Abstract
Objective
HLA-DRB1 alleles associated with risk of rheumatoid arthritis (RA) encode similar HLA-DRβ(1 sequences referred to as the “shared epitope” (SE). The most common SE sequences are QKRAA and QRRAA. A substantial number of RA patients, nevertheless, lack the SE. Bi-directional fetal-maternal trafficking results in long-term persistence of fetal cells in the mother and maternal cells in her offspring, referred to as microchimerism (Mc). We asked whether RA patients who lack the SE can acquire the SE through Mc.
Methods
We developed specific real-time quantitative PCR (qPCR) assays for the SE encoded sequences QKRAA and QRRAA. DNA extracted from peripheral blood mononuclear cells was tested with the SE-specific qPCR assays. A total of 86 subjects who were negative for the SE were studied, 52 women with RA and 34 healthy women.
Results
Mc with the SE was found significantly more often in RA patients than controls, odds ratio 4.1, 95% CI 1.6-10.0, p=0.003. Concentrations of SE Mc were also significantly higher among RA patients than controls, p=0.002. When analyzed separately for SE type, the prevalence of QKRAA Mc in RA vs. healthy women respectively was 17% vs. 3% (9/52 vs. 1/34, p=0.03) and of QRRAA 40% vs. 18% (21/52 vs. 6/34, p=0.04). Mc concentrations were also higher in RA than healthy subjects for QKRAA (p=0.03) and QRRAA (p=0.03).
Conclusion
Results indicate RA patients who genotypically lack the SE can acquire the SE as persistent Mc from maternal-fetal cell exchange and suggest SE-encoding Mc could be a risk factor for RA.
Keywords: microchimerism, shared epitope, rheumatoid arthritis, HLA
Introduction
HLA-DRB1*04 is increased in rheumatoid arthritis (RA) patients but consists of numerous alleles only some of which are RA-associated whereas others are neutral or even RA-protective (1,2). In Caucasian RA patients HLA-DRB1*0401 is most common followed by DRB1*0404, *0405, *0408 and other uncommon alleles. In some populations DRB1*0101 and DRB1*1402 are RA-associated. HLA-DRB1*0401 encodes the sequence “QKRAA” and the other above alleles encode “QRRAA” in the DRβ1 third hypervariable region. These similar sequences are referred to as the RA “shared epitope” (SE). In a few populations HLA-DRB1*10, which encodes an additional arginine, is also RA-associated (1). While RA patients frequently have one or two copies of the SE, a substantial minority and in some populations even a majority lack the SE (1,3). Long-term persistence of maternal cells in her progeny and fetal cells in women, referred to as microchimerism (Mc) (4), raises the question whether individuals who lack RA-associated HLA molecules can acquire them through Mc.
In a recent study subjects lacking HLA-DRB1*04 were tested for DRB1*04 Mc and subjects lacking HLA-DRB1*01 tested for DRB1*01 Mc (5). A significant increase of DRB1*04 Mc and DRB1*01 Mc was found in RA patients vs. controls; however, only some DRB1*04 and DRB1*01 alleles encode the SE while others do not (e.g. DRB1*0402, DRB1*0403, DRB1*0103). Therefore whether increased Mc in RA patients is due to the SE is not certain (other than one case that was sequenced). We developed quantitative PCR (qPCR) assays for the specific SE sequences QKRAA and QRRAA and studied SE-negative RA patients and controls for SE-positive Mc.
Patients and Methods
Study subjects, specimens, DNA extraction
Women with RA (n=52) met American College of Rheumatology criteria (6) and healthy women (n=34) had no autoimmune disease. 88% of RA and 91% of healthy women were Caucasian; others were Asian (n=3 RA, n=1 healthy), African American (n=2 RA and n=2 healthy), Native American (n=1 RA) and 1 unknown (healthy). Age range was 35 to 73 (mean 51) for RA and 35 to 69 (mean 42) for healthy women. 86% of RA and 94% of healthy women had at least one birth (mean parity 2.5 and 1.8 respectively). The institutional review board approved the study; all participants provided informed consent.
Peripheral blood mononuclear cells (PBMC) were isolated from whole blood (in ACD) by density gradient centrifugation, genomic DNA extracted under a biosafety hood using Wizard® Genomic DNA Purification Kit (Promega, Madison, WI) and resuspended in water. The mean total number of cell equivalents tested was 11.3×104 for RA and 12.5×104 for healthy women.
HLA genotyping
Specific HLA-DRB1, DQA1 and DQB1 alleles were determined as previously described (7).
Development of SE-specific real-time qPCR assays
qPCR assays targeting QKRAA and QRRAA were developed with the forward primer complementary to SE-encoding DRB1 alleles (DRB1*0401, *0404, *0405, *0408, *0101 and *1402) from codons 63 to 70 within exon 2 of the HLA-DRB1 gene, and the reverse primer from codons 81 to 74 corresponding to SE-encoding alleles as well. An artificial nucleotide mismatch was introduced into the reverse primer for enhanced specificity. The TaqMan minor groove binder probe was chosen to be specific for QKRAA or QRRAA from codons 70 to 74.
HLA-DRB1*15 shares the last four nucleotides with SE-encoding DRB1 alleles at forward and reverse primers binding sites. Therefore, qPCR sensitivity for SE Mc detection would be reduced in DRB1*15-positive subjects because background DNA (DRB1*15) competes with microchimeric DNA for primers, even though the TaqMan probe is highly specific for SE sequences. To overcome this problem, an inhibitory oligonucleotide was designed with the 3′ end labeled with biotin, complementary to codons 66-72 of DRB1*15 and with a higher melting temperature compared to the primers. During the qPCR annealing step the inhibitory oligonucleotide preferentially attaches to background DNA (DRB1*15) and suppresses binding of the forward primer to the background. No chain extension occurs for the inhibitory oligonucleotide after binding to the template because its 3′ terminal has been modified with biotin. The specific inhibitory oligonucleotide did not interfere with amplification of SE-positive Mc within a non-DRB1*15 background. A single SE-positive cell equivalent could be detected in a background of 10,000 cells genome equivalents (sensitivity: 0.01%).
Reactions were set up in 50 μL with 5 μL DNA from PBMC and tested 12 DNA aliquots with the QKRAA or QRRAA assay for each subject. The overall qPCR approach was similar to that previously reported (8) and included QKRAA or QRRAA-specific calibration curves and standard β-globin curves curves run simultaneously with each assay. Results were expressed as genome equivalents of Mc per 1,000,000 subject′s cell equivalents (gEq/mil). Specificity of SE qPCR assays was established by testing against an extensive panel of controls consisting of well-defined HLA cell lines from the 13th International HLA Working Group.
Statistical analysis
The outcome for analysis was disease status (RA vs. healthy women) and predictors of interest prevalence and concentration of Mc. Differences in characteristics between groups were assessed via t-test for continuous variables and Chi-squared test for categorical factors. Comparisons of Mc concentrations were carried out using linear regression models applied to ranks of Mc values. For Mc prevalence, each subject′s outcome was dichotomized to positive (>0) or negative (0) to compare proportions of subjects with a positive Mc result via logistic regression analysis. Results for the QKRAA and QRRAA sequences were combined for the primary analysis, and each then analyzed separately. Variables evaluated for confounding included age, parity at blood draw and total DNA cell equivalents tested. A confounder was defined by a difference of 10% or more in the estimated coefficient of interest between the multivariable model including the factor and the model without it. For both the linear and logistic regression models of repeated measures per subject, generalized estimating equations were used to obtain robust standard error estimates.
Results
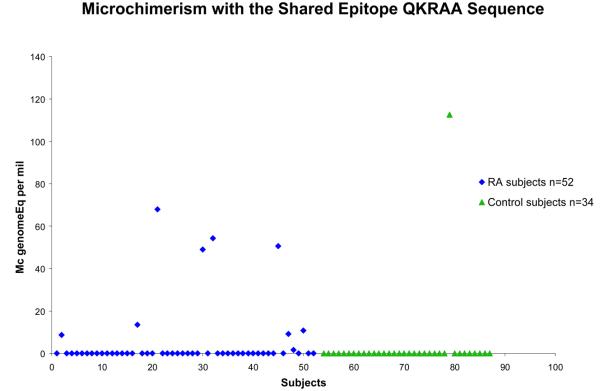
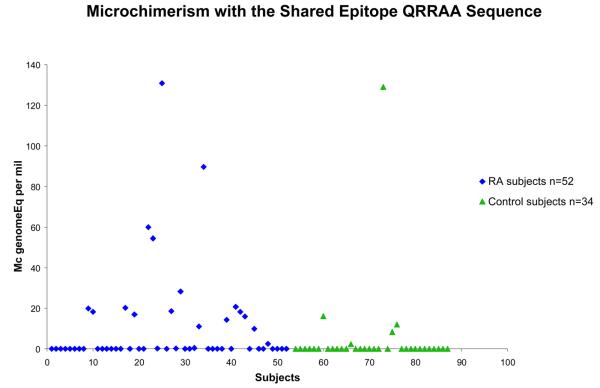
86 women who had no SE allele were studied, 52 with RA and 34 healthy. All subjects had an observation for both of the SE-specific assays, QKRAA and QRRAA. The odds ratio for detection of Mc with the SE, either QKRAA or QRRAA, was 4.1, 95% confidence interval 1.6-10.0, p=0.003 after adjustment for age at draw date (Table 1). When analyzed separately by SE type, SE Mc positivity was significantly RA-associated for both QKRAA and QRRAA. For QKRAA 9 of 52 (17%) RA patients were positive compared to 1 of 34 (3%) healthy women, p=0.03 (Figure 1). For QRRAA 21 of 52 (40%) RA patients were positive compared to 6 of 34 (18%) healthy women, p=0.04 (Figure 2). The prevalence of Mc with QRRAA was approximately twice that of Mc with QKRAA in RA patients.
Table 1.
Prevalence of Microchimerism with the SE in Healthy Women and Women with RA
| Any SE | Study subjects | Number of Observations |
Prevalence N (%) |
Adjusted* OR (95% CI) |
p-value |
|---|---|---|---|---|---|
| Healthy women | 68 | 7 (10) | 1.0 | – | |
| Women with RA | 104 | 30 (29) | 4.1 (1.6 – 10.0) | 0.003 | |
| SE type | |||||
| QKRAA | Healthy women | 34 | 1 (3) | 1.0 | – |
| Women with RA | 52 | 9 (17) | 11.5 (1.3-102.5) | 0.03 | |
| QRRAA | Healthy women | 34 | 6 (18) | 1.0 | – |
| Women with RA | 52 | 21 (40) | 3.1 (1.1-9.2) | 0.04 |
Adjusted for age at draw date.
Figure 1.
Increased prevalence and concentration of Mc encoding the SE sequence QKRAA in women with RA (◆) and healthy women (▲). Concentrations are expressed as the genome equivalent number of microchimeric cells per million subject′s cells.
Figure 2.
Increased prevalence and concentration of Mc encoding the SE sequence QRRAA in women with RA (◆) and healthy women (▲). Concentrations are expressed as the genome equivalent number of microchimeric cells per million subject′s cells.
The quantity of SE Mc (Mc concentration) was higher in RA patients than healthy women. The ranked values of Mc were significantly higher among women with RA than healthy women (p=0.002) after adjustment for age at draw date. When analyzed separately by SE type, the ranked values of QKRAA Mc were significantly higher among women with RA compared to healthy women (p=0.03) in a model adjusted for age at draw date. Ranked values, similarly were significantly higher for QRRAA Mc (p=0.03).
Development of SE-specific qPCR assays also permitted testing subjects who had DRB1*04 or *01 but did not have a SE-encoding allele. Among 10 such RA patients four had DRB1*0402, three DRB1*0403, two DRB1*0407 and one DRB1*0411. One of 10 was positive for QKRAA and QRRAA and five were positive for QRRAA only. Three healthy women had DRB1*04 with non SE-encoding alleles, one each with DRB1*0402, *0403 and *0407; all tested negatively for SE Mc. Four RA patients who had HLA-DRB1*0102 were tested for Mc with QKRAA. All subjects had positive results. These results would have further increased the significance of our findings but were not included in the analysis because DRB1*0102 has the QRRAA SE sequence but, to our knowledge, unlike DRB1*0101 a significant difference of DRB1*0102 has not been reported in RA patients compared to controls.
HLA genotypes with the sequence “DERAA” are thought to be RA-protective (2). There was no suggestion of a difference in SE Mc results if a subject had DERAA. Among RA patients tested for QKRAA 4 of 9 (44%) with positive results had 1 or 2 copies of DERAA compared to 16 of 43 (37%) with negative results. Among RA patients tested for QRRAA, 8 of 21 (38%) with positive results had 1 or 2 copies of DERAA compared to 12 of 31 (39%) with negative results. Similarly, there was no suggestion that the DERAA sequence impacted SE Mc in healthy subjects.
The most common sources of naturally acquired Mc are maternal Mc acquired during fetal life and, in women, from prior pregnancies (4). Most of our study subjects were parous women. Among RA women tested for QKRAA 89% (8 of 9) with positive results and 88% (38 of 43) with negative results were parous. Among RA patients tested for QRRAA 90% (19 of 21) with positive and 87% (27 of 31) with negative results were parous. Among healthy women 1 was positive for QKRAA and 6 for QRRAA, all of whom were parous. The mean parity in RA women positive vs. negative for SE Mc was also similar (2.6 vs. 2.5 for QKRAA; 2.5 vs. 2.5 for QRRAA). Mean time from last birth to Mc testing in parous RA women for those testing positive vs. negative respectively was 18.2 vs. 25.3 years for QKRAA and 24.2 vs. 24.1 years for QRRAA. Births occurred before RA onset for all but one RA patient who was gravid before and parous after RA onset and two who had births only after onset. Results did not materially differ excluding these subjects. For healthy women only 1 had QKRAA Mc (parity 1 vs. 1.9 for those negative) and for with QRRAA Mc mean parity was 2.2 and 2.0 for those without QRRAA Mc. Very few women in our study were nulliparous as noted above.
Some RA patients were taking disease modifying antirheumatic drugs (DMARDs) at the time of the blood draw. Overall 40% (20 of 50, 2 unknown) were taking at least one DMARD including hydroxychloroquine, gold, methotrexate, sulfsalazine and one patient aziothioprine. Others were taking nonsteroidal anti-inflammatory medications, low dose prednisone or no medications. (Samples were obtained before the widespread use of other DMARDs.) Of RA patients testing positive for QKRAA Mc, 33% were taking at least one DMARD compared to 41% of those testing negative. Of RA patients testing positive for QRRAA Mc, 52% were taking at least one DMARD compared to 31% of patients testing negative.
Discussion
Depending upon the population studied 25% to 75% of RA patients lack the SE sequence. An increase of HLA-DRB1*04 Mc in DRB1*04-negative RA patients vs. controls and DRB1*01 Mc in DRB1*01-negative patients vs. controls was recently reported (5). The specific HLA alleles of the Mc, however, were not determined other than for one subject. This important observation first raised the question of whether SE Mc might sometimes contribute to RA-risk. We developed specific qPCR assays for the SE sequences QKRAA and QRRAA and tested PBMC from SE-negative subjects. SE Mc was significantly increased in RA patients with an overall odds of SE Mc approximately four times that of a healthy subject. QKRAA Mc was less frequent than QRRAA Mc although the magnitude of the difference between RA patients and healthy subjects was greater for QKRAA than QRRAA. Moreover, Mc concentrations for both QKRAA and QRRAA Mc were significantly higher in RA patients than healthy subjects.
The most common Mc source is maternal-fetal cell exchange during pregnancy. Fetal Mc can also be accrued from pregnancy resulting in a birth, miscarriage or elective termination (9). Mc can also be acquired from a recognized or sometimes unrecognized twin (4). Another potential source, although not yet demonstrated, is from an older sibling transferred by the mother to a later birth order child. Blood transfusion can also sometimes result in Mc (4). While the origin of SE Mc in our study is not known the most common sources are maternal Mc and/or fetal Mc. That maternal Mc could affect RA-risk is supported by reports describing an increased frequency of DRB1*04-positive and/or SE-positive mothers in DRB1*04-negative and/or SE-negative RA patients (10). This phenomenon, while reported in some and not other studies (reviewed in 11), is referred to as the non-inherited maternal antigen or “NIMA” effect. A protective “NIMA” effect has also recently been reported (12).
Fetal Mc is commonly accrued from pregnancy. Our study investigated women with known pregnancy history, with almost all pregnancies before RA onset. Most women with SE Mc were parous but Mc-negative women were similarly parous. In other studies we found that parous women had decreased RA-risk compared to nulliparous women, but benefit was only evident for younger women and with more recent births (13). In the current study selection of study subjects was based on absence of the SE sequence, women were older, time from births longer and there were very few nulliparous women. While other studies will be necessary to address any potential role of fetal Mc in RA a reasonable hypothesis is that there are trade-offs for women, with protection in younger women who acquire fetal Mc with the protective DERAA sequence but risk for women who harbor SE-containing Mc over the long-term.
A strength of our study is quantitative assays were developed that specifically identified SE Mc. A limitation is transfusion history was unknown, although it is unlikely that a difference in transfusions could explain our results. Another consideration is DMARD use, which could affect detection of Mc, although most patients in our study, with and without Mc, were not taking DMARDS. Finally, our findings do not address cause and effect. While it is of special interest that Mc measured in these studies is with RA-risk associated HLA alleles, to better address cause and effect a prospective study of incident cases and controls would be needed.
A question of interest for future studies is whether Mc can have an additive or synergistic effect for individuals who have one copy of the SE. Particularly strong RA-risk has been reported for the genotype combination DRB1*0401,*0404 (14). By analogy it may be asked whether similar combinations with Mc are particularly disadvantageous, for example when the genotype includes DRB1*0404 and a woman acquires fetal Mc with DRB1*0401.
In conclusion, the biological significance of Mc is not yet known, but it is likely that Mc has both beneficial and detrimental consequences and that the HLA specificity of the Mc is a key contributor to the outcome (15). The current studies raise the question whether SE Mc becomes a “mini-dose” in the spectrum of RA-risk, which has been considered to increase according to whether an individual′s HLA-genotype contains 0, 1 or 2 SE copies. Additional studies are needed to elucidate the role of different Mc types in RA since different consequences may also accrue from fetal than maternal Mc as the former is acquired during adult life and the latter during fetal life.
Acknowledgements
We are grateful for the participation of RA patients and healthy volunteers. We also thank our study coordinators, Kathy Vickers, Jennifer Brackensick, Dawn Stief and Darlene Segerstrom.
Supported by NIH grants AI45659 and AI41721
Footnotes
There are no financial conflicts for any authors
References
- 1.Winchester R. The molecular basis of susceptibility to rheumatoid arthritis. Adv Immunol. 1994;56:389–466. doi: 10.1016/s0065-2776(08)60456-3. [DOI] [PubMed] [Google Scholar]
- 2.Van der Helm-van Mil A, Huizinga T, Schreuder G, Breedveld F, de Vries RRP, Toes R. An independent role of protective HLA class II alleles in rheumatoid arthritis severity and susceptibility. Arthritis Rheum. 2005;52:2637–44. doi: 10.1002/art.21272. [DOI] [PubMed] [Google Scholar]
- 3.Hughes LB, Morrison D, Kelley JM, Padilla M, Vaughan L, Westfall A, et al. The HLA-DRB1 shared epitope is associated with susceptibility to rheumatoid arthritis in African Americans through European genetic admixture. Arthritis Rheum. 2008;58:349–58. doi: 10.1002/art.23166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gammill H, Nelson JL. Naturally acquired microchimerism. Int J Dev Bio. 2010;54:531–43. doi: 10.1387/ijdb.082767hg. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rak JM, Maestroni L, Balandraud N, Guis S, Boudinet H, Guzian MC, et al. Transfer of shared epitope through microchimerism in women with rheumatoid arthritis. Arthritis and Rheumatism. 2009;60:73–80. doi: 10.1002/art.24224. [DOI] [PubMed] [Google Scholar]
- 6.Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31:315–24. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 7.Nelson JL, Gillespie KM, Lambert NC, Stevens AM, Loubiere LS, Rutledge JC, et al. Maternal microchimerism in peripheral blood in type 1 diabetes and pancreatic islet β cell microchimerism. Proc Natl Acad Sci. 2007;104:1637–42. doi: 10.1073/pnas.0606169104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lambert NC, Erickson TD, Yan Z, Pang JM, Guthrie KA, Furst DE, et al. Quantification of maternal microchimerism by HLA-specific real-time polymerase chain reaction: studies of healthy women and women with scleroderma. Arthritis Rheum. 2004;50:906–14. doi: 10.1002/art.20200. [DOI] [PubMed] [Google Scholar]
- 9.Yan Z, Lambert NC, Guthrie KA, Porter AJ, Loubiere LS, Madeleine MM, et al. Male microchimerism in women without sons: Quantitative assessment and correlation with pregnancy history. Am J Med. 2005;118:899–906. doi: 10.1016/j.amjmed.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 10.Harney S, Newton J, Milicic A, Brown MA, Wordsworth BP. Non-inherited maternal HLA alleles are associated with rheumatoid arthritis. Rheumatol. 2003;42:171–4. doi: 10.1093/rheumatology/keg059. [DOI] [PubMed] [Google Scholar]
- 11.Guthrie KA, Tishkevich NR, Nelson JL. Non-inherited maternal human leukocyte antigen alleles in susceptibility to familial rheumatoid arthritis. Ann Rheum Dis. 2009;68:107–9. doi: 10.1136/ard.2008.092312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feitsma A, Worthington J, van der Helm-van Mil A, Plant D, Thomson W, Ursum J, et al. Protective effect of noninherited maternal HLA-DR antigens on rheumatoid arthritis development. Proceedings of the National Academy of Sciences. 2007;104(50):19966–70. doi: 10.1073/pnas.0710260104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guthrie KA, Dugowson C, Voigt LF, Koepsell TD, Nelson JL. Does pregnancy provide vaccine-like protection against rheumatoid arthritis? Arthritis Rheum. 2010;62:1842–8. doi: 10.1002/art.27459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wordsworth P, Pile KD, Buckely JD, Lanchbury JSS, Ollier B, Lathrop M, Bell JI. HLA heterozygosity contributes to susceptibility to rheumatoid arthritis. Am J Hum Genet. 1992;51:585–591. [PMC free article] [PubMed] [Google Scholar]
- 15.Nelson JL. Naturally acquired microchimerism: For better or for worse. Arthritis Rheum. 2009;60:5–7. doi: 10.1002/art.24217. [DOI] [PMC free article] [PubMed] [Google Scholar]