Abstract
Mouse innate-like B cells are a heterogeneous collection of multifunctional cells that control infection, play housekeeping roles, contribute to adaptive immunity, and suppress inflammation. We show that, amongst leukocytes, chemokine internalisation by the D6 receptor is a unique and universal feature of all known innate-like B cell populations and, to our knowledge, the most effective unifying marker of these cells. Moreover, we identify novel D6active B1 cell subsets, including those we term B1d, which lack CD5 and CD11b but exhibit typical B1 cell properties, including spontaneous ex vivo production of IgM, interleukin-10, and anti-phosphorylcholine antibody. The unprecedented opportunity to examine D6 on primary cells has allowed us to clarify its ligand specificity and show that, consistent with a scavenging role, D6 internalises chemokines but cannot induce Ca2+ fluxes or chemotaxis. Unexpectedly, however, D6 can also suppress the function of CXCR5, a critical chemokine receptor in innate-like B cell biology. This is associated with a reduction in B1 cells and circulating class-switched anti-phosphorylcholine antibody in D6-deficient mice. Thus, we identify a unifying marker of innate-like B cells; describe novel B1 cell subsets; reveal a dual role for D6; and provide the first evidence of defects in resting D6-deficient mice.
Introduction
There are three mature B cell types in mice: follicular (FO), marginal zone (MZ), and B11. Despite distinct ontology, localisation, and immunophenotype, MZ and B1 B cells (collectively termed innate-like B cells (IBCs)) are functionally-related with restricted BCR repertoires enriched for certain host, commensal- and pathogen-associated antigens (Ags) (e.g. phosphorylcholine (PC)). Antibody (Ab) generation by IBCs occurs without T cell help, somatic hypermutation or affinity maturation, and class-switching is limited. After infection, IBCs rapidly generate large amounts of low affinity IgM, IgA and IgG3, supplementing pre-existing ‘natural’ Ab (derived mainly from IBCs2), and ensuring early protection against infection3-5. Natural Ab also aids important housekeeping functions, including apoptotic debris removal, lipoprotein sequestration6, and microflora regulation2. In addition to generating Abs, IBCs can transport Ag to follicular DCs, present Ag to naïve T cells7,8, and produce interleukin-10 (IL-10), a key factor in B cell-mediated immunosuppression9. Thus, IBCs are multifunctional cells with critical roles in homeostasis and immune responses.
Panels of Abs are needed to identify IBC subsets. Anti-CD21 and -CD23 distinguish MZ B cells (CD19+CD21hiCD23lo) from FO B cells (CD19+CD21intCD23hi), and provide some resolution of B cell progenitors (e.g. MZ B cell progenitors (MZP) are in the CD19+CD21hiCD23hi subset)1. B1 cells, abundant in body cavities, are categorised as CD19+CD11b+CD23− and subdivided into developmentally and functionally distinct B1a (CD5+) and B1b (CD5−) subsets. Other body cavity B cells are usually classified as FO B cells, but CD11b− B1 cells exist (e.g. B1c, CD19+CD11b−CD5+)10 arising earlier in ontogeny and showing greater reconstitution potential than CD11b+ B1 cells11. B1 cells undergo phenotypic changes when they leave body cavities: splenic B1 cells are CD19+IgMhiCD23loCD5+, but B1 cells elsewhere are difficult to detect12. Some proteins (e.g. CD9, CD43) are elevated on peritoneal cavity (PerC) B1a/b cells13,14, but do not allow definitive B1 cell identification in PerC, let alone elsewhere. Pan-IBC markers would have considerable utility and provide unifying insights into IBCs.
Functionally-distinct lymphocyte subsets carry specific chemokine receptor repertoires15. Trafficking of IBCs differs markedly from FO B cells, and their Ab-secreting capacity and survival is intimately linked to migration and adhesion7,12,16-19. Like FO B cells, IBCs are dependent on CXCR5 ligand, CXCL13, which controls departure from the MZ7,20, and B1 entry/exit from the PerC12,18,21. However, CXCR5, and other IBC receptors (CXCR4, CCR7)22-24, are present throughout the B cell compartment15. Mouse splenic B cells express transcripts for the atypical chemokine receptor D625,26. In transfected cell lines, D6 binds many CC chemokines but cannot drive chemotaxis or activate signalling pathways used by other chemokine receptors27,28. However, it constitutively traffics via the surface of these cell lines to facilitate chemokine scavenging29,30. This is thought to underpin indispensable roles for D6 in regulating inflammation, and associated pathology, in mice27,28,31,32. D6 also induces β-arrestin redistribution from cytoplasm to membrane33,34 which, in our hands, is driven by chemokine-independent D6 phosphorylation and dispensable for scavenging34. It might instead help regulate co-expressed chemokine receptors. Notably, however, D6 function on primary cells has not been investigated. Moreover, although D6 protein is expressed by lymphatic endothelial cells, trophoblasts, and some leukocytes in humans25,35-37, little is known about expression in mice, hampered by lack of reliable Abs.
Here we use fluorescent chemokines to provide unrivalled insight into active D6 protein expression by mouse leukocytes. Remarkably, D6 activity is a restricted and distinguishing feature of IBCs and is, to our knowledge, the best unifying marker of these cells. Detailed fractionation of body cavity B cells identifies novel subsets of conventional B1 cells; defines a new CD11b− B1 cell subset, termed B1d; and provides a rigorous definition of body cavity FO B cells (CD19+CD11b−CD5−CD23+D6negative). We also provide the first ligand specificity profile of D6 on primary cells; reveal a dual role for D6 on IBCs; and show that D6-deficient mice carry defects in their B1 cell compartment.
Materials and Methods
Mice
Wild-type (WT) and D6-deficient mice38 (C57BL/6J (F11)) were housed under specific pathogen-free conditions. Studies were approved by ethical review and licensed by the UK Home Office. Age-matched females (8–12wk) were used.
Cell isolation and culture
Cells were flushed from body cavities with PBS/2mM EDTA (ethylenediaminetetraacetic acid). Lymphoid tissue suspensions were prepared by mechanical disruption and passed through 40μm nylon mesh. Blood was collected by cardiac puncture. Red cells were lysed using 0.8% NH4Cl/10mM EDTA. Where appropriate, cells were cultured in complete medium (RPMI1640, penicillin/streptomycin, 4mM L-glutamine, 10% fetal calf serum (FCS)). FACS-sorted B cells were cultured for 3d at 2×105 cells/well in 200μl of media.
Chemokine uptake assay
Cells (≤2×106/well) were cultured (37°C, 60min) in 50μl of complete medium (+20mM HEPES (N-2-hydroxyethylpiperazine-N’-2-ethanesulfonic acid) pH7.4) containing 25nM AlexaFluor647 (AF647)-coupled human CCL2 or CCL22 (CCL2AF647 or CCL22AF647 (Almac)) +/−250nM unlabeled mouse chemokine (Peprotech) then briefly washed before immunostaining.
Flow cytometry and FACS
Cells were pre-incubated with Fc block (BD Biosciences) in FACS buffer (PBS, 1% FCS, 0.02% sodium azide, 5mM EDTA) and labeled with fluorescent anti-mouse Abs including: CD19 (ID3), CD11b (M1/70), CD5 (53-7.3), CD23 (B3B4) (BD Biosciences, various fluorochromes); CXCR5-biotin (2G8), CD49-biotin (R1-2), CD29-biotin (Ha2/5), CD21-PE (phycoerythrin) (7G6), IgM-PE (II/41) (BD Biosciences); CD9-APC (allophycocyanin) (KMC8), CD43-biotin (R2/60), B220-AF647 (RA3-6B2) (eBiosciences): isotype controls (BD Biosciences or eBioscience). Streptavidin-APC (BD Biosciences) detected biotinylated Abs. Dead cells were labeled using DAPI (4,6 diamidino-2-phenylindole) (Invitrogen) or Viaprobe (BD Biosciences). With goat anti-mouse D6 or isotype control (Abcam), cells were blocked with Fc block and donkey serum, and PE-conjugated donkey anti-goat IgG (Abcam) used for detection. Ab-labeled cells were analysed using FACScalibur or MACSQuant cytometers, or sorted on a FACS ARIA. Data were analysed using FlowJo (Treestar) with populations defined by size, viability, and ‘fluorescence minus one’ isotype controls.
Ca2+ Flux
PerC cells (4×106/ml) were labeled in calcium buffer (145mM NaCl, 5mM KCl, 1mM MgCl2, 10mM HEPES, 1mM CaCl2, 0.18% Glucose, 0.2% BSA (bovine serum albumin), pH7.4) with Fura-Red (4μM) and Fluo-4 (2μM) (Invitrogen) (37°C, 30min), washed twice, resuspended in calcium buffer, incubated (RT, 30min), then stained with Abs. Samples were incubated (37°C, 10min) before flow cytometry. Baseline fluorescence was determined (30-45s); CXCL13 (to 100nM) and/or CCL22 (to 50nM) added; then ionomycin (to 1μM). [Ca2+] is expressed as Fluo-4/Fura-Red mean fluorescence intensity ratio. Data were analysed using FlowJo.
Quantitative PCR
RNA was extracted using RNeasy columns with DNAse treatment (Qiagen), cDNA generated (AffinityScript (Stratagene)) and QPCR done using D6-specific Taqman assay (mm0044555_m1) on a Prism 7900HT (Applied Biosystems). GAPDH-specific probes were used to normalise D6 expression. Analysis used the relative quantitation ΔΔ−2 CT method to give an RQ (fold change) value, with splenic FO or PerC CD11b−CD5− B cells as calibrators set to 1.
Chemotaxis assay
PerC cells were resuspended in chemotaxis buffer (RPMI1640; 0.5% BSA), and B cell subset composition determined by FACS. 5×105 cells were put in upper chambers of 24-well Transwell plates (3μm filters (Corning)) above 600μl of chemotaxis buffer (+/−chemokine (Peprotech)) and incubated (37°C, 5% CO2, 3h). Migrated cells were stained with Abs, enumerated by FACS (using CountBrite™ beads (Invitrogen)), and converted into percent of input for each subset.
ELISAs
IL-10 was quantified using a mouse IL-10 ELISA (Insight). For serum Ig, wells were coated with 5μg/ml anti-mouse IgM, IgG3 or IgA polyclonal antibody (Bethyl), washed (PBS/0.05% Tween), blocked with PBS/3% BSA, and sera or supernatants added. After 1h, wells were washed, and captured Ag detected using peroxidase-conjugated anti-mouse IgM (Paris Anti-corps), IgG3, or IgA (Bethyl). ELISAs were developed using Sureblue™ (Insight). Total Ig was quantified relative to a Mouse Reference Serum (Bethyl). For PC-specific Ab, plates were coated with PC-conjugated BSA (Biosearch), normalising with a reference serum.
Microscopy
FACS-sorted cells were cytospun onto Polysine™ slides (VWR International), stained (hematoxylin/eosin), and analysed on an Axiostar Plus microscope with Axiovision software.
Statistics
Data were analysed using unpaired Student T tests or ANOVA in GraphPad PRISM 4 software.
Results
D6-mediated chemokine uptake is restricted to MZ B cells in spleen
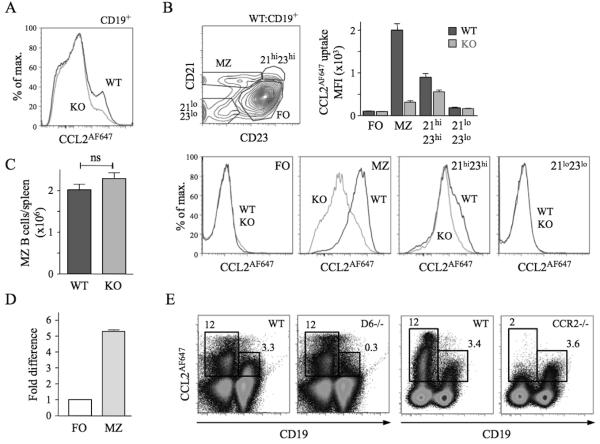
CCL2AF647-loaded splenocytes were analysed by flow cytometry (Figure 1). Nearly all D6-dependent CCL2AF647 uptake by CD19+ cells was by MZ B cells. The CD21hiCD23hi B cell population (which contains MZPs) also showed activity (~20%) (Figure 1B). Similar results were seen when CCL22AF647 was used (data not shown). CCL2AF647 uptake by WT MZ B cells was blocked by unlabeled CCL3 or CCL22, and CCL22AF647 uptake was inhibited by CCL2, consistent with D6’s unique ability to bind CCL2, 3 and 22 (data not shown). MZ B cell abundance was unaffected by D6 deletion (Figure 1C). D6 transcripts were more abundant in MZ B cells than FO B cells (Figure 1D), consistent with microarray data26. A commercially-available anti-D6 polyclonal Ab, reported to preferentially bind MZ B cells26, stained all B cells strongly (vs. isotype controls) but, importantly, this was unaffected by D6 deficiency (Supplementary Figure 1A). CCL2AF647 uptake, with appropriate controls, was the only effective way of detecting mouse D6 protein.
Figure 1. MZ B cells express active D6.
(A) Overlaid CCL2AF647 uptake histograms of WT (black) and KO (grey) CD19+ splenocytes. (B) Dotplot showing fractionation of WT CD19+ splenocytes using anti-CD21 and -CD23, and overlaid CCL2AF647 uptake histogram plots from WT (black) and KO (grey) subsets. Bar graph shows average Mean Fluorescence Intensity (MFI) (+/−SEM) of CCL2AF647 uptake by each subset. (C) Mean number (+/− SEM) of MZ B cells (CD19+CD23loCD21hi) per WT and KO spleen (n=18 per strain). ns, not significant (D) D6 mRNA expression by FACS-purified WT splenic FO and MZ B cells. FO cells were set to 1. (E) CCL2AF647 uptake profiles of D6-deficient (D6−/−) and CCR2-deficient (CCR2−/−) CD19+ and CD19− splenocytes alongside WT samples prepared in parallel. Boxes highlight cells with specific CCL2AF647 uptake based on controls performed in the absence of CCL2AF647. Flow cytometry profiles are representative of data generated from >3 mice per genotype per experiment, with experiments repeated at least 3 times.
Many WT non-B cells internalised CCL2AF647. This was unaffected by D6 deletion, or inclusion of unlabeled CCL22, but lost when CCR2 was absent (Figure 1E; data not shown). Uptake by CCR2-deficient MZ B cells was comparable to WT, and blocked by unlabeled CCL12 (CCR2/D6 ligand) (Figure 1E; Supplementary Figure 1C). CCL2AF647 uptake by WT non-B cell splenic subsets (CD4+, CD8+, or CD4+CD25+ T cells; monocytes; NK cells; macrophages (MOs); γδ T cells) was substantially reduced in equivalent CCR2-deficient populations, but unaffected by D6 deletion (data not shown). Thus, D6 activity is restricted to MZ B cells and CCR2 does not contribute to their CCL2AF647 uptake.
All conventional body cavity B1 cells show robust D6 activity
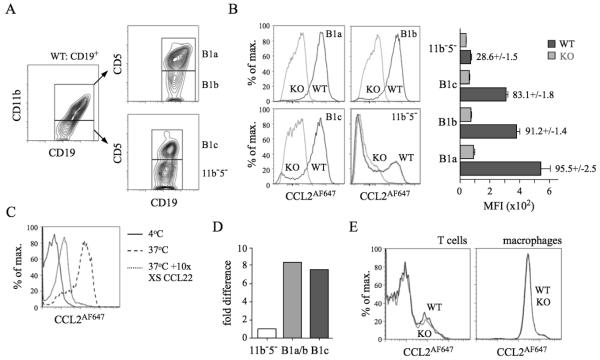
PerC B cells were subdivided by CD11b/CD5 expression into B1a, B1b, B1c, and CD11b−CD5− cells (Figure 2A). Strikingly, nearly all WT B1a, B1b and B1c cells, and a subset of CD11b−CD5− B cells, internalised large amounts of CCL2AF647 (Figure 2B) or CCL22AF647 (data not shown) unlike D6-deficient counterparts. Uptake by WT CD11b+ B cells was blocked by excess unlabeled CCL22, and not observed at 4°C, when receptor internalisation is prevented (Figure 2C). D6 transcripts were more abundant in CD11b+ B cells and B1c cells compared with CD11b−CD5− B cells (Figure 2D). The polyclonal anti-mouse D6 Ab showed no D6-dependent binding to B1a/b cells (Supplementary Figure 1B). All PerC non-B cells lacked D6 activity: some T cells, and all MOs, internalised CCL2AF647, but this was unaffected by D6 deletion (Figure 2E), or inclusion of unlabeled CCL22 (data not shown). Uptake by WT and D6-deficient T cells was blocked by CCL2 or CCL12 (Supplementary Figure 2), and lost from CCR2-deficient T cells (data not shown). CCL2 or CCL12 partially prevented uptake by MO, suggesting CCR2-dependent and D6/CCR2-independent mechanisms (Supplementary Figure 2). Pleural cavity (PleC) B1a, B1b, and B1c cells, and a CD11b−CD5− B cell subset, showed strong D6-dependent CCL2AF647 uptake (Supplementary Figure 3). PleC T cells and MOs had no D6 activity. Thus, D6 activity is a universal property of conventional body cavity B1 cells in resting mice.
Figure 2. All conventional PerC B1 cells show D6-dependent CCL2AF647 uptake.
(A) Flow cytometric fractionation of WT PerC CD19+ cells using anti-CD11b and -CD5. (B) Overlaid CCL2AF647 uptake profiles of WT (black) and D6-deficient (KO; grey) PerC CD19+ cell subsets. Bar graph shows average Mean Flourescence Intensity (MFI) (+/−SEM) of CCL2AF647 uptake by each subset. (C) Overlaid flow cytometry plots of CCL2AF647 uptake by WT CD19+CD11b+ PerC cells after incubation at 4°C (black), or at 37°C in the presence (grey) or absence (dashed) of 10-fold excess of unlabeled CCL22. (D) Expression of D6 mRNA in FACS-sorted B cell populations (pooled from 4 WT mice). ‘B1a/b’ are CD19+CD11b+ cells. Expression by CD19+CD11b−CD5− (11b−5−) cells was set to 1. (E) Overlaid CCL2AF647 uptake profiles of WT (black) and KO (grey) PerC T cells (CD19−CD11b−CD5hi) and MOs (CD19−CD11b+CD5−). Flow cytometry profiles are representative of data generated from >3 mice per genotype per experiment, with experiments repeated on at least 3 occasions.
CD23+ body cavity B1 cells with elevated expression of FO B cell markers
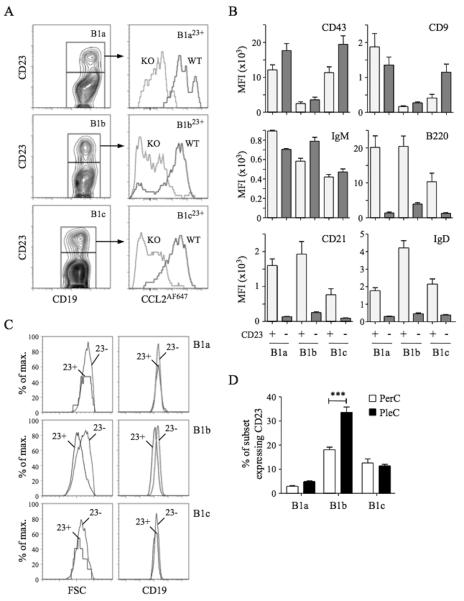
D6 activity amongst CD11b−CD5− body cavity B cells suggested the presence of B1 cells. CD23 is on most FO B cells but thought to be absent from B1 cells, so we used anti-CD23 to try to identify FO B cells amongst CD11b−CD5− B cells. CD11b−CD5− PerC B cells could be subdivided into CD23+ and CD23− populations (see below) but, unexpectedly, CD23+ B1a, B1b and B1c cells (termed B1a23+, B1b23+ and B1c23+) were also present, representing 5-22% of each subset (Figure 3). CD23+ B1 cells showed strong D6-dependent CCL2AF647 uptake and expressed abundant CD43, CD9 and IgM, but, surprisingly, also had high levels of B220, CD21 and IgD, typically seen as FO B cell properties. These were not B1/FO doublets because, as in all flow cytometry, we gated for small, live cells prior to analysis (which successfully removed CD19+CD5hi B/T conjugates (Supplementary Figure 4)). Moreover, CD23+ B1 cells were not bigger than CD23− counterparts and showed equivalent anti-CD19 fluorescence. In fact, B1b23+ cells were smaller than CD23− B1b cells, and expressed less CD19. CD23+ B1 cells with strong D6 activity were also present in PleC, where a larger proportion of B1b cells were CD23+ (Figure 3D). Thus, body cavity B1 cells, expressing B1 markers (CD11b, CD5, CD43, CD9, IgM) and with strong D6 activity, can have high surface expression of proteins typically associated with FO B cells.
Figure 3. Characterisation of D6active body cavity CD23+ B1 cells.
(A) Left panels: Sub-fractionation of WT PerC B1a (CD19+CD11b+CD5+), B1b (CD19+CD11b+CD5−) and B1c (CD19+CD11b−CD5+) cells by anti-CD23. Right panels: Overlaid CCL2AF647 uptake profiles of WT (black) and D6-deficient (KO, grey) PerC CD23+ B1 cell subsets. (B) Surface immunophenotype of WT PerC CD23+ (light grey) and CD23− (dark grey) B1 cell subsets. Data show average Mean Fluorescence Intensity (+/−SEM). (C) Forward scatter (FSC) and surface CD19 of WT PerC CD23+ (23+) and CD23− (23−) B1 cell subsets. (D) Mean percentage (+/−SEM) of each B1 cell subset expressing surface CD23 in WT PerC and PleC lavages (n=5). ***p<0.001. Flow cytometry profiles are representative of data from >3 mice per genotype per experiment, with experiments repeated on at least 3 occasions.
D6 activity identifies B1 cells amongst body cavity CD11b−CD5− B cells
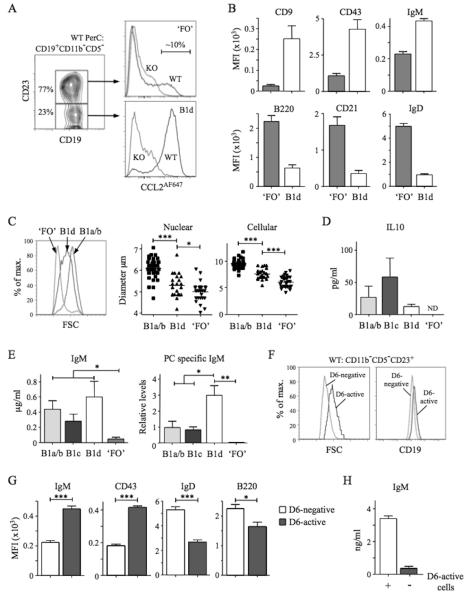
PerC CD11b−CD5− B cells were divisible into CD23− and CD23+ subsets (Figure 4A). Most triple negative B cells (CD11b−CD5−CD23−) and some CD11b−CD5−CD23+ cells showed robust D6-dependent CCL2AF647 uptake. Amongst PleC CD11b−CD5− B cells, >95% CD23− cells and ~15% CD23+ cells had D6 activity (data not shown). The D6 activity of CD11b−CD5−CD23− B cells implied that they were B1 cells: we provisionally termed them B1d. CD11b−CD5−CD23+ cells were predominantly D6negative so we called them ‘FO’ B cells (quotation marks reflecting the presence of some D6active putative B1 cells). Consistent with these definitions, B1d cells had more surface CD19, CD9, CD43 and IgM than ‘FO’ B cells, and less B220, CD21 and IgD (Figure 4A-B). They were, on average, larger than ‘FO’ B cells but smaller than B1a/b cells (Figure 4C). In addition to lacking CD11b, B1d carried less IgM, CD29 and CD49 than B1b, while lack of CD5, and less CD9 and CD43, distinguished them from B1a and B1c (data not shown). Characteristically, cultured B1a/b cells spontaneously secreted IL-10 and IgM enriched with anti-PC specificities (Figure 4D-E). B1c and B1d cells, but not ‘FO’ B cells, also produced substantial quantities of IL-10 and IgM. PC-specific IgM was produced by B1a/b and B1c cells, but not ‘FO’ B cells, and, notably, was most abundant in B1d cultures. Thus, B1d and B1c cells, a major fraction of PerC CD11b− B cells, have robust IBC-like activity ex vivo.
Figure 4. Identification and characterisation of PerC B1d and B1d23+ cells.
(A) Left panel: Sub-fractionation of WT PerC CD19+CD11b−CD5− cells by anti-CD23. The abundance of the two subsets is shown as the mean percentage of total CD19+CD11b−CD5− cells. Right panels: Overlaid CCL2AF647 uptake profiles of WT (black) and D6-deficient (KO, grey) CD23+ and CD23− subsets, referred to as ‘FO’ and B1d, respectively. Proportion of D6active ‘FO’ B cells is indicated. (B) Surface immunophenotype of WT PerC ‘FO’ and B1d B cells. Data show average Mean Fluorescence Intensity (+/−SEM). (C) Comparison of size of WT PerC ‘FO’, B1d and CD11b+ B1 (B1a/b) cells by forward scatter (FSC) (left panel), and microscopic measurements of nuclear and cellular diameter (middle and right panels) of FACS-sorted PerC cells. (D-E) Production of IL-10, IgM, and anti-PC Ab by FACS-sorted, cultured CD11b+ B1 (B1a/b), B1c, B1d, and ‘FO’ cells. Data show mean (+/−SEM) of data generated from 3 repeat experiments. ND, not detected. (F) Overlaid forward scatter (FSC) (left panel) and surface CD19 (right panel) of WT PerC D6- active and D6-negative CD19+CD11b−CD5−CD23+ ‘FO’ cells. (G) Surface immunophenotype of WT PerC D6-active and D6-negative CD19+CD11b−CD5−CD23+ ‘FO’ cells (average Mean Fluorescence Intensity (+/−SEM)). (H) Production of IgM by FACS-sorted, cultured CD19+CD11b−CD5−CD23+ cells in the absence (−) or presence (+) of their D6-active subset. Flow cytometry profiles are representative of data from at least 3 mice per genotype per experiment, with experiments repeated on at least 3 occasions. *p<0.05; **p<0.01; ***p<0.001.
The existence of CD23+ B1a, B1b and B1c cells suggested that D6active PerC ‘FO’ B cells (Figure 4A) could be CD23+ B1d cells (B1d23+). Indeed, like B1d cells, they were larger than D6negative CD11b− CD5−CD23+ cells, and expressed more CD19, IgM and CD43 (Figure 4F-G). Like B1a23+, B1b23+ and B1c23+ cells, B1d23+ cells carried more surface IgD and B220 than B1d cells, although still less than D6negative CD11b−CD5−CD23+ cells (Figure 4G). Moreover, removal of D6active cells from CD11b−CD5−CD23+ cells substantially reduced their spontaneous, low-level IgM production (Figure 4H). Thus, D6 activity defines phenotypically distinct cells amongst the CD11b−CD5−CD23+ population that resemble CD23+ variants of all other B1 cell types.
In summary, D6 activity is not limited to conventional B1 cells, identifying novel B1 subsets amongst CD11b−CD5− B cells. We define nine distinct body cavity B cell subsets in naïve mice: eight show clear evidence of B1 cell identity and represent >75% of the B cell compartment (Supplementary Table 1). Unlike existing markers, D6 activity is a specific unifying feature of these cells.
D6active B cells in omentum, spleen, blood, gut-associated lymphoid tissue (GALT), and intestine
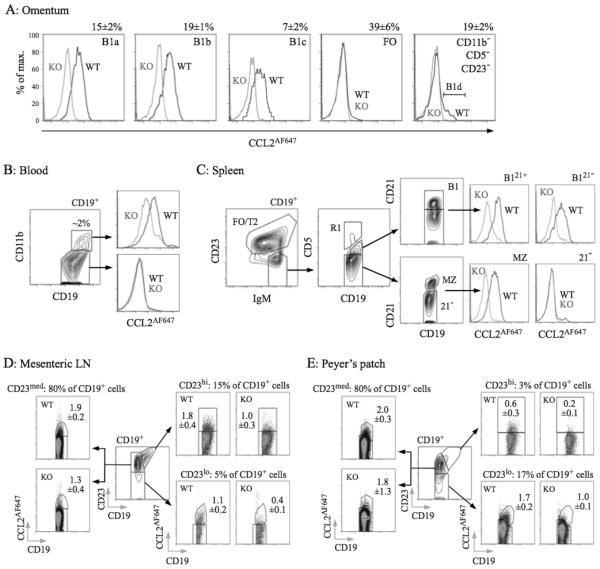
B1 cells are thought to enter and exit the PerC via the omentum, circulate in blood, and reside in other lymphoid tissues, including spleen12,18,21,39. They also enter the intestinal lamina propria and GALT, contributing to commensal-induced IgA generation2,16,19. We compared CCL2AF647 uptake by WT and D6-deficient B cells from these tissues/fluids (Figure 5), confirming D6 activity by competing uptake into WT B cells with unlabeled CCL22 (data not shown). Blood-borne CD11b+ B cells, omental B1a, B1b, and B1c cells (which were almost exclusively CD23−), and splenic B1 cells (CD19+IgMhiCD23loCD5+)12 all had D6 activity (Figure 5A-C). Anti-CD21 identified two clear splenic B1 subsets, and D6active MZ B cells amongst IgMhiCD23loCD5− cells (Figure 5C). As expected for IBCs, splenic B1 and MZ B cells were larger and expressed less IgD than FO B cells (Supplementary Figure 5). Circulating CD11b− B cells, and IgMhiCD23loCD5−CD21− (enriched for T1 cells) and IgM+CD23+ splenic B cells, were D6negative (Figure 1B-C and data not shown). D6active CD11b−CD5− B cells (B1d/B1d23+) were undetectable in spleen or blood. In omentum <10% of CD11b−CD5−CD23− B cells were D6active and CD11b−CD5−CD23+ B cells were almost exclusively D6negative (Figure 5A). This could be due to loss of D6 activity, but more likely reflects increased abundance of CD23− FO B cells outside the body cavities. Likewise, D6 activity was rare in LN and Peyer’s patch B cells (uniformly CD11b−CD5−), even in CD23lo fractions. Nonetheless, although B cells in skin-draining LNs lacked D6 activity (data not shown), it was detectable amongst GALT B cells, particularly those in mesenteric LN (MesLN), but also Peyer’s patch (Figure 5D-E). D6active B cells were also found in bone marrow, colon32, and small intestine, and all had surface immunophenotypes consistent with an IBC identity (data not shown). Thus, along with MZ B cells and body cavity B1 cells, D6 activity is associated with IBCs, and B cells with IBC-like immunophenotypes, at other locations in resting mice.
Figure 5. D6active B1 cells in omentum, blood, spleen, MesLN, and Peyer’s patch.
(A) Overlaid CCL2AF647 uptake profiles for WT (black) and D6-deficient (KO, grey) omental CD19+ B cell subsets (identified as in PerC lavages). Numbers above plots show proportion of retrieved CD19+ cells in each subset in WT mice (mean+/−SEM). B1d cells (CD19+CD11b−CD5−CD23−D6active) are indicated in the far right plot. (B) Contour plot showing CD11b expression on CD19+ blood cells, with proportion of CD19+ CD11b+ cells indicated. Overlaid CCL2AF647 uptake profiles are shown in the right panels (WT, black; KO, grey). (C) Contour plots showing gating of CD19+ splenocytes for FO/T2 cells, MZ B cells, and B1 cells (fractionated into CD21+ (B121+) and CD21− (B121−) subsets). Events in gate R1 (CD5hi) are rare T/B cell conjugates, and were excluded. The CD19+IgMhiCD23loCD5−CD21− population, containing T1 progenitors, are labeled ‘21−’. The overlaid histograms show CCL2AF647 uptake profiles for WT (black) and KO (grey) subsets. (D-E) CD19+ cells in MesLN and pooled Peyer’s patches fractionated into CD23lo, CD23med and CD23hi subsets (central dotplot in each figure). Subset size, as proportion of retrieved WT CD19+ cells, is indicated above each CCL2AF647 uptake dotplot. Mean (+/−SEM) of CCL2AF647-positive cells (as a proportion of retrieved CD19+ cells) is indicated on each plot. All data are representative of experiments repeated 2-5 times.
B1 cell D6 does not elicit Ca2+ fluxes or induce chemotaxis
Our limited insight into the molecular biology of mouse D6 comes from in vitro studies of immortalised cell lines expressing high levels of exogenous D6. IBCs present a unique opportunity to examine D6 on primary cells. First, we explored ligand selectivity by assessing if previously assigned D6 ligands block CCL2AF647 uptake by B1 cells (Figure 6A). CCL2, 3, 4, 5, 11, 17 and 22 did, but unexpectedly, CCL7 did not and CCL8 was unable to block D6- or CCR2-mediated CCL2AF647 uptake (data not shown). Also, despite not being thought of as a CCR2 ligand, CCL3 partially blocked CCR2-mediated CCL2AF647 uptake by PerC T cells (Figure 6A), splenic Ly6Chi monocytes (Supplementary Figure 6) and other splenic subsets, even at low concentrations (12.5nM). Next, we examined signalling and chemotaxis (Figure 6B-E). CCL22 was used: PerC B1a/b cells lack CCR4 (data not shown), so D6 is their only CCL22 receptor. Positive controls (ionomycin and CXCL13 (non-D6 ligand)) generated robust Ca2+ fluxes in WT and D6-deficient B1a/b cells, while MOs only responded to ionomycin. CCL22 did not affect Ca2+ levels in either cell type, or alter CXCL13 responses in B cells. Prolonged CCL22 pre-incubation did not dampen B1a/b CXCL13 responses. Similar unresponsiveness was seen with CCL2 or 3 (data not shown). CCL2, 3, 4, 5 or 22 were unable to induce Ca2+ fluxes in WT splenic B cells: CXCL13 gave strong responses (data not shown). WT PerC B cells migrated towards CXCL13 but not CCL22 or CCL3 (Figure 6E). CCL22 and CCL3 were bioactive because they could block D6-mediated CCL2AF647 uptake and induce migration through receptors other than D6 (data not shown). Thus, D6 on B1 cells internalises its ligands, but cannot induce Ca2+ fluxes or cell migration after chemokine engagement. In addition, flow cytometry using a panel of Abs (including IgM, IgD, B220, CXCR4, CXCR5, CD1d, 9, 11a, 11b, 19, 21, 23, 29, 43, 49, 69, 86) did not reveal any significant differences between WT and D6-deficient body cavity B cells (data not shown). These observations are consistent with the designation of D6 as a chemokine scavenger.
Figure 6. Chemokine binding and signalling properties of D6 on PerC B1 cells.
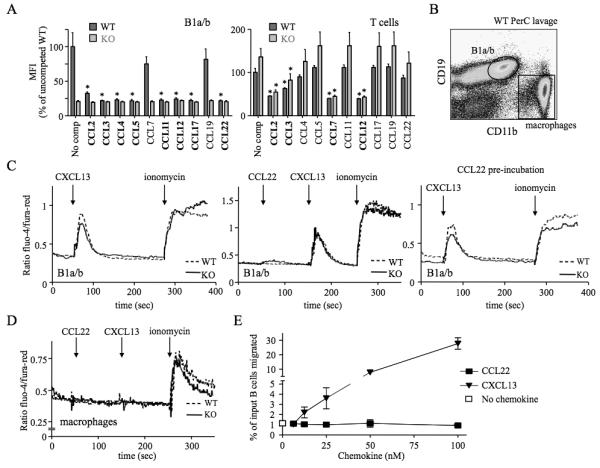
(A) CCL2AF647 uptake by WT and D6-deficient (KO) PerC B1a/b (CD19+CD11b+) or T cells (CD19−CD11b−CD5+) in the presence of a 10-fold molar excess of the indicated unlabeled chemokines. ‘No comp’; CCL2AF647 uptake in the absence of competitor. The average Mean Fluorescence Intensity (MFI) of competed samples is shown as percent (+/−SEM) of the average MFI of uncompeted WT uptake. Asterisks mark chemokines that significantly reduced CCL2AF647 uptake by WT or KO cells (p<0.01). (B) Gates used to gauge Ca2+ fluxes in B1a/b and MOs. (C-D) Representative Ca2+ flux profiles of Fluo-4/Fura-Red-loaded WT and KO PerC B1a/b cells (C) and MOs (D). The points of chemokine (50-100nM) and ionomycin addition are indicated with arrows. Cells in the far right panel of C were incubated in 100nM CCL22 for 1h at 37°C before analysis. (E) Migration of WT B cells from PerC lavages to CCL22 or CXCL13. Migration in media alone is indicated by the unfilled square. The mean number of live cells (+/−SEM) migrated from 4 replicate wells is presented as percent of input. All data are representative of 3 or more independent experiments.
D6 deficiency modifies B1 cells, the B1 cell compartment and anti-PC abundance
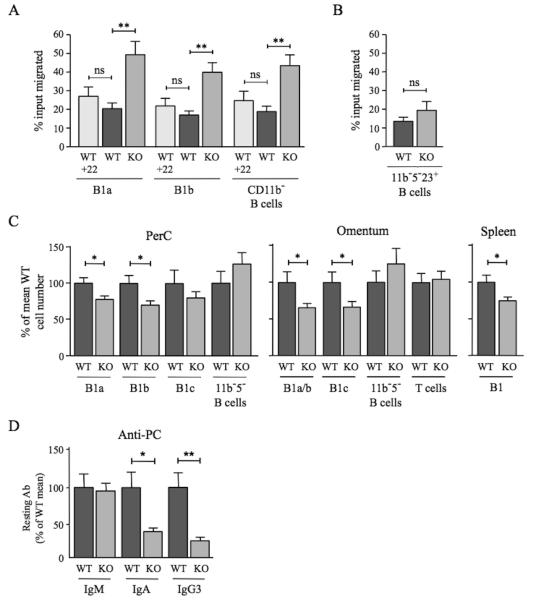
All existing in vitro and in vivo data, including those above, point to a scavenging role for D6. However, D6 drives β-arrestin redistribution in cell lines33,34 so could regulate responses through other chemokine receptors. Thus, we directly compared the migratory responses of D6-deficient and WT PerC B cells towards CXCL13. Interestingly, there was a substantial increase in the migration of D6-deficient B1a, B1b, and CD11b− B cells (i.e. B1c, B1c23+, B1d, B1d23+ and FO B cells) towards 100nM CXCL13 when compared with WT (Figure 7A). D6 engagement failed to modify the response of WT B1 cells to 100nM CXCL13, since addition of CCL22 (Figure 7A) or CCL3 (data not shown) to upper or lower chambers of chemotaxis wells, or pre-incubation with these chemokines (data not shown), did not significantly alter migration. D6 deficiency had no significant impact on basal motility, or migration to 50nM CXCL13, but it was associated with enhanced migration towards 200nM CXCL13 (Supplementary Figure 7A). PerC CD5−CD11b−CD23+ B cell chemotaxis towards CXCL13 was unaffected by deletion of D6 (Figure 7B), and D6-deficient inguinal LN B cells were not more responsive than those from WT animals (data not shown). Consistent with previous studies24, CXCL12 was a much less effective PerC B cell attractant than CXCL13. Its ability to stimulate B1a and B1b cell migration was unaffected by D6 deletion (Supplementary Figure 7B). Thus, D6 can specifically suppress B1 cell migration towards CXCL13.
Figure 7. Impact of D6 deficiency on B1 cell motility, B1 cell abundance, and serum anti-PC levels.
(A) WT and D6-deficient (KO) PerC B cells that migrated in response to 100nM CXCL13 (plus 50nM CCL22 where indicated (+22)) were enumerated by flow cytometry after staining with antibodies against CD19, CD11b and CD5. Data are representative of at least three repeat experiments. (B) Proportion of WT and KO PerC CD5−CD11b−CD23+ B cells migrating towards 100nM CXCL13. A repeat experiment generated similar results. In A and B, data are presented as the mean number of live cells migrated (as percent of input) (+/−SEM) from at least four replicates. (C) Number of cells in the indicated B cell subsets in PerC lavage, omentum and spleen of WT and D6-deficient (KO) mice as a percent (+/−SEM) of WT (WT average set to 100%). PerC, 26 mice per genotype; omentum, 20 mice per genotype; spleen, 6 mice per genotype. ‘11b−5− B cells’ were CD19+CD11b−CD5−; omental T cells were FSCloSSCloCD19−CD11b− CD23−CD5hi. (D) Mean quantity of anti-PC Abs (+/−SEM) in the serum of resting WT and KO mice (9 per genotype). Mean WT levels set to 100%. Similar data were generated in a repeat experiment. *p<0.05; **p<0.01; ***p<0.001; ns, not significant.
B1 cell motility regulates PerC cellularity and Ab production against host/commensal Ag12,16,19,22. Indeed, fewer cells were retrieved from the PerC of D6-deficient mice compared with WT. This was due to a significant reduction in B1a and B1b cells: MOs, T cells and other B cell subsets were unaffected (Figure 7C and data not shown). D6-deficient mice also had fewer omental CD11b+ B cells and B1c cells, and fewer splenic B1 cells (Figure 7C). Splenic FO B cell and T cell numbers were normal (data not shown). Furthermore, B1 cell deficiency was accompanied by a significant reduction in circulating class-switched anti-PC Abs (Figure 7D), despite normal levels of PC-specific IgM, and total IgM, IgA and IgG3 (data not shown). Thus, D6 deficiency alters the chemotactic responsiveness of B1 cells, reduces the size of the B1 cell compartment, and suppresses constitutive production of anti-PC Abs.
Discussion
Comprehensive dissection of B cell populations in resting mice reveals D6 activity to be a feature of all conventional IBCs (i.e. MZ B cells, B1a, B1b). It also identifies B1 cells in blood, spleen, and omentum, and amongst body cavity CD11b− B cells (B1c and B1d). By extension, D6active B cells in intestinal lamina propria32 and GALT are presumably IBCs. In contrast, we did not detect D6 activity on (i) non-B cells, (ii) bone marrow B cell precursors or B1 progenitors (CD19+B220lo/−veLin−)40, or (iii) CD138+ plasma cells (data not shown). Activation of BCR and/or TLRs fails to induce D6 activity on splenic or PerC FO B cells, although this can, in some cases, reduce B1 or MZ B cell D6 activity (data not shown). Nonetheless, D6-mediated chemokine uptake is clearly a unique and specific property of IBCs in resting mice. In fact, to our knowledge, it is the best unifying marker of these cells. It showed far broader expression amongst IBCs than any surface marker we examined, which could not even identify all B1 cells in body cavities, let alone elsewhere. For example, anti-CD9 or -CD43 did not distinguish PerC B1b and B1d cells from FO B cells (Supplementary Figure 8). Fc receptor-like protein FCRL5 (FcRH3) might be as widely expressed as D6 activity amongst IBCs41. It is absent from splenic FO B cells, but detected on MZ B cells, MZPs, and B220+CD5+ and B220loCD5− PerC B cells. Some PerC ‘FO’ B cells (defined as B220hiCD5−) are also FCRL5+. These might be B1b23+, B1d or B1d23+ cells. It will be interesting to examine FCRL5 expression by splenic B1 cells and other IBCs outside body cavities, and directly compare distribution of FCRL5 and D6 activity amongst B cells.
We were surprised to find B1 cells (B1a-d23+) with elevated expression of ‘typical’ FO B cell markers, but this is not without precedent. Bw cells, the dominant PerC B cell subset in outbred mice, are CD5−CD11b+B220hiIgMhiIgDhiCD43−CD9−42, similar to B1b23+ cells (CD5−CD11b+B220hiIgMhi IgDhiCD43loCD9lo). Bw cells are enriched for autoAbs and anti-PC specificities, features of IBCs. We predict they will have robust D6 activity.
The precise interrelationship between body cavity B1 cell subsets remains to be defined. PerC CD11b− B1 cells arise earlier in B1 ontogeny, show greater reconstitution potential, and can act as CD11b+ B1 cell progenitors11. Based on immunophenotypes, it is possible that B1c and B1d cells are progenitors for B1a and B1b cells, respectively. Interestingly, there are rare D6negative cells within body cavity B1c and B1d subsets. Since MZPs only carry weak D6 activity (Figure 1B) and transitional B cells are D6negative, we speculate that these cells are more primitive B1 progenitors.
Immunophenotyping, D6 activity, and ex vivo functional studies support using CD19+CD11b−CD5−CD23+D6negative as a rigorous definition of body cavity FO B cells. This has implications for previous work. For example, PerC ‘FO’ B cells are reported to be (i) expanded in CCR7-deficient mice23, (ii) B1-like and capable of giving rise to B1b43, or (iii) capable of specific PerC homing44, based on B220hi, B220+CD11b−CD5− or CD19+B220hiCD23+ immunophenotypes, respectively. These populations contain substantial numbers of B1 cells. Similarly, gene-deficient mice with reported defects in FO or B1 B cell development were analysed using what was, in retrospect, imprecise phenotyping. BAFF depletion, or deletion of BAFF or BAFF-R, is reported to cause profound FO B cell deficiency, with B1 cells either unaffected45, reduced in PerC only46, or reduced in spleen only47. In each case, a different, and inadequate, B1 cell identification method was used. Likewise, IL7-deficiency is reported to only affect FO B cells, but the observed 75% loss of CD11b− PerC B cells might include B1 cells48.
Fluorescent chemokine uptake is currently the only way of effectively detecting D6 protein in mice. It also provides unrivalled sensitivity for CCR2 detection (Figure 1), outperforming the anti-CCR2 Abs we have tested, including the widely-used mAb MC2149 (data not shown). Our study highlights the importance of using primary cells to define receptor specificity. Mouse D6 is promiscuous, but CCL7 and CCL8 are not ligands. CCL7 induces migration via CCR1-315: by avoiding D6-mediated scavenging it might be uniquely placed to drive protracted periods of recruitment during inflammation. Mouse CCL8, typically classified as a CCR2 ligand, did not block uptake by CCR2. In fact, it has recently emerged that mouse CCL8 is a ligand for CCR8, not CCR2, with CCL12 being the likely orthologue of human CCL8 (a bona fide human CCR2 ligand)50. Unexpectedly, CCL2AF647 uptake via CCR2 was blocked by CCL3, contrary to the accepted view that it does not bind CCR215. CCL3 competition was previously used to infer D6 usage during CCL2AF647 uptake25: these data need careful re-evaluation. CCL22 competition alone reliably confirms D6 usage, while CCL7 competition is an effective CCR2 discriminator.
IBC D6 internalises chemokines, but cannot induce Ca2+ fluxes or chemotaxis. This is the first examination of D6 on primary cells, and provides critical support for the scavenger model of D6 function. D6-mediated chemokine scavenging, like IL-10 release, might contribute to the anti-inflammatory properties of IBCs and experiments are underway to explore this possibility. Understanding where and when D6-mediated scavenging occurs requires consideration of the microanatomical limitations of IBCs and their behaviour during inflammation. Peritonitis or splenic inflammation is typically accompanied by the rapid departure and differentiation of large numbers of IBCs from the PerC or MZ, respectively. However, IBC compartments repopulate as inflammation subsides and homeostasis is restored, and we predict that it will be during this period that chemokine modulation by IBC D6 will be important.
D6 deficiency leads to three IBC phenotypes: (i) enhanced B1 cell responses to CXCL13, (ii) reduction in B1 cells, and (iii) lower levels of serum anti-PC Abs. These phenotypes might be linked because the migratory properties of B1 cells profoundly influence their survival and Ab-secreting capacity12,16,18,19. Resting CXCL13- and CXCR5-deficient mice have few body cavity and omental B1 cells12,22,23, and reduced serum IgM specific for PC, despite normal levels of total IgM, IgA and IgG312. The phenotypes in D6-deficient animals are less dramatic, but the enhanced sensitivity of their B1 cells to CXCL13 might alter their recirculation and distribution to perturb access to niches required for optimal survival, expansion, and/or differentiation. CXCL13-deficient mice also show poor responses to exogenous T-independent Ags12. Although D6 deficiency had no obvious impact on Ab responses to standard intraperitoneal doses of conventional T-independent Ags (data not shown), more detailed investigations are underway (varying Ag dose and route of delivery) to reveal any critical roles for D6 during IBC responses.
The mechanisms responsible for the specific enhanced CXCL13 responsiveness of D6-deficient B1 cells are unclear. D6 deletion did not affect surface expression of CXCR5, before or after exposure to CXCL13 (data not shown), and WT B1 cell responses to CXCL13 were not altered by D6 ligands, consistent with the inability of D6 to transduce signals. Interestingly, D6-mediated β-arrestin redistribution occurs in the absence of chemokines, and is unaffected by chemokine exposure33,34. β-arrestins are critical regulators of G-protein coupled receptors, capable of switching signalling from G-protein-dependent to - independent pathways, and their activity is profoundly influenced by subcellular localisation. Their presence at B1 cell surfaces, driven by D6 expression, might alter signalling, and subsequent migratory responses, mediated by robust CXCR5 activation. In our hands, β-arrestin redistribution relies on D6 phosphorylation, yet neither β-arrestins nor D6 phosphorylation is required for scavenging34. Thus, by controlling D6 phosphorylation, IBCs might be able to rapidly fine-tune responsiveness to CXCL13 without affecting scavenging.
IBCs are the sole D6active leukocyte subset detectable in resting mice, and D6 activity is a unifying feature of this heterogeneous B cell population. Our study reveals, in unprecedented detail, the remarkable complexity of the body cavity B1 cell compartments, and identifies novel B1 cell subsets (B1d and CD23+ B1a-d). Through chemokine scavenging and suppression of CXCL13 responsiveness, D6 plays a dual role on IBCs, and this controls the cellularity of the IBC compartment and its ability to generate anti-PC Abs. It is now critical to define if, and how, IBCs contribute to the well-documented ability of D6 to suppress tissue inflammation.
Supplementary Material
Acknowledgements
Work was funded by Arthritis Research UK (C.A.H.H., I.B.M., C.S.G., R.J.B.N.), Biotechnology and Biological Sciences Research Council (C.A.H.H., C.S.G., R.J.B.N.), Medical Research Council (Y.B., R.J.B.N.), and Wellcome Trust (L.F., R.K., R.J.B.N.). R.J.B.N. acknowledges Dr Wilson’s support.
Footnotes
Conflict-of-interest disclosure: The authors have no competing financial interests.
References
- 1.Allman D, Pillai S. Peripheral B cell subsets. Curr Opin Immunol. 2008;20:149–157. doi: 10.1016/j.coi.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fagarasan S, Kawamoto S, Kanagawa O, Suzuki K. Adaptive immune regulation in the gut: T cell-dependent and T cell-independent IgA synthesis. Annu Rev Immunol. 2010;28:243–273. doi: 10.1146/annurev-immunol-030409-101314. [DOI] [PubMed] [Google Scholar]
- 3.Haas KM, Poe JC, Steeber DA, Tedder TF. B-1a and B-1b cells exhibit distinct developmental requirements and have unique functional roles in innate and adaptive immunity to S. pneumoniae. Immunity. 2005;23:7–18. doi: 10.1016/j.immuni.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 4.Baumgarth N, Herman OC, Jager GC, Brown LE, Herzenberg LA, Chen J. B-1 and B-2 cell-derived immunoglobulin M antibodies are nonredundant components of the protective response to influenza virus infection. J Exp Med. 2000;192:271–280. doi: 10.1084/jem.192.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alugupalli KR, Gerstein RM, Chen J, Szomolanyi-Tsuda E, Woodland RT, Leong JM. The resolution of relapsing fever borreliosis requires IgM and is concurrent with expansion of B1b lymphocytes. J Immunol. 2003;170:3819–3827. doi: 10.4049/jimmunol.170.7.3819. [DOI] [PubMed] [Google Scholar]
- 6.Binder CJ, Horkko S, Dewan A, et al. Pneumococcal vaccination decreases atherosclerotic lesion formation: molecular mimicry between Streptococcus pneumoniae and oxidized LDL. Nat Med. 2003;9:736–743. doi: 10.1038/nm876. [DOI] [PubMed] [Google Scholar]
- 7.Cinamon G, Zachariah MA, Lam OM, Foss FW, Jr., Cyster JG. Follicular shuttling of marginal zone B cells facilitates antigen transport. Nat Immunol. 2008;9:54–62. doi: 10.1038/ni1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Attanavanich K, Kearney JF. Marginal zone, but not follicular B cells, are potent activators of naive CD4 T cells. J Immunol. 2004;172:803–811. doi: 10.4049/jimmunol.172.2.803. [DOI] [PubMed] [Google Scholar]
- 9.Bouaziz JD, Yanaba K, Tedder TF. Regulatory B cells as inhibitors of immune responses and inflammation. Immunol Rev. 2008;224:201–214. doi: 10.1111/j.1600-065X.2008.00661.x. [DOI] [PubMed] [Google Scholar]
- 10.Hastings WD, Gurdak SM, Tumang JR, Rothstein TL. CD5+/Mac-1- peritoneal B cells: a novel B cell subset that exhibits characteristics of B-1 cells. Immunol Lett. 2006;105:90–96. doi: 10.1016/j.imlet.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 11.Ghosn EE, Yang Y, Tung J, Herzenberg LA, Herzenberg LA. CD11b expression distinguishes sequential stages of peritoneal B-1 development. Proc Natl Acad Sci U S A. 2008;105:5195–5200. doi: 10.1073/pnas.0712350105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ansel KM, Harris RB, Cyster JG. CXCL13 is required for B1 cell homing, natural antibody production, and body cavity immunity. Immunity. 2002;16:67–76. doi: 10.1016/s1074-7613(01)00257-6. [DOI] [PubMed] [Google Scholar]
- 13.Wells SM, Kantor AB, Stall AM. CD43 (S7) expression identifies peripheral B cell subsets. J Immunol. 1994;153:5503–5515. [PubMed] [Google Scholar]
- 14.Won WJ, Kearney JF. CD9 is a unique marker for marginal zone B cells, B1 cells, and plasma cells in mice. J Immunol. 2002;168:5605–5611. doi: 10.4049/jimmunol.168.11.5605. [DOI] [PubMed] [Google Scholar]
- 15.Rot A, von Andrian UH. Chemokines in innate and adaptive host defense: basic chemokinese grammar for immune cells. Annu Rev Immunol. 2004;22:891–928. doi: 10.1146/annurev.immunol.22.012703.104543. [DOI] [PubMed] [Google Scholar]
- 16.Fagarasan S, Shinkura R, Kamata T, et al. Alymphoplasia (aly)-type nuclear factor kappaB-inducing kinase (NIK) causes defects in secondary lymphoid tissue chemokine receptor signaling and homing of peritoneal cells to the gut-associated lymphatic tissue system. J Exp Med. 2000;191:1477–1486. doi: 10.1084/jem.191.9.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu TT, Cyster JG. Integrin-mediated long-term B cell retention in the splenic marginal zone. Science. 2002;297:409–412. doi: 10.1126/science.1071632. [DOI] [PubMed] [Google Scholar]
- 18.Ha SA, Tsuji M, Suzuki K, et al. Regulation of B1 cell migration by signals through Toll-like receptors. J Exp Med. 2006;203:2541–2550. doi: 10.1084/jem.20061041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kunisawa J, Kurashima Y, Gohda M, et al. Sphingosine 1-phosphate regulates peritoneal B-cell trafficking for subsequent intestinal IgA production. Blood. 2007;109:3749–3756. doi: 10.1182/blood-2006-08-041582. [DOI] [PubMed] [Google Scholar]
- 20.Cinamon G, Matloubian M, Lesneski MJ, et al. Sphingosine 1-phosphate receptor 1 promotes B cell localization in the splenic marginal zone. Nat Immunol. 2004;5:713–720. doi: 10.1038/ni1083. [DOI] [PubMed] [Google Scholar]
- 21.Berberich S, Forster R, Pabst O. The peritoneal micromilieu commits B cells to home to body cavities and the small intestine. Blood. 2007;109:4627–4634. doi: 10.1182/blood-2006-12-064345. [DOI] [PubMed] [Google Scholar]
- 22.Hopken UE, Achtman AH, Kruger K, Lipp M. Distinct and overlapping roles of CXCR5 and CCR7 in B-1 cell homing and early immunity against bacterial pathogens. J Leukoc Biol. 2004;76:709–718. doi: 10.1189/jlb.1203643. [DOI] [PubMed] [Google Scholar]
- 23.Hopken UE, Winter S, Achtman AH, Kruger K, Lipp M. CCR7 regulates lymphocyte egress and recirculation through body cavities. J Leukoc Biol. 2010;87:671–682. doi: 10.1189/jlb.0709505. [DOI] [PubMed] [Google Scholar]
- 24.Foussat A, Balabanian K, Amara A, et al. Production of stromal cell-derived factor 1 by mesothelial cells and effects of this chemokine on peritoneal B lymphocytes. Eur J Immunol. 2001;31:350–359. doi: 10.1002/1521-4141(200102)31:2<350::aid-immu350>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 25.McKimmie CS, Fraser AR, Hansell C, et al. Hemopoietic cell expression of the chemokine decoy receptor D6 is dynamic and regulated by GATA1. J Immunol. 2008;181:8171–8181. doi: 10.4049/jimmunol.181.11.8170-a. [DOI] [PubMed] [Google Scholar]
- 26.Kin NW, Crawford DM, Liu J, Behrens TW, Kearney JF. DNA microarray gene expression profile of marginal zone versus follicular B cells and idiotype positive marginal zone B cells before and after immunization with Streptococcus pneumoniae. J Immunol. 2008;180:6663–6674. doi: 10.4049/jimmunol.180.10.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mantovani A, Bonecchi R, Locati M. Tuning inflammation and immunity by chemokine sequestration: decoys and more. Nat Rev Immunol. 2006;6:907–918. doi: 10.1038/nri1964. [DOI] [PubMed] [Google Scholar]
- 28.Graham GJ. D6 and the atypical chemokine receptor family: novel regulators of immune and inflammatory processes. Eur J Immunol. 2009;39:342–351. doi: 10.1002/eji.200838858. [DOI] [PubMed] [Google Scholar]
- 29.Fra AM, Locati M, Otero K, et al. Cutting edge: scavenging of inflammatory CC chemokines by the promiscuous putatively silent chemokine receptor D6. J Immunol. 2003;170:2279–2282. doi: 10.4049/jimmunol.170.5.2279. [DOI] [PubMed] [Google Scholar]
- 30.Weber M, Blair E, Simpson CV, et al. The chemokine receptor D6 constitutively traffics to and from the cell surface to internalize and degrade chemokines. Mol Biol Cell. 2004;15:2492–2508. doi: 10.1091/mbc.E03-09-0634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vetrano S, Borroni EM, Sarukhan A, et al. The lymphatic system controls intestinal inflammation and inflammation-associated Colon Cancer through the chemokine decoy receptor D6. Gut. 2010;59:197–206. doi: 10.1136/gut.2009.183772. [DOI] [PubMed] [Google Scholar]
- 32.Bordon Y, Hansell CA, Sester DP, Clarke M, Mowat AM, Nibbs RJ. The atypical chemokine receptor D6 contributes to the development of experimental colitis. J Immunol. 2009;182:5032–5040. doi: 10.4049/jimmunol.0802802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galliera E, Jala VR, Trent JO, et al. beta-Arrestin-dependent constitutive internalization of the human chemokine decoy receptor D6. J Biol Chem. 2004;279:25590–25597. doi: 10.1074/jbc.M400363200. [DOI] [PubMed] [Google Scholar]
- 34.McCulloch CV, Morrow V, Milasta S, et al. Multiple roles for the C-terminal tail of the chemokine scavenger D6. J Biol Chem. 2008;283:7972–7982. doi: 10.1074/jbc.M710128200. [DOI] [PubMed] [Google Scholar]
- 35.Madigan J, Freeman DJ, Menzies F, et al. Chemokine scavenger D6 is expressed by trophoblasts and aids the survival of mouse embryos transferred into allogeneic recipients. J Immunol. 2010;184:3202–3212. doi: 10.4049/jimmunol.0902118. [DOI] [PubMed] [Google Scholar]
- 36.Nibbs RJ, Kriehuber E, Ponath PD, et al. The beta-chemokine receptor D6 is expressed by lymphatic endothelium and a subset of vascular tumors. Am J Pathol. 2001;158:867–877. doi: 10.1016/s0002-9440(10)64035-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinez de la Torre Y, Buracchi C, Borroni EM, et al. Protection against inflammation- and autoantibody-caused fetal loss by the chemokine decoy receptor D6. Proc Natl Acad Sci U S A. 2007;104:2319–2324. doi: 10.1073/pnas.0607514104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jamieson T, Cook DN, Nibbs RJ, et al. The chemokine receptor D6 limits the inflammatory response in vivo. Nat Immunol. 2005;6:403–411. doi: 10.1038/ni1182. [DOI] [PubMed] [Google Scholar]
- 39.Yang Y, Tung JW, Ghosn EE, Herzenberg LA, Herzenberg LA. Division and differentiation of natural antibody-producing cells in mouse spleen. Proc Natl Acad Sci U S A. 2007;104:4542–4546. doi: 10.1073/pnas.0700001104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Montecino-Rodriguez E, Leathers H, Dorshkind K. Identification of a B-1 B cell-specified progenitor. Nat Immunol. 2006;7:293–301. doi: 10.1038/ni1301. [DOI] [PubMed] [Google Scholar]
- 41.Won WJ, Foote JB, Odom MR, Pan J, Kearney JF, Davis RS. Fc receptor homolog 3 is a novel immunoregulatory marker of marginal zone and B1 B cells. J Immunol. 2006;177:6815–6823. doi: 10.4049/jimmunol.177.10.6815. [DOI] [PubMed] [Google Scholar]
- 42.Thiriot A, Drapier AM, Vieira P, et al. The Bw cells, a novel B cell population conserved in the whole genus Mus. J Immunol. 2007;179:6568–6578. doi: 10.4049/jimmunol.179.10.6568. [DOI] [PubMed] [Google Scholar]
- 43.Hastings WD, Tumang JR, Behrens TW, Rothstein TL. Peritoneal B-2 cells comprise a distinct B-2 cell population with B-1b-like characteristics. Eur J Immunol. 2006;36:1114–1123. doi: 10.1002/eji.200535142. [DOI] [PubMed] [Google Scholar]
- 44.Berberich S, Dahne S, Schippers A, et al. Differential molecular and anatomical basis for B cell migration into the peritoneal cavity and omental milky spots. J Immunol. 2008;180:2196–2203. doi: 10.4049/jimmunol.180.4.2196. [DOI] [PubMed] [Google Scholar]
- 45.Shulga-Morskaya S, Dobles M, Walsh ME, et al. B cell-activating factor belonging to the TNF family acts through separate receptors to support B cell survival and T cell-independent antibody formation. J Immunol. 2004;173:2331–2341. doi: 10.4049/jimmunol.173.4.2331. [DOI] [PubMed] [Google Scholar]
- 46.Gavin AL, Duong B, Skog P, et al. deltaBAFF, a splice isoform of BAFF, opposes full-length BAFF activity in vivo in transgenic mouse models. J Immunol. 2005;175:319–328. doi: 10.4049/jimmunol.175.1.319. [DOI] [PubMed] [Google Scholar]
- 47.Scholz JL, Crowley JE, Tomayko MM, et al. BLyS inhibition eliminates primary B cells but leaves natural and acquired humoral immunity intact. Proc Natl Acad Sci U S A. 2008;105:15517–15522. doi: 10.1073/pnas.0807841105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carvalho TL, Mota-Santos T, Cumano A, Demengeot J, Vieira P. Arrested B lymphopoiesis and persistence of activated B cells in adult interleukin 7(−/−) mice. J Exp Med. 2001;194:1141–1150. doi: 10.1084/jem.194.8.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mack M, Cihak J, Simonis C, et al. Expression and characterization of the chemokine receptors CCR2 and CCR5 in mice. J Immunol. 2001;166:4697–4704. doi: 10.4049/jimmunol.166.7.4697. [DOI] [PubMed] [Google Scholar]
- 50.Islam SA, Chang DS, Colvin RA, et al. Mouse CCL8, a CCR8 agonist, promotes atopic dermatitis by recruiting IL-5+ T(H)2 cells. Nat Immunol. 2011;12:167–177. doi: 10.1038/ni.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.