Abstract
A fine mapping study in the HNF1B gene at 17q12 among two study populations revealed a second prostate cancer locus, ~26 kb centromeric to the first known locus (rs4430796); these are separated by a recombination hotspot. A SNP in the second locus (rs11649743) was confirmed in five additional populations, and P=1.7×10−9 for an allelic test in the seven combined studies. The association at each SNP remains significant after adjusting for the other SNP.
A locus in the HNF1B gene at 17q12 was initially reported to be associated with prostate cancer risk in a genome-wide association study (GWAS) in Iceland1 and was later replicated in two additional GWAS in the UK2 and in the United States3, as well as in two confirmation studies from our group4–5. We found several SNPs at 17q12, including rs4430796, to be strongly associated with prostate cancer risk in a population based case-control study from Sweden (CAPS) (P = 6.0 × 10−7)4 and in a hospital based case-control study from Johns Hopkins Hospital (JHH) (P = 5.1 × 10−4)5. To assess whether there are additional independent prostate cancer risk variants in the flanking region, we performed a fine mapping analysis of the 17q12 genomic region in the CAPS and JHH study populations. This approach was motivated by the characterization of the 8q24 prostate cancer association where at least two additional independent loci were subsequently discovered6–9 in the flanking region after the first locus at 8q24 was identified from a GWAS10 and an admixture mapping study11.
Our region of interest (33,111,655-33,189,279, Build 35) for this fine mapping effort spanned ~80kb and included the entire HNF1B gene and ~10 kb upstream and downstream of the gene. A total of 41 tagging SNPs in the region of interest were identified based on the HapMap Phase II data and genotyped using Sequenom iPLEX in CAPS (2,899 prostate cancer patients and 1,722 controls) and JHH (1,527 prostate cancer patients and 482 controls). A detailed description of the study subjects in these two populations is presented in Supplementary methods and Supplementary Tables 1&2. The average genotype call rate for these SNPs was 98.3% and the average concordance rate was 99.8% among 100 duplicated quality control samples. All of the SNPs were in Hardy-Weinberg equilibrium (P ≥ 0.05) among each of the control groups. We also imputed 23 SNPs in the region using the computer program IMPUTE12. A posterior probability of 0.90 was used to call the imputed genotypes. Pair-wise linkage disequilibrium (LD) among these 64 SNPs in controls was estimated and haplotype blocks were inferred using Haploview13. Heat maps for pair-wise LD (D′) of these SNPs in the entire fine mapping region in CAPS and JHH are presented in Supplementary Figure 1.
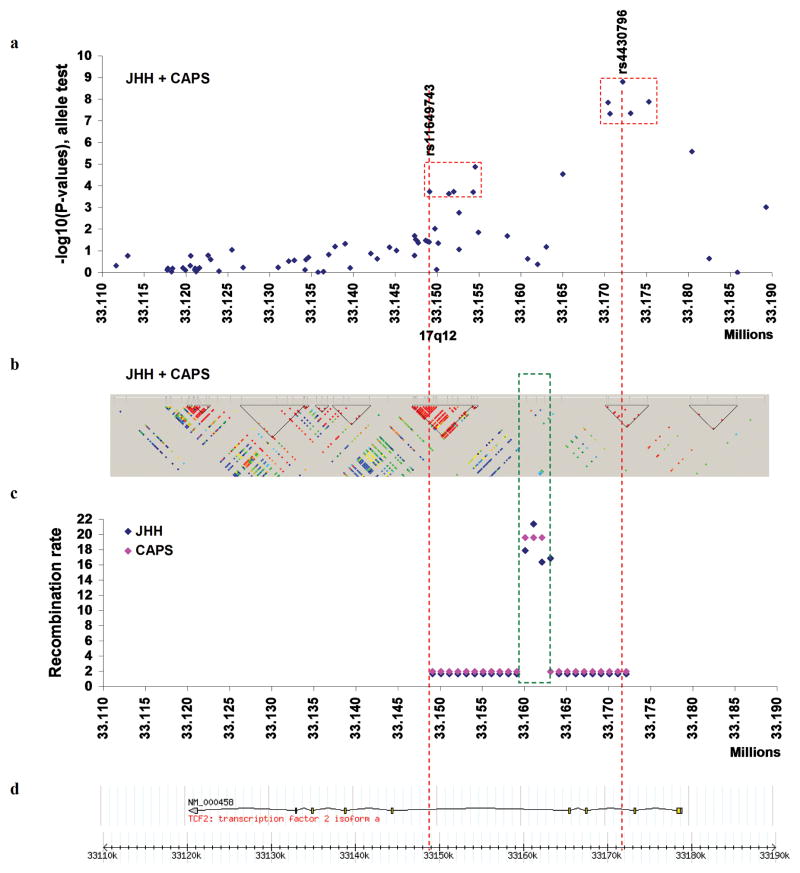
Allele frequency differences between cases and controls in CAPS and JHH were tested for these 64 SNPs using a chi-square test with 1-degree of freedom (df) (Supplementary Table 3a–b). We also performed a combined allelic test for CAPS and JHH study populations using the Cochran-Mantel-Haenszel test (Figure 1a). Two separate clusters of prostate cancer associated SNPs were found; one was the previously identified region between 33,170,413 and 33,175,629 (first locus), including rs4430796, and the second was a new region between 33,149,092 and 33,154,541 (second locus), ~26 kb centromeric to the first locus. These two loci were in different haplotype blocks estimated from control subjects in CAPS and JHH (Supplemental method), and the two blocks were separated by five SNPs with low pair-wise LD (Figure 1b). We also estimated the recombination rate between these two loci using 18 consecutive SNPs in the interval (bounded by rs4430796 at the first locus and rs11649743 at the second locus) among control subjects using SequenceLDhot software14. Strong evidence for a recombination hotspot between the two loci at 33,160,000-33,163,000 was found in JHH (P = 1.2 × 10−15) and in CAPS (P = 5.2 × 10−6) (Figure 1c). This is consistent with the known recombination hotspot at 33,162,001 in the HapMap data (Release 21, Phase I & II). These data strongly suggest that the two loci are genetically independent. The first locus spans a region between introns 1 and 2 of the HNF1B gene, and the second locus resides within intron 4 of the gene (Figure 1d).
Figure 1. A schematic view of genetic association between SNPs at 17q12 and prostate cancer risk in two study populations.
(a) Combined allele tests for 64 SNPs at 17q12 and prostate cancer risk among a total of 4,456 prostate cancer cases and 2,357 controls from a population-based case-control study in CAPS and JHH. Two separate clusters of prostate cancer associated SNPs were found (red dotted boxes). (b) Inferred haplotype blocks of these 64 SNPs were estimated from the control subjects in CAPS and JHH using the Haploview computer program13. These two loci were in different haplotype blocks. (c) Recombination hotspot estimated using 18 consecutive SNPs (bounded by rs4430796 at the first locus and rs11649743 at the second locus) among control subjects of CAPS and JHH using SequenceLDhot software15. Strong evidence for a recombination hotspot between the two loci was found (green dotted box). (d) Genomic view at 17q12 where the only gene (HNF1B) in this region is shown.
To replicate the associations at these two loci, we utilized data from the publicly available National Cancer Institute Cancer Genetic Markers of Susceptibility (CGEMS) study. SNPs rs4430796 at locus 1 and rs11649743 at locus 2 were genotyped in five study populations of CGEMS, including PLCO, CPS-II, HPFS, FPCC, and ATBC(Supplementary method)6. These two SNPs were selected to represent these two loci. Individual genotypes of PLCO were obtained through an approved application and summary genotypes of the remaining four populations were downloaded from the public CGEMS website http://cgems.cancer.gov/data/. The alleles at 4430796 and 11649743 that were associated with increased prostate cancer risk in CAPS and JHH were both higher in cases than controls in each of these five populations (Table 1). The differences were statistically significant (P < 0.05) for four of the five populations for each SNP. We performed combined allelic association tests among all seven study populations using a Cochran-Mantel-Haenszel test. Highly significant associations were found for rs4430796 (P = 5.0 × 10−20) and for rs11649743 (P = 1.7 × 10−9) among a total of 9,572 cases and 7,421 controls of European descent; both reached genome-wide significance level. There was no evidence for heterogeneity in allelic associations among these study populations using a Breslow-Day test for homogeneity (P = 0.30 for rs4430796 and P = 0.99 for rs11649743),
Table 1.
Association of prostate cancer risk with SNPs rs4430796 at locus 1 and rs11649743 at locus 2
| Risk Allel | Genotype counts
|
Allele frequency
|
P * | P^ | Heterozygotes
|
Homozygotes
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | OR | 95% CI | OR | 95% CI | ||||||||
| rs4430796 | GG | AG | AA | GG | AG | AA | AG vs GG | AA vs GG | |||||||
|
|
|
|
|||||||||||||
| CAPS | A | 446 | 1355 | 1073 | 316 | 883 | 509 | 0.61 | 0.56 | 7.5E-07 | 1.09 | 0.92–1.29 | 1.49 | 1.25–1.79 | |
| JHH | A | 254 | 779 | 488 | 106 | 253 | 120 | 0.58 | 0.51 | 7.0E-04 | 1.28 | 0.98–1.68 | 1.70 | 1.25–2.30 | |
| ATBC | A | 87 | 395 | 419 | 136 | 431 | 335 | 0.68 | 0.61 | 3.4E-06 | 1.43 | 1.06–1.94 | 1.96 | 1.44–2.65 | |
| FPCC | A | 149 | 308 | 163 | 161 | 309 | 148 | 0.51 | 0.49 | 0.28 | 1.08 | 0.82–1.42 | 1.19 | 0.87–1.63 | |
| HPFS | A | 113 | 289 | 179 | 153 | 300 | 138 | 0.56 | 0.49 | 7.6E-04 | 1.30 | 0.97–1.75 | 1.76 | 1.26–2.44 | |
| PLCO | A | 254 | 522 | 345 | 257 | 529 | 262 | 0.54 | 0.50 | 0.01 | 1.00 | 0.81–1.23 | 1.33 | 1.05–1.69 | |
| ACS | A | 357 | 843 | 516 | 434 | 850 | 434 | 0.55 | 0.50 | 1.2E-04 | 1.21 | 1.02–1.43 | 1.45 | 1.20–1.75 | |
| ALL | A | 1660 | 4491 | 3183 | 1563 | 3555 | 1946 | 0.58 | 0.53 | 5.0E-20 | 0.30 | 1.16 | 1.07–1.26 | 1.50 | 1.37–1.64 |
|
|
|
|
|||||||||||||
| rs11649743 | AA | AG | GG | AA | AG | GG | AG vs AA | GG vs AA | |||||||
| CAPS | G | 115 | 895 | 1842 | 92 | 587 | 1009 | 0.80 | 0.77 | 4.2E-04 | 1.22 | 0.91–1.64 | 1.16 | 0.63–2.17 | |
| JHH | G | 40 | 395 | 1055 | 14 | 139 | 317 | 0.84 | 0.82 | 0.19 | 0.99 | 0.53–1.88 | 1.61 | 0.88–2.95 | |
| ATBC | G | 18 | 219 | 690 | 27 | 250 | 644 | 0.86 | 0.83 | 0.02 | 1.31 | 0.70–2.45 | 1.72 | 0.97–3.06 | |
| FPCC | G | 20 | 191 | 445 | 32 | 211 | 413 | 0.82 | 0.79 | 0.03 | 1.45 | 0.80–2.62 | 1.45 | 0.79–2.65 | |
| HPFS | G | 19 | 159 | 418 | 27 | 174 | 410 | 0.83 | 0.81 | 0.17 | 1.30 | 0.70–2.43 | 1.90 | 1.18–3.07 | |
| PLCO | G | 28 | 361 | 777 | 47 | 359 | 687 | 0.82 | 0.79 | 0.02 | 1.69 | 1.03–2.76 | 1.35 | 0.92–1.98 | |
| ACS | G | 48 | 495 | 1216 | 62 | 546 | 1167 | 0.83 | 0.81 | 0.02 | 1.17 | 0.79–1.74 | 1.45 | 1.23–1.71 | |
| ALL | G | 288 | 2715 | 6443 | 301 | 2266 | 4647 | 0.83 | 0.80 | 1.7E-09 | 0.99 | 1.28 | 1.07–1.52 | 1.50 | 1.26–1.77 |
P *: Based on allelic test assuming multiplicative model. The combined tests are based on Mantel-Haenszel test test.
P ^: Breslow-Day test for homogeneity
To infer the mode of inheritance of these two SNPs, we fit four genetic models in the combined data from CAPS, JHH, and PLCO where individual genotype data and age information is available using a logistic regression analysis and adjusting for age (5-year group) and study population. These four models include a 2-df general model, and 1-df additive, dominant, and recessive models. The most parsimonious model, defined by lowest AIC value, was a recessive model for rs4430796 and an additive model for rs11649743 in the combined analysis (Supplementary Table 4). However, several other models had similar AIC values, reflecting a lack of statistical power to distinguish different genetic models in this analysis.
We tested the independence of prostate cancer associations for these two SNPs by including both SNPs (assuming a general model at each SNP) in a logistic regression model in the combined data from CAPS, JHH, and PLCO. The prostate cancer association at each SNP remained significant after adjusting for the other SNP, age (5-year group), and study population (Supplementary Table 5); suggesting prostate cancer associations at these two loci are independent. We then tested joint effects of these two SNPs on prostate cancer association by estimating Odds Ratios (ORs) for prostate cancer for carriers of nine combined genotypes (unconstrained model) using a logistic regression model and adjusting for age and study. Using men who were homozygous for non-risk alleles at both SNPs as a reference group, men who were homozygous for risk alleles at both SNPs had the highest risk in the combined study (OR=2.00, 95% CI: 1.37–2.91) (Supplementary Table 6).
We inferred haplotypes for 18 consecutive SNPs that are bounded by rs4430796 at the first locus and rs11649743 at the second locus in the CAPS and JHH using PHASE15. More than 32 haplotypes with frequencies of 1% and higher were inferred, reflecting a recombination hotspot between the two loci (Supplementary Table 7). Three haplotypes that contain risk alleles of both rs4430796 and rs11649743 (ID: 1, 2, and 20) had higher frequencies in cases than controls (nominal P < 0.05); however, the results were not consistent in these two populations. These results suggested that the observed associations at the two independent loci are unlikely due to a single long range haplotype that connects these two alleles.
These two SNPs were not associated with aggressiveness of prostate cancer in CAPS, JHH, and PLCO (Supplementary Table 8). Although these two SNPs were associated with PSA levels in controls in CAPS, no associations were found in controls of JHH (Supplementary Table 9). Additional studies are needed to dissect associations of these SNPs with prostate cancer risk and PSA levels.
While risk allele of rs4430796 was significantly higher in 364 cases (0.38) than 364 controls of African ancestry in JHH (P = 0.04)5, the frequency of the risk allele for rs11649743 was not significantly different between cases (0.934) and controls (0.935).
In summary, using data from our study populations and the publicly available data generated by CGEMS, we provide evidence of a second independent prostate cancer risk locus at 17q12. Interestingly, both these loci are located within the HNF1B gene. Sequencing the exons of HNF1B in 200 men with prostate cancer revealed no mutations that could account for the original gene association observed in an Icelandic study population1. Additional studies will be necessary to uncover the causal mechanisms underlying the independent as well as possible interaction effects of these variants on prostate cancer risk.
Supplementary Material
Acknowledgments
Funding
The study is supported by National Cancer Institute (CA106523, CA106523, and CA95052 to J.X., CA112517, CA58236 to W.B.I., CA86323 to AWP, Department of Defense grant PC051264 to J.X), as well as support from Swedish Cancer Society (Cancerfonden) and Swedish Research Council to H.G.
The authors thank all the study subjects who participated in this study. We also thank the Regional Cancer Registries and the CAPS-steering committee including Dr. Eberhart Varenhorst. We acknowledge the support of William T. Gerrard, Mario Duhon, Jennifer and John Chalsty, and David Koch.
The authors also thanks for the National Cancer Institute Cancer Genetic Markers of Susceptibility Initiative (CGEMS) for making the data available publicly.
The authors take full responsibility for the study design, data collection, analysis and interpretation of the data, the decision to submit the manuscript for publication, and the writing of the manuscript.
Footnotes
Competing interest statement
The authors declare that they have no competing financial interests.
Author Contributions
J.X., W.B.I., H.G. J.C., D.D. and J.M.T. designed and directed the study. S.L.Z. directed genotyping. J.S., F.W., F.C.H., S.T.K., Y.Z., L.D. J.S. J.L. and J.X. conducted data analysis. S.D.I., K.E.W., B.J.T., A.W.P., P.C.W. and W.B.I involved in the sample collection at Johns Hopkins Hospital. P.S., H.O.A., J.A., A.E.J, and H.G. involved in the sample collection in Sweden. G.L. performed laboratory analyses. A.R.T. and T.S.A. provided administrative and technical support for the study.
References
- 1.Gudmundsson J, et al. Nat Genet. 2007;39:977–83. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 2.Eeles RA, et al. Nat Genet. 2008;40:316–21. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 3.Thomas G, et al. Nat Genet. 2008;40:310–5. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 4.Zheng SL, et al. N Engl J Med. 2008;358:910–9. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]
- 5.Sun J, et al. Prostate. 2008 In press. [Google Scholar]
- 6.Yeager M, et al. Nat Genet. 2007;39:645–9. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 7.Gudmundsson J, et al. Nat Genet. 2007;39:631–7. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 8.Haiman CA, et al. Nat Genet. 2007;39:638–44. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng SL, et al. JNCI. 2007;99:1525–33. doi: 10.1093/jnci/djm169. [DOI] [PubMed] [Google Scholar]
- 10.Amundadottir LT, et al. Nat Genet. 2006;38:652–8. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 11.Freedman ML, et al. Proc Natl Acad Sci U S A. 2006;103:14068–73. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marchini J, et al. Am J Hum Genet. 2006;78:437–50. doi: 10.1086/500808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barrett JC, et al. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 14.Fearnhead P, et al. Bioinformatics. 2006;22:3061–6. doi: 10.1093/bioinformatics/btl540. [DOI] [PubMed] [Google Scholar]
- 15.Stephens M, et al. Am J Hum Genet. 2001;68:978–89. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.