Abstract
Following fertilization the transition from a highly differentiated oocyte to a totipotent 2-cell embryo requires two unique mitotic cell cycles. The first cell cycle is characterized by a prolonged G1 phase, DNA replication (S phase) that occurs separately in the female and male pronuclei, and a short G2 phase that occur in the absence of cell growth. During the second cell cycle, G1 is short whereas G2 is prolonged and occurs concurrently with zygotic genome activation, which is essential for progression past the 2-cell stage. CDC14B, a dual specificity phosphatase that counteracts cyclin dependent kinase 1 (CDK1/CDC2A) action, regulates mitosis in somatic cells and prevents premature meiotic resumption in mouse oocytes. It is not known if CDC14B plays a role during the unique mitotic cell cycles of preimplantation development. We report that CDC14B is present in mouse embryos and localizes to mitotic centrosomes and spindles. overexpressing CDC14B in 1-cell embryos results in 40% and 60% of the embryos arresting at the 1- and 2-cell stages, respectively. embryos arrested at the 1-cell stage contained reduced CDC2A activity, whereas embryos arrested at the 2-cell stage were in G2 and failed to activate the zygotic genome. In contrast, overexpressing CDC14B in meiotically-incompetent oocytes, which are arrested in a G2-like state and are transcriptionally active, does not repress global transcription. these data suggest that CDC14B is a negative regulator of the 1-to-2-cell transition and of zygotic genome activation in mouse embryogenesis.
Keywords: preimplantation embryo, mitosis, phosphatase, zygotic genome activation, CDC14
Introduction
The first major transition during preimplantation development is the maternal-to-zygotic transition, in which maternal transcripts from the egg that direct early development are replaced by transcripts expressed from the zygotic genome.1 In mouse embryos, the first two mitotic cell cycles are prolonged and each last approximately 20 h. The first mitosis (M phase) is almost twice as long as the second M phase and this increase may be due to a transient metaphase arrest that is independent of the spindle assembly checkpoint.2 Gap phases of the second cell cycle differ from those of the first cell cycle because G1 is extremely short (1–2 h)3 and G2 prolonged (12–16 h).4-7 It is during this unusually long G2 phase when the major wave of zygotic genome activation (ZGA) occurs in mouse.1 The subsequent embryonic cell cycles are more similar in duration to one another and to that of somatic cells.
The mitotic cell cycle is driven by cyclin dependent kinases (CDKs) that function in a complex with cyclin subunits. CDK activity is high during DNA replication (S phase) and M phase and is low during the gap phases (G1/G2). In yeast, exit from M phase is regulated by cell division cycle homolog 14 (CDC14), a dual-specificity phosphatase that counteracts CDK activity by preferentially dephosphorylating CDK substrates.8,9
Mammals contain two CDC14 homologs, CDC14A and CDC14B, which are required for multiple cell cycle events. The requirement for CDC14B is not clear because one group that deleted the Cdc14b coding region in somatic cells reported no observable phenotype in cells lacking CDC14B,10 whereas other groups that reduce the amount of this phosphatase by RNAi strategies find that it is required to bundle and stabilize microtubules,11 maintain proper numbers of centrioles,12 and activate the G2 DNA damage checkpoint.13 Recently, we demonstrated that CDC14B plays a meiosis-specific role in mouse oocytes because CDC14B prevents meiotic resumption by maintaining low cyclin B1 (CCNB1) levels and perhaps regulates spindle dynamics during meiosis I.14
Because the role of CDC14B in the oocyte differs from its role in somatic cells,14 we explored whether this difference also extends to the first embryonic cell cycles that differ from subsequent mitotic cell cycles. We report that CDC14B is expressed in mouse preimplantation embryos and localizes to mitotic centrosomes and spindles. Overexpressing CDC14B in 1-cell embryos prevents development to the blastocyst stage due to embryonic arrest at the 1- and 2-cell stages. One-cell stage arrested embryos contain reduced CDC2A activity suggesting that this block is due to a cell cycle defect. In contrast, embryos arrested at the 2-cell stage fail to activate the zygotic genome and arrest in G2. Finally, we observe defects in the timing of nucleologenesis and a reduced number of nucleoli in arrested 2-cell embryos.
Results
Temporal-spatial expression patterns of CDC14B during mouse preimplantation development
Before conducting studies to address the role of CDC14B during mouse preimplantation development, we first characterized its temporal and spatial expression pattern. Similar to many maternally-expressed mRNAs,15,16 Cdc14b mRNA degradation was initiated with the onset of oocyte maturation and continued to decline during early preimplantation development (Fig. 1A). Between the 2- and 8-cell stages the relative abundance of Cdc14b mRNA increased, which is likely due to transcription of the zygotic genome.17 In contrast, the relative abundance of CDC14B protein did not change significantly between metaphase of meiosis II and blastocyst stages (Fig. 1B), indicating that CDC14B protein is stable during preimplantation embryonic development.
Figure 1.
expression of CDC14B during mouse preimplantation development. (A) Relative amounts of Cdc14b mRNA during preimplantation development were determined by qRt-pCR. In each experiment, 50 oocytes or embryos were used to isolate mRNA. this experiment was repeated three times and the data are expressed as mean ± standard deviation relative to the value obtained for GV oocytes. (B) Western blot detection of CDC14B in different stages of preimplantation embryogenesis. the membrane was stripped and re-probed with an anti-β-tubulin antibody for loading standard. Note that much less total protein was loaded in the blastocyst (BL) lane as indicated by the loading control. this experiment was conducted three times with a total of 30 embryos per stage. GV, germinal vesicle (prophase-arrested oocyte); MII, metaphase II; 1C, 1-cell embryo; 2C, 2-cell embryo; 8C, 8-cell embryo; BL, blastocyst.
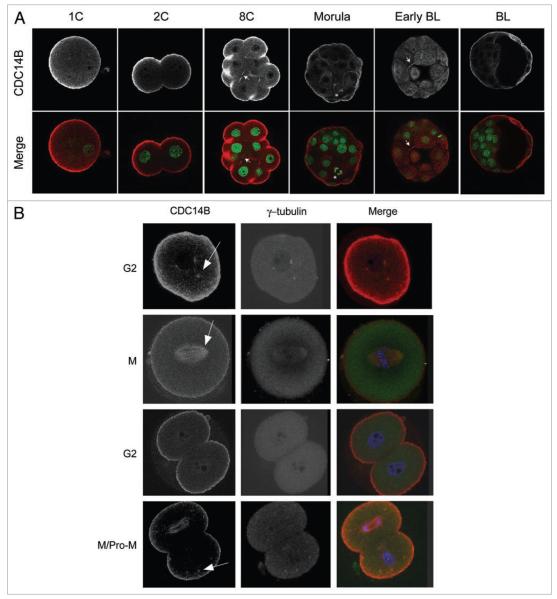
We previously demonstrated that CDC14B colocalizes with the cytoplasmic microtubule network during prophase of meiosis I (MI) and with MI and meiosis II (MII) spindles in mouse oocytes.14 We used immunocytochemistry to determine the localization of CDC14B during preimplantation development. Similar to its localization in oocytes, CDC14B localized to the cortex except at regions of cell-cell apposition. Furthermore CDC14B exhibited a meshwork-like pattern throughout the embryo, which is likely the microtubule network, during all embryonic stages examined (Fig. 2A). We also observed CDC14B staining at the midbody in blastomeres from 8-cell and early blastocyst stages and on the mitotic spindle in a blastomere that was undergoing mitosis in a morula stage embryo.
Figure 2.
temporal-spatial localization of CDC14B during mouse preimplantation embryonic development. embryos at the indicated stages were fixed in 3.7% paraformaldehyde prior to immunocytochemical detection. (A) Localization of CDC14B during preimplantation embryogenesis. In the merged images CDC14B is red and DNA is green. the arrows point to localization at midbodies and the asterisk indicates localization at a mitotic spindle. (B) Localization of CDC14B during the cell cycle of the first two embryonic divisions. One or two-cell embryos were collected at specific times after hCG injection as described in the methods section. In merged images CDC14B is red, γ-tubulin is green and DNA is blue. In the 2-cell embryo in the last row, the top blastomere is in anaphase and the lower blastomere is in prometaphase (pro-M). the arrows indicate co-localization at microtubule organizing centers (MtoCs).
To more carefully examine the localization of CDC14B during the first unique mitotic cell cycles, we examined the localization of CDC14B during the first two G2 and M phases. We found that CDC14B co-localized with γ-tubulin during the first G2 phase and the second prometaphase stage and with the mitotic spindle during the first and second M phases (Fig. 2B). No signal was observed when the primary antibody was omitted (data not shown).
Overexpression of CDC14B causes mitotic arrest at the 1- and 2-cell stages
Overexpression of human CDC14B in interphase-arrested somatic cells causes aberrant microtubule bundling and stabilization18,19 and delays meiotic resumption when overexpressed in prophase-arrested oocytes.14 We evaluated the consequences of overexpressing CDC14B during preimplantation development by microinjecting 1-cell embryos with Cdc14b cRNA. As a control, we injected embryos with Gfp cRNA. We found by immunocytochemistry and immunoblot analyses that overexpression of Cdc14b cRNA increased the endogenous pool of CDC14B protein by approximately 2-fold when compared to Gfp cRNA-injected controls (Fig. 3C). Furthermore, we detected the overexpressed protein in the same subcellular locations as the endogenous protein.
Figure 3.
overexpression of CDC14B causes mitotic arrest at the one or two-cell embryonic stage. 1-cell embryos were microinjected with the Cdc14b or Gfp cRNA and incubated in KSOM. (A) Injected embryos were incubated for the indicated amount of time and were classified according to their developmental stage. These experiments were repeated 5 times, and the error bars indicate S.E.M. (B) Bright field images of microinjected embryos 96 h after eCG injection. (C) CDC14B overexpression was confirmed by immunocytochemistry (top) and immunoblotting (bottom). To estimate the level of overexpression for the immunocytochemistry, the average intensities of CDC14B were compared between injected and non-injected 2-cell embryos using Image J software. Image J was also used to normalize the β-tubulin signals in the immunoblot to estimate the amount of CDC14B overexpression compared to non-injected embryos at the stage.
Overexpressing CDC14B caused mitotic arrest (Fig. 3A) as 40% and 60% of the embryos arrested at the 1- and 2-cell stages, respectively. In contrast, the non-injected embryos and those injected with Gfp cRNA progressed to the blastocyst stage by 96 h post eCG. At this time point the embryos overexpressing CDC14B remained arrested at either the 1- or the 2-cell stage and did not display any sign of fragmentation or loss of viability (Fig. 3B). A similar phenomenon was observed when embryos were treated with roscovitine, a specific cyclin dependent kinase 1 (CDK1/CDC2A) inhibitor, which also caused an arrest at the 2-cell stage (data not shown).
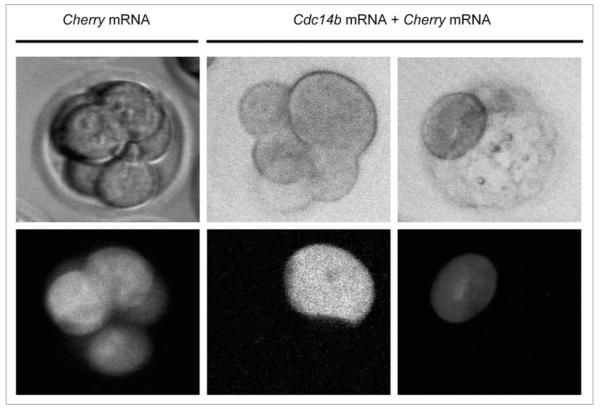
To determine if the observed arrest phenotype was specific to the first mitotic division or if overexpression of CDC14B could also cause mitotic arrest in the second division, we microinjected one blastomere of a 2-cell embryo with a mixture of Cdc14b and mCherry cRNAs and allowed development to the blastocyst stage. As a control, we microinjected one blastomere of 2-cell embryos with mCherry cRNA alone. We observed mitotic arrest in the single blastomere overexpressing CDC14B in contrast to the non-injected blastomere that continued development to the blastocyst stage despite having one arrested blastomere that is larger in size (Fig. 4). Furthermore, microinjecting mCherry alone did not cause mitotic arrest.
Figure 4.
overexpression of CDC14B caused mitotic arrest in one blastomere of the 2-cell embryo. one blastomere of a 2-cell embryo was microinjected with a mixture of Cdc14b and mCherry cRNAs and incubated for a time in which non-injected 2-cell embryos had reached either the 8-cell or blastocyst stages. As a control, we injected one blastomere of 2-cell embryos with mCherry cRNA alone (left). these live-cell images were obtained using a spinning disc confocal microscope that is equipped with an environmental chamber to support life. this experiment was repeated three times with a total of 20 embryos.
We next asked whether CDC14B overexpression in a blastomere from an embryo that is no longer undergoing the unique first two cell cycles would have that same inhibitory affect. Accordingly, we overexpressed CDC14B in a blastomere from an 8-cell embryo and found that this blastomere also failed to divide even though the non-injected blastomeres continued dividing and formed a blastocyst (data not shown). These data suggest that overexpression of CDC14B during any preimplantation development stage can cause cell cycle arrest.
Because depletion of CDC14B via RNAi induced meiotic resumption in mouse oocytes,14 we examined the effect of knocking down CDC14B in mouse embryos by microinjecting 1-cell embryos with Cdc14b double strand RNA. Despite a ~60% reduction of CDC14B in oocytes, we did not observe any significant reduction in the amount of CDC14B protein in the microinjected embryos, suggesting that CDC14B protein becomes more stable during mouse embryo development (data not shown). These data are also consistent with the observation that CDC14B protein is stable during preimplantation embryogenesis despite the fact that the transcript is degraded between the 1-cell and 8-cell stages (Fig. 1A and B). The inability to reduce CDC14B in embryos precluded assessing the requirement for CDC14B during embryonic development.
Embryos overexpressing CDC14B that are arrested at the 1-cell stage have low CDC2A activity
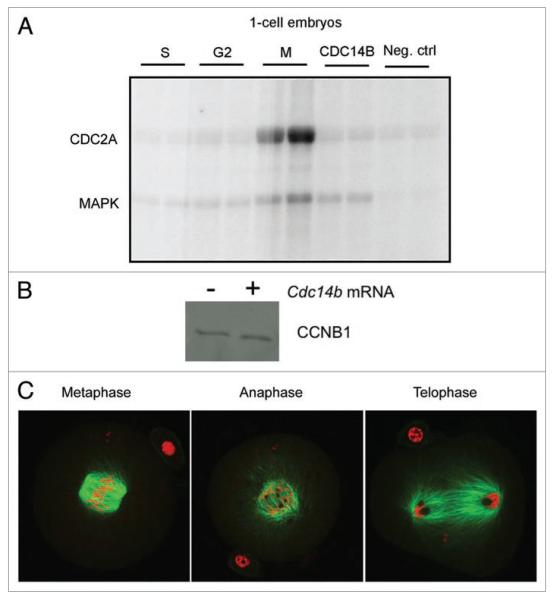
CDC2A activity fluctuates during the cell cycle in germ and somatic cells, peaking during S and M phases.20-22 Overexpression of CDC14B reduces CDC2A activity in prophase-arrested oocytes by promoting the degradation of cyclin B1 (CCNB1) via activation of the CDH1 regulatory subunit of the anaphase-promoting complex (APC).14 To investigate the molecular basis by which CDC14B causes mitotic arrest in embryos, we measured the level of CDC2A activity by performing in vitro kinase assays with single embryos injected with either Gfp or Cdc14b cRNA. Embryos overexpressing CDC14B that were arrested at the 1-cell stage had CDC2A activity levels comparable to levels of control embryos in G2 phase of the cell cycle (Fig. 5A), suggesting that these embryos were arrested in G2 phase and that CDC14B may be a negative regulator of CDC2A during the first mitotic cell cycle as it is in oocytes.
Figure 5.
embryos overexpressing CDC14B that are arrested at the 1-cell stage have low CDC2A activity. (A) one-cell embryos were collected at the indicated cell cycle stage and CDC2A and MApK activities for histone H1 or MBp, respectively, were measured by in vitro kinase assays. The graph represents quantification of the band intensity as measured in Image J for CDC2A of three independent experiments. (B) Immunoblotting analysis of levels of CCNB1 protein between control embryos in G2 phase to 1-cell arrested embryos that were overexpressing CDC14B. this experiment was conducted two times. (C) embryos overexpressing CDC14B that did not contain visible pronuclear membranes were fixed in 3.7% paraformaldehyde and stained with an antibody against α-tubulin to visualize the spindles and mounted in propidium iodide to visualize the DNA. the images were obtained on a laser-scanning confocal microscope.
In human cell lines CDC14B triggers the G2 DNA damage checkpoint by dephosphorylating CDH1, thus promoting CDH1 binding to the APC.13 We previously demonstrated that CDC14B regulates CCNB1 turnover in prophase-arrested oocytes through regulating the CDH1.14 To assess whether this mechanism also exists in the first G2 phase we compared the amount of CCNB1 protein by immunoblot analysis between control embryos in G2 phase and 1-cell arrested embryos that were overexpressing CDC14B. We found that the arrested embryos contained similar amounts of CCNB1 as control embryos (Fig. 5B) suggesting that decreased amounts of CCNB1 due to excess CDC14B is not the cause of the G2 arrest in 1-cell embryos as it is in the oocyte. Moreover, reduction of either the CDH1 and CDC20 regulatory subunits of the APC by morpholino or siRNA injection, respectively, did not rescue the cell cycle block caused by CDC14B overexpression (data not shown).
During the course of these overexpression experiments we found that some 1-cell embryos did not have visible pronuclear membranes, suggesting that they were in M phase. We fixed these embryos and stained the spindles with an antibody against α-tubulin and visualized the DNA with propidium iodide (Fig. 5C). Embryos that were in metaphase of the first cell cycle contained DNA that was aligned on the metaphase plate and spindles that had lateral microtubules projections. Interestingly, embryos that were in either anaphase or telophase contained dark circular areas adjacent to segregating chromosomes that did not stain with the tubulin antibody or propidium iodide. These areas appear similar in morphology to nucleoli suggesting that 2-cell embryo nucleologenesis is occurring prior to completion of chromosome segregation and cytokinesis.
Embryos overexpressing CDC14B that are arrested at the 2-cell stage have decreased global transcription levels
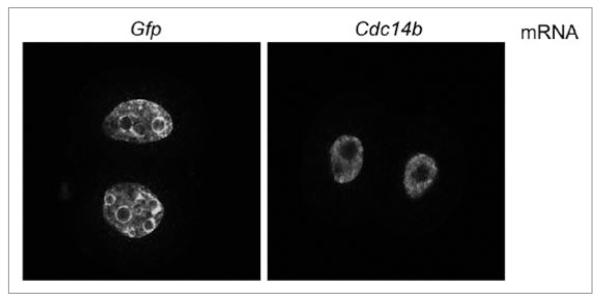
To determine what stage of the second cell cycle the embryos arrested, we first assessed whether they underwent DNA replication by measuring BrdU incorporation. Embryos overexpressing CDC14B incorporated BrdU and contained visible nuclear membranes (Fig. 6), suggesting they were arrested in G2. Furthermore, we observed a difference in the number of nucleoli in 2-cell-arrested embryos that overexpress CDC14B compared to control injected embryos. On average the control 2-cell embryos contained 11 nucleoli whereas the embryos overexpressing CDC14B contained three nucleoli.
Figure 6.
embryos overexpressing CDC14B that arrest at the 2-cell embryonic stage complete the second S phase. embryos overexpressing CDC14B or GFp were incubated with BrdU for 1 h as described in materials and methods. Distribution of incorporated BrdU in the pronuclei was observed by laser-scanning confocal microscopy following the immunostaining with anti-BrdU antibody.
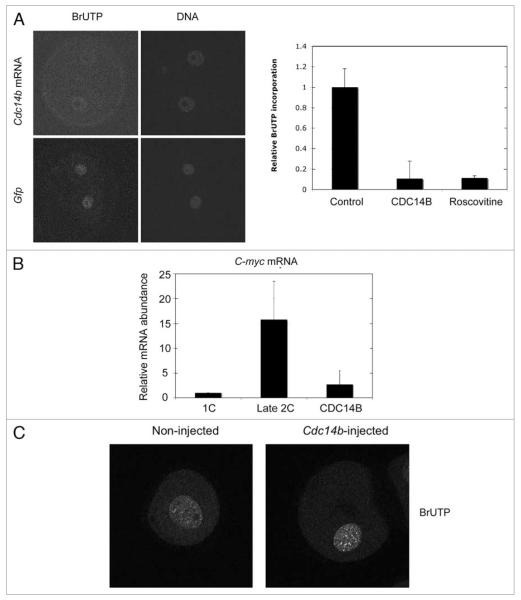
The G2 phase of the second cell cycle is prolonged and during this phase the zygotic genome is activated. Because inhibition of transcription causes cleavage arrest in 2-cell embryos,23 we next evaluated whether embryos arrested at the 2-cell stage have undergone zygotic genome activation (ZGA). First we measured the global increase in transcription using a transcription run-on assay that monitors BrUTP incorporation.24 We found a significant decrease in BrUTP incorporation (~90% decrease) in the embryos injected with Cdc14b cRNA that were arrested at the 2-cell stage when compared to late 2-cell embryos injected with Gfp cRNA (Fig. 7A). This decrease in the overall transcription was confirmed at the gene expression level with the analysis of one transcript, C-myc, whose expression increases during ZGA (Fig. 7B). These data suggest that 2-cell embryos that overexpress CDC14B fail to activate the zygotic genome. This phenomenon seems specific to embryos because overexpression of CDC14B in meiotically-incompetent oocytes that are transcriptionally-active did not inhibit transcription, when evaluated by the same assay (Fig. 7C).
Figure 7.
two-cell embryos overexpressing CDC14B fail to activate the zygotic genome. BrUtp incorporation of 2-cell embryos (A) or meioticallyincompetent oocytes (C) microinjected with Cdc14b or Gfp cRNAs. (A) the left panels show examples of images obtained by laser-scanning confocal microscopy and the graph on the right is the quantification of the nuclear anti-BrUTP signals from 3 experiments as determined by Image J. (B) The decrease in the overall transcription in 2-cell embryos was confirmed at the gene expression level with the analysis of one zygotically-expressed transcript, C-myc, by qRt-pCR analysis. (C) Laser-scanning confocal microscope images of anti-BrUtp staining in meiotically-incompetent oocytes either injected Cdc14b cRNA or not injected as a control. The amount of BrUTP incorporation was quantified using Image J as in (A).
Nuclear translocation and carboxyl-terminal domain (CTD) phosphorylation of RNA polymerase II delineate the two phases of zygotic gene activation in mammalian embryos.25 The CTD contains a heptad repeat (YSPTSPS) where phosphorylation of serine 5 by CDK7 is essential for its function in transcriptional elongation. Because serine 5 is contained within a CDK consensus sequence and CDC14B preferentially dephosphorylates sites phosphorylated by CDKs, we investigated whether the levels of phosphorylated RNA polymerase II were affected in the arrested 2-cell embryos overexpressing CDC14B using immunocytochemistry. We found no significant difference in the intensity of the nuclear immunofluorescent signal using a phospho-specific antibody against serine 5 of the CTD between embryos overexpressing CDC14B and Gfp-injected control embryos that were fixed at the late 2-cell stage (data not shown) suggesting that the genome activation defect occurs despite the presence of active RNA polymerase II in the 2-cell nuclei.
Discussion
We demonstrate for the first time a role for CDC14B during mouse preimplantation development. We found that overexpression of CDC14B in 1-cell embryos causes a block in G2 of either the first or second cell cycle (Fig. 3A). Overexpression of CDC14B in prophase-arrested oocytes delays meiotic resumption,14 a G2/M-like transition, suggesting that there may be some similar CDC14B functions shared between meiotic maturation and the first mitotic cell cycles. In human tissue culture cell lines, CDC14B is required to activate the G2 DNA damage checkpoint.13 During both maintenance of the prophase arrest and activation of the G2 DNA damage checkpoint, CDC14B activates the CDH1 (FZR1) regulatory component of the APC to reduce CDC2A activity via reduction of available CCNB1. In contrast, the embryonic G2 arrest observed in these experiments appears to not be due to APC activation because reduction of CDH1 (or CDC20, the other known regulatory subunit) fails to alleviate the G2 arrest. Collectively, these data suggest that CDC14B plays a general inhibitory trend during G2 phase of all cell cycles examined thus far although the mechanism by which this occurs in preimplantation embryos differs from that in mitotic and meiotic cells.
We found that embryos blocked at the 2-cell stage failed to activate the zygotic genome (Fig. 7A and B). Although there is a minor wave of zygotic gene transcription in the 1-cell embryo, the first cell cycle cleavage can occur in its absence.23,26 In contrast, there is an absolute requirement for zygotic gene transcription in the 2-cell embryo for cleavage to the 4-cell stage. This inhibitory affect on transcription appears to be specific to the embryo because overexpression of CDC14B in incompetent oocytes that are transcriptionally active does not alter global transcription levels (Fig. 7C). There are no known roles for CDC14B regulating gene transcription in mitotic cells. In budding yeast, however, Cdc14 inhibits RNA Polymerase I, the polymerase responsible for transcribing ribosomal DNA (rDNA), during anaphase.27 Although preimplantation embryos utilize maternal stores of ribosomes and largely do not transcribe rDNA until the 8-cell stage28,29 there is evidence that RNA Polymerase I is active in 1-cell embryos.30 These data suggest that despite the similar inhibitory function on the cell cycle during G2 phase, the CDC14B’s inhibitory role on transcription appears specific to the 2-cell embryo. Although a block in the cell cycle just prior to genome activation is a formal possibility, the permanence of this block is more indicative of a global transcription defect. Surprisingly, we found that phosphorylated (active) RNA Polymerase II was in the nucleus of the blocked 2-cell embryos (data not shown). Exploring the mechanism by which CDC14B regulates this major wave of embryonic transcription should shed light on how this developmental transition is controlled.
In mammalian preimplantation embryos, nucleoli are not fully functional prior to the blastocyst stage and are referred to as nucleolar precursor bodies (NPBs).31 NPBs are composed of a network of densely packed filaments32 and the first indication of nucleolar activation during nucleologenesis occurs in 2-cell embryos when a fibrillan center forms in the NPBs cortex.33 In human somatic cells grown in culture, CDC14B localizes to the nucleolus for much of the cell cycle and co-localizes with centrosomes during M phase.12,18,19 During the first embryonic cell cycles we did not detect CDC14B in nucleoli. Similarly, we did not detect CDC14B in nucleoli of oocytes,14 indicating that its localization is more similar to that in the oocyte that undergoes meiosis than of that in somatic cells that are mitotically cycling.
Although we did not detect CDC14B in nucleoli of preimplantation embryos, we did observe alterations in nucleoli in embryos that overexpress this phosphatase. Embryos in the first anaphase and telophase contained circular zones in close proximity to their chromosomes that did not stain with propidium iodide and are reminiscent of NPBs (Fig. 5C). These observations indicate that the nucleologenesis cycle is unlinked to the cell cycle because these embryos have not yet completed cytokinesis have the nuclear appearance normally found in G1 of the second cell cycle. Moreover, we found that embryos blocked at the 2-cell stage contained fewer NPBs than control embryos in G2 phase, further suggesting some role in either nucleologenesis or in promoting nucleolar fusion. Interestingly, 2-cell embryos that fail to cleave but are not dying contain 1–2 large NPBs compared to 11–13 smaller NPBs in embryos that proceed into the second cleavage.34 These collective observations suggest that the morphology of nuclei in preimplantation embryos may be linked to the transcriptional status of the embryo, therefore further exploration on CDC14B’s role during nucleologenesis will shed light on how the NPBs and cell cycle regulators are connected to zygotic genome activation.
Similar to its localization during oocyte maturation,14 CDC14B localizes to the cortex of the embryo and to microtubules of the mitotic spindle and midbody (Fig. 2A and B). Furthermore, the localization at the cortex excludes regions of cell-cell apposition. Several maternal effect genes including Mater (NLRP5), Floped (OOEP), TLE6 and Filia localize and form a subcortical maternal complex that is also excluded from regions of cell-cell apposition in preimplantation embryos.35 Loss of formation of this MDa-sized complex causes a delay in progression to and block at the 2-cell stage. Components of this complex have a variety functions. For example, embryos lacking Filia form abnormal spindles through a failure to localize properly spindle regulators AURKA and PLK1, and fail to activate the spindle assembly checkpoint, which collectively cause aneuploidy in filia-/- embryos.36 Because deletion of many of the complex components causes a block at the 2-cell stage it is possible that they may also play a role in zygotic genome activation. The mechanism by which proteins localized at the cortex control a nuclear event, however, is unknown. Determining how CDC14B is negatively regulating zygotic genome activation may help explain how this complex is regulated and how it communicates with the nucleus during this critical developmental transition.
In summary, by overexpressing CDC14B we have uncovered three potential novel functions of the phosphatase during mouse preimplantation embryogenesis. Similar to other cell types, CDC14B appears to have an inhibitory cell cycle function during G2 phase; however the mechanism by which this block is induced appears to be different in mouse embryos. We also demonstrated that CDC14B overexpression inhibits zygotic genome activation. Currently, there are no reports indicating that CDC14B regulates transcription in any other cell type suggesting that its inhibitory function is unique to embryos. This may be a mechanism by which a developmentally compromised embryo prevents further development. Finally, we also uncovered defects in nucleoli of these embryos. Little is known about the regulation of nucleologenesis in early embryos and these data suggest that the cell cycle machinery may regulate this process. Deciphering the mechanism by which CDC14B regulates these processes will further our understanding of how the mitotic cell cycle in somatic cells evolves from the first unique embryonic cell cycle.
Materials and Methods
Collection cells and microinjection
One-cell embryos were collected from eCG-primed CF-1 female mice mated with B6D2F1/J males (Jackson Laboratory) as previously described.37 One-cell, 2-cell, 4-cell, 8-cell, and blastocysts that developed in vivo were collected from ampullae, or flushed from either oviducts or uteri at 20–21, 41–44, 60–61, 68, 75–77 and 92–96 h post-eCG, respectively.
One-cell embryos were injected in bicarbonate-free Whitten38 medium containing 25 mM HEPES (pH 7.2) and 0.01% PVA with 7 pl of cRNA at 2 μg/μl or 0.15 μg/μl of double-strand RNA at 106 copies/μl as previously described (Kurasawa S 1989). Morpholinos (GeneTools) were used at 1.5 μM and are the sequences of them are the same as previously described.39 The Cdc20 siRNA (Applied Biosystems) was used at 50 μM and was the same sequence as previously described.40 cRNA-injected embryos were held in Chatot, Ziomek, and Bavister (CZB) medium41 in an atmosphere of 5% CO2/5% O2/90% N2 for 3 h and then transfer to KSOM.42 To collect different cell cycle phases from 1-cell and 2-cell embryos in vivo we used the method described previously.43 All animal experiments were approved by the Institutional Animal Use and Care Committee and were consistent with NIH guidelines.
Immunocytochemistry
Embryos were fixed in 3.7% paraformaldehyde in PBS for 1 h at room temperature, permeabilized in PBS containing 0.1% Triton X-100 plus 0.3% BSA for 15 min at room temperature and rinsed through 3 drops of blocking solution (0.3% BSA plus 0.01% Tween-20 in PBS) prior to blocking for 15 min. In a humidified chamber, embryos were incubated in blocking solution containing primary antibody for 1 h at room temperature. The following dilutions were used: CDC14B (Abcam ab26194, 1:100), γ-tubulin (Sigma T6557, 1:100), α-tubulin-FITC (Abcam ab11303; 1:200). After washing, secondary antibodies were applied for 1 h. Secondary antibodies (Jackson) were Cy5-conjugated anti-chicken and FITC-conjugated anti-mouse IgG (Southern Biotech). DNA was detected by mounting the cells in VectaShield containing 3 μg/ml propidium iodide or 1 μM of SYTOX Green (Molecular Probes, Eugene, OR). Fluorescence was detected on a Leica TCS SP laser scanning confocal microscope with Leica Confocal Software and images were processed using Photoshop software (Adobe Systems, Inc.,). Images were viewed with a 40X oil immersion objective. In Figure 4, the embryos were visualized under 20X using a Leica DMI4000B spinning disc confocal microscope.
Immunoblotting
Samples stored at −80°C were thawed on ice, diluted in 2X Laemmli sample buffer44 and loaded on 8% polyacrylamide gels. Samples were electrophoresed at 15 mA, transferred to polyvinylidene fluoride (PVDF) membrane (Millipore) and either blocked overnight in PBS (pH 7.5) plus 0.1% (v/v) Tween-20 and 2% Blocking Agent (ECL Advance, GE Healthcare) at 4°C (CDC14B, TUBB) or for 2 h at room temperature (CCNB1). Detection of CDC14B and TUBB were achieved by probing the membrane with anti-CDC14B or anti-β-tubulin (Sigma T4026) at 1:5,000 for 1 h at room temperature. CCNB1 was detected by probing the membrane overnight at 4°C with an anti-CCNB1 antibody (Abcam ab72; 1:500). After washing, the membrane was incubated with secondary anti-chicken (CDC14B) or anti-mouse antibodies (Amersham 1:200,000) for 1 h at room temperature. Secondary antibodies were detected with chemiluminescence (Amersham ECL Advance).
Real-time PCR
Total RNA from 50 MII eggs or embryos was extracted using the Absolutely RNA Microprep Kit (Stratagene, La Jolla, CA). The reverse transcription reaction, primed with random hexamers, was performed using Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA) following the manufacturer’s instructions. Total RNA isolated was reverse transcribed in a 20 μl reaction volume. The resulting cDNA was quantified by real time PCR (qRT-PCR) using the ABI Taqman Assay-on-demand probe/primer sets for Cdc14b and C-myc. One embryo equivalent of cDNA was used for each real-time PCR reaction and analyzed in duplicate. Unless otherwise stated, quantification was normalized to Gfp mRNA and fold change in expression was determined using the comparative Ct method.
In vitro synthesis of cRNA
Cdc14b was cloned into the pIVT expression vector as previously described.14,45 Plasmids containing Cdc14b, eGfp or mCherry sequences were linearized, in vitro transcribed using a mMessage mMachine kit (Ambion) and RNA was purified using a RNeasy Mini Kit (Qiagen).
Dual kinase assay
CDC2A and MAPK1 activities, as assessed by their ability to phosphorylate histone H1 and myelin basic protein (MBP), respectively, were both assayed in single embryos as previously described.46 Images were detected using a Typhoon 9410 PhosphorImager (GE Healthcare) and quantified with Image J software (NIH).
Detection of transcription
BrUTP incorporation assays were performed as previously described.23 Fluorescence was detected on a Leica TCS SP laser-scanning confocal microscope. The intensity of fluorescence was quantified using NIH Image J software (National Institutes of Health) by averaging signal intensities from three regions in each of the 2-cell embryo nuclei and subtracting the average background signal measured in the cytoplasm of the embryo.
Detection of DNA synthesis
Embryos were labeled with 10 μM bromodeoxyuridine (BrdU) for 30 min and then washed with phosphate-buffered saline (PBS) containing 0.3% bovine serum albumin and then fixed with 3.7% paraformaldehyde. The DNA was denatured by incubating the embryos with 2 N HCl at 37°C for 1 h and finally the sample was neutralized by addition of 0.1 M borate buffer, pH 8.5, for 15 min. The incorporated BrdU was detected as previously described.23 The embryos were mounted in VectaShield (Vector Laboratories, Burlingame, CA) and observed on a Leica TCS SP laser-scanning confocal microscope.
Acknowledgements
We express our gratitude to Drs. Paula Stein and Francesca Duncan for their technical advice. This work was supported by a grant from the National Institutes of Health (HD22681) to R.M.S. K.S. was supported by an NRSA Fellowship (HD055822) from the NIH. M.G.B. was supported by the Fogarty International Center Fellowship Program 5D43TW000671 from the National Institutes of Health.
Abbreviations
- APC
anaphase-promoting complex
- BrdU
bromodeoxyuridine
- BrUTP
5-bromouridine 5′-triphosphate
- CTD
carboxyl-terminal domain
- MI
meiosis I
- MII
meiosis II
- NPBs
nucleolar precursor bodies
- ZGA
zygotic genome activation
References
- 1.Flach G, Johnson MH, Braude PR, Taylor RA, Bolton VN. The transition from maternal to embryonic control in the 2-cell mouse embryo. EMBO J. 1982;1:681–6. doi: 10.1002/j.1460-2075.1982.tb01230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sikora-Polaczek M, Hupalowska A, Polanski Z, Kubiak JZ, Ciemerych MA. The first mitosis of the mouse embryo is prolonged by transitional metaphase arrest. Biol Reprod. 2006;74:734–43. doi: 10.1095/biolreprod.105.047092. [DOI] [PubMed] [Google Scholar]
- 3.Gamow EI, Prescott DM. The cell life cycle during early embryogenesis of the mouse. Exp Cell Res. 1970;59:117–23. doi: 10.1016/0014-4827(70)90630-0. [DOI] [PubMed] [Google Scholar]
- 4.Sawicki W, Abramczuk J, Blaton O. DNA synthesis in the second and third cell cycles of mouse preimplantation development. A cytophotometric study. Exp Cell Res. 1978;112:199–205. doi: 10.1016/0014-4827(78)90540-2. [DOI] [PubMed] [Google Scholar]
- 5.Luthardt FW, Donahue RP. DNA synthesis in developing two-cell mouse embryos. Dev Biol. 1975;44:210–6. doi: 10.1016/0012-1606(75)90389-9. [DOI] [PubMed] [Google Scholar]
- 6.Molls M, Zamboglou N, Streffer C. A comparison of the cell kinetics of pre-implantation mouse embryos from two different mouse strains. Cell Tissue Kinet. 1983;16:277–83. [PubMed] [Google Scholar]
- 7.Molls M, Pon A, Streffer C, van Beuningen D, Zamboglou N. The effects of lead and X-rays, alone or in combination, on blastocyst formation and cell kinetics of preimplantation mouse embryos in vitro. Int J Radiat Biol Relat Stud Phys Chem Med. 1983;43:57–69. doi: 10.1080/09553008314550051. [DOI] [PubMed] [Google Scholar]
- 8.Visintin R, Craig K, Hwang ES, Prinz S, Tyers M, Amon A. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol Cell. 1998;2:709–18. doi: 10.1016/s1097-2765(00)80286-5. [DOI] [PubMed] [Google Scholar]
- 9.Jaspersen SL, Charles JF, Tinker-Kulberg RL, Morgan DO. A late mitotic regulatory network controlling cyclin destruction in Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:2803–17. doi: 10.1091/mbc.9.10.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berdougo E, Nachury MV, Jackson PK, Jallepalli PV. The nucleolar phosphatase Cdc14B is dispensable for chromosome segregation and mitotic exit in human cells. Cell Cycle. 2008;7:1184–1190. doi: 10.4161/cc.7.9.5792. [DOI] [PubMed] [Google Scholar]
- 11.Cho HP, Liu Y, Gomez M, Dunlap J, Tyers M, Wang Y. The dual-specificity phosphatase CDC14B bundles and stabilizes microtubules. Mol Cell Biol. 2005;25:4541–51. doi: 10.1128/MCB.25.11.4541-4551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu J, Cho HP, Rhee DB, Johnson DK, Dunlap J, Liu Y, et al. Cdc14B depletion leads to centriole amplification, and its overexpression prevents unscheduled centriole duplication. J Cell Biol. 2008;181:475–83. doi: 10.1083/jcb.200710127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bassermann F, Frescas D, Guardavaccaro D, Busino L, Peschiaroli A, Pagano M. The Cdc14B-Cdh1-Plk1 axis controls the G2 DNA-damage-response checkpoint. Cell. 2008;134:256–67. doi: 10.1016/j.cell.2008.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schindler K, Schultz RM. CDC14B acts through FZR1 (CDH1) to prevent meiotic maturation of mouse oocytes. Biol Reprod. 2009;80:795–803. doi: 10.1095/biolreprod.108.074906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Su YQ, Sugiura K, Woo Y, Wigglesworth K, Kamdar S, Affourtit J, et al. Selective degradation of transcripts during meiotic maturation of mouse oocytes. Dev Biol. 2007;302:104–17. doi: 10.1016/j.ydbio.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schultz RM. Regulation of zygotic gene activation in the mouse. Bioessays. 1993;15:531–8. doi: 10.1002/bies.950150806. [DOI] [PubMed] [Google Scholar]
- 17.Schultz RM. The molecular foundations of the maternal to zygotic transition in the preimplantation embryo. Hum Reprod Update. 2002;8:323–31. doi: 10.1093/humupd/8.4.323. [DOI] [PubMed] [Google Scholar]
- 18.Mailand N, Lukas C, Kaiser BK, Jackson PK, Bartek J, Lukas J. Deregulated human Cdc14A phosphatase disrupts centrosome separation and chromosome segregation. Nat Cell Biol. 2002;4:317–22. doi: 10.1038/ncb777. [DOI] [PubMed] [Google Scholar]
- 19.Kaiser BK, Zimmerman ZA, Charbonneau H, Jackson PK. Disruption of centrosome structure, chromosome segregation and cytokinesis by misexpression of human Cdc14A phosphatase. Mol Biol Cell. 2002;13:2289–300. doi: 10.1091/mbc.01-11-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masui Y, Clarke HJ. Oocyte maturation. Int Rev Cytol. 1979;57:185–282. doi: 10.1016/s0074-7696(08)61464-3. [DOI] [PubMed] [Google Scholar]
- 21.Forsburg SL, Nurse P. Cell cycle regulation in the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe. Ann Rev Cell Biol. 1991;7:227–56. doi: 10.1146/annurev.cb.07.110191.001303. [DOI] [PubMed] [Google Scholar]
- 22.Hampl A, Eppig JJ. Analysis of the mechanism(s) of metaphase I arrest in maturing mouse oocytes. Development. 1995;121:925–33. doi: 10.1242/dev.121.4.925. [DOI] [PubMed] [Google Scholar]
- 23.Aoki F, Worrad DM, Schultz RM. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev Biol. 1997;181:296–307. doi: 10.1006/dbio.1996.8466. [DOI] [PubMed] [Google Scholar]
- 24.Inoue A, Nakajima R, Nagata M, Aoki F. Contribution of the oocyte nucleus and cytoplasm to the determination of meiotic and developmental competence in mice. Human Reprod. 2008;23:1377–84. doi: 10.1093/humrep/den096. [DOI] [PubMed] [Google Scholar]
- 25.Bellier S, Chastant S, Adenot P, Vincent M, Renard JP, Bensaude O. Nuclear translocation and carboxylterminal domain phosphorylation of RNA polymerase II delineate the two phases of zygotic gene activation in mammalian embryos. EMBO J. 1997;16:6250–62. doi: 10.1093/emboj/16.20.6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aoki F, Hara KT, Schultz RM. Acquisition of transcriptional competence in the 1-cell mouse embryo: requirement for recruitment of maternal mRNAs. Mol Reprod Dev. 2003;64:270–4. doi: 10.1002/mrd.10227. [DOI] [PubMed] [Google Scholar]
- 27.Clemente-Blanco A, Mayan-Santos M, Schneider DA, Machin F, Jarmuz A, Tschochner H, et al. Cdc14 inhibits transcription by RNA polymerase I during anaphase. Nature. 2009;458:219–22. doi: 10.1038/nature07652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeng F, Schultz RM. Gene expression in mouse oocytes and preimplantation embryos: use of suppression subtractive hybridization to identify oocyte- and embryo-specific genes. Biol Reprod. 2003;68:31–9. doi: 10.1095/biolreprod.102.007674. [DOI] [PubMed] [Google Scholar]
- 29.Zeng F, Baldwin DA, Schultz RM. Transcript profiling during preimplantation mouse development. Dev Biol. 2004;272:483–96. doi: 10.1016/j.ydbio.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 30.Nothias JY, Miranda M, DePamphilis ML. Uncoupling of transcription and translation during zygotic gene activation in the mouse. EMBO J. 1996;15:5715–25. [PMC free article] [PubMed] [Google Scholar]
- 31.Baran V, Vesela J, Rehak P, Koppel J, Flechon JE. Localization of fibrillarin and nucleolin in nucleoli of mouse preimplantation embryos. Mol Reprod Dev. 1995;40:305–10. doi: 10.1002/mrd.1080400306. [DOI] [PubMed] [Google Scholar]
- 32.Hillman N, Tasca RJ. Ultrastructural and autoradiographic studies of mouse cleavage stages. Am J Anat. 1969;126:151–73. doi: 10.1002/aja.1001260203. [DOI] [PubMed] [Google Scholar]
- 33.Geuskens M, Alexandre H. Ultrastructural and autoradiographic studies of nucleolar development and rDNA transcription in preimplantation mouse embryos. Cell Differ. 1984;14:125–34. doi: 10.1016/0045-6039(84)90037-x. [DOI] [PubMed] [Google Scholar]
- 34.Bogolyubova IO, Bogoliubova NA, Bogolyubov DS, Parfenov VN. Nuclear structure in early mouse embryos: A comparative ultrastructural and immunocytochemical study with special emphasis on the “2-cell block in vitro”. Tissue Cell. 2006;38:389–98. doi: 10.1016/j.tice.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 35.Li L, Baibakov B, Dean J. A subcortical maternal complex essential for preimplantation mouse embryogenesis. Dev Cell. 2008;15:416–25. doi: 10.1016/j.devcel.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng P, Dean J. Role of Filia, a maternal effect gene, in maintaining euploidy during cleavage-stage mouse embryogenesis. Proc Natl Acad Sci USA. 2009;106:7473–8. doi: 10.1073/pnas.0900519106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Temeles GL, Ram PT, Rothstein JL, Schultz RM. Expression patterns of novel genes during mouse preimplantation embryogenesis. Mol Reprod Dev. 1994;37:121–9. doi: 10.1002/mrd.1080370202. [DOI] [PubMed] [Google Scholar]
- 38.Whitten W. Nutrient requirements for the culture of preimplantion mouse embryo in vitro. Adv Biosci. 1971;6:129–39. [Google Scholar]
- 39.Reis A, Chang HY, Levasseur M, Jones KT. APCcdh1 activity in mouse oocytes prevents entry into the first meiotic division. Nat Cell Biol. 2006;8:539–40. doi: 10.1038/ncb1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amanai M, Shoji S, Yoshida N, Brahmajosyula M, Perry AC. Injection of mammalian metaphase II oocytes with short interfering RNAs to dissect meiotic and early mitotic events. Biol Reprod. 2006;75:891–8. doi: 10.1095/biolreprod.106.054213. [DOI] [PubMed] [Google Scholar]
- 41.Chatot CL, Ziomek CA, Bavister BD, Lewis JL, Torres I. An improved culture medium supports development of random-bred 1-cell mouse embryos in vitro. J Reprod Fertil. 1989;86:679–88. doi: 10.1530/jrf.0.0860679. [DOI] [PubMed] [Google Scholar]
- 42.Ho Y, Wigglesworth K, Eppig JJ, Schultz RM. Preimplantation development of mouse embryos in KSOM: augmentation by amino acids and analysis of gene expression. Mol Reprod Dev. 1995;41:232–8. doi: 10.1002/mrd.1080410214. [DOI] [PubMed] [Google Scholar]
- 43.Artus J, Cohen-Tannoudji M. Cell cycle regulation during early mouse embryogenesis. Mol Cell Endocrinol. 2008;282:78–86. doi: 10.1016/j.mce.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 44.Laemmli UK, Quittner SF. Maturation of the head of bacteriophage T4 IV. The proteins of the core of the tubular polyheads and in vitro cleavage of the head proteins. Virology. 1974;62:483–99. doi: 10.1016/0042-6822(74)90409-7. [DOI] [PubMed] [Google Scholar]
- 45.Igarashi H, Knott JG, Schultz RM, Williams CJ. Alterations of PLCbeta1 in mouse eggs change calcium oscillatory behavior following fertilization. Dev Biol. 2007;312:321–30. doi: 10.1016/j.ydbio.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsafriri A, Chun SY, Zhang R, Hsueh AJ, Conti M. Oocyte maturation involves compartmentalization and opposing changes of cAMP levels in follicular somatic and germ cells: studies using selective phosphodiesterase inhibitors. Dev Biol. 1996;178:393–402. doi: 10.1006/dbio.1996.0226. [DOI] [PubMed] [Google Scholar]