction. Immunity. 2007;27:763–774. doi: 10.1016/j.immuni.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chung B, Aoukaty A, Dutz J, Terhorst C, Tan R. Signaling lymphocytic activation molecule-associated protein controls NKT cell functions. J. Immunol. 2005;174:3153–3157. doi: 10.4049/jimmunol.174.6.3153. [DOI] [PubMed] [Google Scholar]
- 46.Nichols KE, Hom J, Gong SY, Ganguly A, Ma CS, Cannons JL, Tangye SG, Schwartzberg PL, Koretzky GA, Stein PL. Regulation of NKT cell development by SAP, the protein defective in XLP. Nat. Med. 2005;11:340–345. doi: 10.1038/nm1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pasquier B, Yin L, Fondanèche MC, Relouzat F, Bloch-Queyrat C, Lambert N, Fischer A, de Saint-Basile G, Latour S. Defective NKT cell development in mice and humans lacking the adapter SAP, the X-linked lymphoproliferative syndrome gene product. J. Exp. Med. 2005;201:695–701. doi: 10.1084/jem.20042432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kovalovsky D, Uche OU, Eladad S, Hobbs RM, Yi W, Alonzo E, Chua K, Eidson M, Kim HJ, Im JS, et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat. Immunol. 2008;9:1055–1064. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kovalovsky D, Alonzo ES, Uche OU, Eidson M, Nichols KE, Sant’Angelo DB. PLZF induces the spontaneous acquisition of memory/effector functions in T cells independently of NKT cell-related signals. J. Immunol. 2010;184:6746–6755. doi: 10.4049/jimmunol.1000776. [DOI] [PubMed] [Google Scholar]
- 50.Weinreich MA, Odumade OA, Jameson SC, Hogquist KA. T cells expressing the transcription factor PLZF regulate the development of memory-like CD8+ T cells. Nat. Immunol. 2010;11:709–716. doi: 10.1038/ni.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verykokakis M, Boos MD, Bendelac A, Kee BL. SAP protein-dependent natural killer T-like cells regulate the development of CD8(+) T cells with innate lymphocyte characteristics. Immunity. 2010;33:203–215. doi: 10.1016/j.immuni.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]