Abstract
Studies on the adherence properties of oral bacteria have been a major focus in microbiology research for several decades. The ability of bacteria to adhere to the variety of surfaces present in the oral cavity, and to become integrated within the resident microbial communities, confers growth and survival properties. Molecular analyses have revealed several families of Gram-positive bacterial surface proteins, including serine-rich repeat, antigen I/II, and pilus families, that mediate adherence to a variety of salivary and oral bacterial receptors. In Gram-negative bacteria, pili, auto-transporters, and extracellular matrix-binding proteins provide components for host tissue recognition and building of complex microbial communities. Future studies will reveal in greater detail the binding pockets for these adhesin families and their receptors. This information will be crucial for the development of new inhibitors or vaccines that target the functional regions of bacterial proteins that are involved in colonization and pathogenesis.
Keywords: bacteria, biofilm(s), microbiology
Introduction
The mouth is an open system, rather like a river, with a continual flow of liquid washing out particles that do not attach and hold fast to surfaces. Many micro-organisms that enter the mouth are immediately trapped by saliva and swallowed. Some come into brief contact with surfaces of hard or soft oral tissues, but do not thrive, because they lack the appropriate machinery to proliferate in the competitive oral biofilm environment. These are transient colonizers, or allochthonous microbiota, that are sometimes detected in taxonomic surveys but play little part in shaping the structure and function of oral microbial communities. The micro-organisms that successfully colonize the oral cavity are specialists, adapted to adhere to oral surfaces and to utilize the nutrients available. Over 600 different species of bacteria are found naturally in the mouth, and an individual may carry 100 or more different species in their mouth at any time (Dewhirst et al., 2010). The oral cavity presents a variety of different niches for micro-organisms, and only a small proportion of the 600 or so natural colonizers are able to adhere primarily to hard or soft tissues. The other bacteria attach to these primary colonizers. Cell-cell binding between micro-organisms is thought to play a key role in integrating secondary colonizers into oral biofilms, and building complex networks of interacting microbial cells (Fig. 1).
Figure 1.
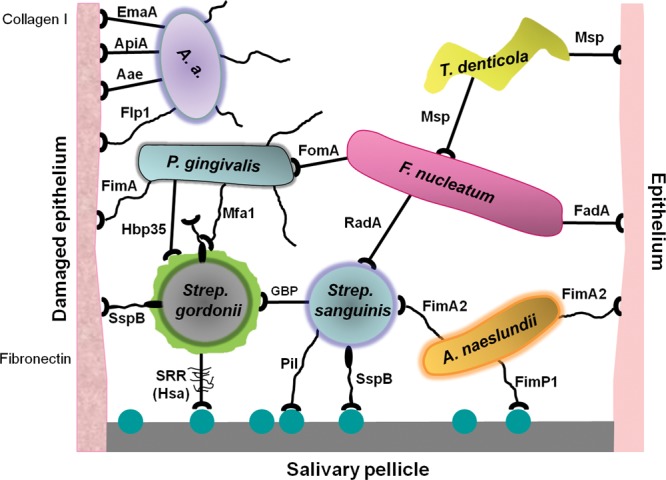
Diagrammatic representation of interactions occurring between oral micro-organisms and with host tissues that contribute to the formation of microbial communities within the human oral cavity. Microbial adhesins are represented as suction cups on stalks that may be flexible (e.g., pilus), while pellicle receptors are represented as molecular spheres. The adhesins are labeled according to the nomenclature in Table 1. Polysaccharides (RPS and glucan) are indicated as surrounding Streptococcus cells. Glucan-binding proteins (GBP) are not included in Table 1, although these play a crucial role in conjunction with glucosyltransferases (GTFs) in S. mutans colonization (Koo et al., 2010).
At the simplest level, bacterial adhesion involves the formation of a substantial number of non-covalent bonds between the bacterial cell surface and a substratum. The bonds result from electrostatic, ionic, or hydrophobic interactions, and these are largely dependent upon the presence of specific adhesin molecules on the bacterial cell surface. Different species or strains of bacteria are by no means equivalent in their capacities to bind host or bacterial ligands. Since there are fundamental differences between the cell envelope structures of Gram-positive and Gram-negative bacteria, the adhesion mechanisms of these two groups of bacteria can be quite distinct. Here, we review recent advances made in understanding the molecular mechanisms of oral bacterial adhesion, with focus on protein adhesins.
Adhesion to Pellicle
The rule for colonization within the oral cavity is simple: stick or be swallowed. Micro-organisms entering the mouth must attach to a surface to evade clearance by salivary flow from the mouth to the digestive tract. Pioneer colonizing micro-organisms have therefore developed capacity to bind constituents of the salivary film (pellicle) that continuously bathes both soft and hard tissues within the mouth. This section highlights some of the major protein adhesin families used by the oral microbiota for pellicle attachment.
Serine-rich Repeat (Srr) Protein Family
The Srr bacterial surface adhesins have the capacity to recognize carbohydrate (saccharide) moieties of glycosylated salivary constituents. The Srr proteins are produced by streptococci, staphylococci, and lactobacilli, and form appendages (fibrils or fimbriae) that extend up to 600 nm from the bacterial cell surface. They are characterized by the presence of multiple serine-rich repeats that can constitute about 80% of the entire protein. High-resolution studies with Streptococcus parasanguinis Srr protein Fap1 (Ramboarina et al., 2010) suggest that the dipeptide S(V/I/E) repeats within the C-terminal region form a super-helical extended stalk that projects the N-terminal protein domain away from the bacterial cell surface. Each protein molecule is approximately 300 nm in length, and it is hypothesized that the molecules might be cross-linked head to tail via N-terminal lysine residues to generate 600 nm or longer fibrils. These could promote greater distance interactions between bacterium and substratum.
Two variants of Srr proteins have been found in oral commensal Streptococcus gordonii, designated Hsa and GspB. Both proteins mediate adhesion to salivary pellicle through recognition of sialic-acid-containing components, including salivary mucin MG2 and salivary agglutinin (Takamatsu et al., 2006). Discrete domains have been identified within these Srr proteins for mediating initial pellicle attachment vs. biofilm development (Wu et al., 1998, 2007; Takamatsu et al., 2006; Ramboarina et al., 2010), indicating the capacity of these adhesins to perform multiple adhesive functions simultaneously.
Further insights from structural studies of Fap1 relate to the influence of environmental factors on bacterial adhesive capabilities. It was found that lowering pH from 8 to 5 resulted in Fap1 conformational changes, and fibril aggregation with adhesive domains clustering uniquely at the tip (Ramboarina et al., 2010). Such changes were associated with a concomitant increase in adhesion of Fap1 to saliva-coated hydroxyapatite (Wu et al., 2007), suggesting a mechanism by which S. parasanguinis might resist competition by more aciduric microbes and survive in the environmental niche.
Antigen I/II Family Polypeptides
To ensure colonization and persistence, bacteria express surface proteins that are capable of recognizing multiple receptors in the oral cavity. Some of the best characterized adhesins are members of the antigen I/II (AgI/II) family of polypeptides (Brady et al., 2010). The Streptococcus mutans AgI/II family polypeptide, variously designated as AgI/II, P1, SpaP, PAc, and AgB, is a protective antigen in experimental dental caries (Taubman and Nash, 2006). These adhesins have been described in virtually all streptococci indigenous to the oral cavity and have been found in pathogenic streptococci. Several AgI/II proteins have been shown to interact with salivary pellicle, specifically targeting innate immunity scavenger receptor glycoprotein-340 (gp-340). This may represent a pattern recognition molecule that is exploited by micro-organisms for colonization of the human host. Streptococcal attachment to fluid-phase gp-340 typically results in bacterial aggregation and clearance from the oral cavity by swallowing. However, this protein is also secreted by epithelial cells and adsorbed onto the surfaces of teeth, where it can promote adherence. Three predominant glycosylation variants of gp-340 have been identified in saliva, designated gp-340 I-III (Eriksson et al., 2007). Interestingly, S. mutans AgI/II protein exhibits significantly higher levels of adhesion to gp-340-I compared with glycoforms II and III, and this correlates with increased caries susceptibility in individuals producing gp-340-I (Jonasson et al., 2007). Thus, the specificity of bacterial adhesin-receptor interactions can influence both oral colonization and disease susceptibility.
AgI/II proteins fold to form elongated fibrillar structures that extend away from the streptococcal cell surface, presenting a potential adhesion domain at the tip (Larson et al., 2010). It is suggested that gp-340-binding sites within the stalk region might be utilized for initial attachment over an approximately 50-nm distance, while the C-terminal region adjacent to the cell surface promotes secondary, closer-range interactions (Fig. 1). High-resolution crystallography of the C-region of S. gordonii SspB (Forsgren et al., 2009) showed this to form two distinct domains, each containing a covalent isopeptide bond between a lysine and an asparagine residue. Such intra-molecular cross-links have been reported in Gram-positive bacterial pili (Kang et al., 2007) and are thought to stabilize elongated structures, potentially enabling streptococci to better resist detachment from oral surfaces. In support of this, it has been demonstrated that AgI/II proteins promote the specific adsorption of salivary proteins to the bacterial cell surface through the provision of enthalpically favorable adsorption sites (Xu et al., 2007a,b). These interactions are driven by short-range, pH-dependent electrostatic forces and are sufficiently strong to withstand variations in shear rate.
Outer Membrane Proteins – FomA
Not all surface proteins that promote oral microbial colonization are exclusive adhesins. One example is FomA, produced by Fusobacterium nucleatum (Fig. 2). The adhesins of Gram-negative bacteria can be divided into two classes: fimbrial adhesins and non-fimbrial adhesins, which include auto-transporters (see later) and outer membrane proteins (OMPs). FomA is a major OMP of F. nucleatum and belongs to a family of Gram-negative porin proteins. These proteins typically contain a β-barrel structure composed of trans-membrane, anti-parallel β-strands that fold back and forth across the lipid bilayer to form a series of surface-exposed loops that surround a central transmembrane channel (Puntervoll et al., 2002). The primary function of porin proteins is to allow for the non-specific diffusion of small solutes across the cell envelope. However, FomA also mediates strong attachment to the salivary constituent statherin (Sekine et al., 2004), a non-glycosylated phosphor-protein, specifically targeting epitope YQPVPE (Nakagaki et al., 2010). Studies are now under way in an attempt to resolve the crystal structure of FomA so that the reciprocal statherin-binding epitope in FomA can be identified.
Figure 2.
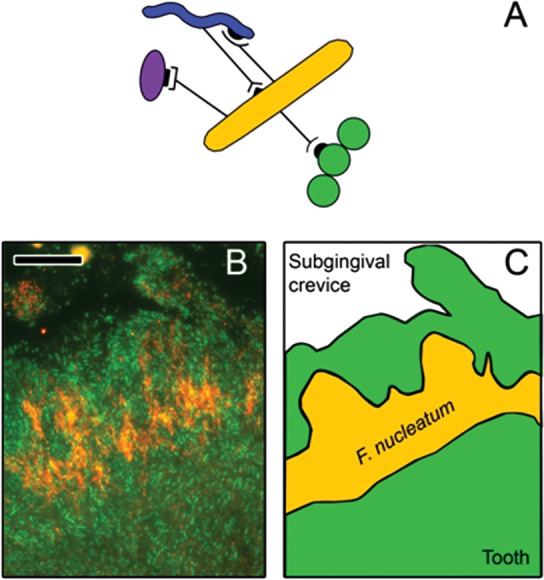
Role of Fusobacterium nucleatum as a bridge between early- and late-colonizers. (A) F. nucleatum (orange) co-aggregates with many different genera, including early-colonizers such as streptococci (green), and with late-colonizers such as A. actinomycetemcomitans (purple) or T. denticola (blue). (B) Analysis of subgingival biofilms by fluorescence in situ hybridization with a specific probe for F. nucleatum (orange) shows that it is localized in the middle layer of the biofilm, between early- and late-colonizers (eubacterial probe; green). (C) The localization of F. nucleatum within the biofilm is highlighted in a cartoon depiction of panel B. Scale bar on panel B is 10 µm. Image B is adapted from Zijnge et al. (2010) with permission.
Adhesion to Host Tissues
The adult oral cavity has a mean surface area of approximately 215 cm2, of which 30% and 50% constitutes keratinized and non-keratinized soft tissues, respectively. The oral epithelium therefore provides an extensive surface to which micro-organisms can attach, with bacteria principally recognizing receptors expressed directly on the epithelial cell surface or components of the underlying extracellular matrix (ECM).
Pili/Fibrillar Proteins
Many of the fibril adhesins involved in binding salivary pellicle, such as Srr and AgI/II proteins, are also implicated in bacterial attachment to host tissues (Table 1). Another major family of bacterial fibrillar proteins are pili. In Gram-positive bacteria, these appendages are formed from the sortase C-mediated polymerization of major (backbone) and minor (ancillary) protein subunits to generate a stalk that extends up to 3 µm from the cell surface, often with an adhesive tip. Pilus structures on the surface of Streptococcus sanguinis are composed of polymers of 3 distinct subunits (PilA, B, C) that promote attachment to fibronectin and epithelial cells (Okahashi et al., 2010). Two pilus loci have also been identified in Actinomyces naeslundii, type 1 (FimP/Q) and type 2 (FimA/B) (Mishra et al., 2007). Type 1 fimbriae promote adhesion of A. naeslundii T14V (renamed A. oris) to proline-rich proteins within salivary pellicle, while FimA of type 2 fimbriae targets Gal-GalNAc-containing structures on the surfaces of host cells and other members of the oral microbiota (Mishra et al., 2010) (Fig. 1).
Table 1.
Functional Properties of Bacterial Protein Adhesins Detailed in This Review
| Protein Group | Protein(s) | Species | Function(s) and/or Substrata |
|---|---|---|---|
| Serine-rich repeat (Srr) family | Fap1 | S. parasanguinis | Salivary pellicle |
| Hsa/GspB | S. gordonii | Salivary pellicle (gp-340); fibronectin; host cells | |
| Antigen I/II family | SpaP | S. mutans | Salivary pellicle; host cells (α5β1 integrins); fibronectin; type I collagen; laminin |
| SspA/SspB | S. gordonii | Salivary pellicle (gp-340); host cells (α5β1 integrins); fibronectin; type I collagen; co-aggregation (Actinomyces spp., P. gingivalis, C. albicans) | |
| Pili/Fimbriae | FadA | F. nucleatum | Epithelial/endothelial cells |
| FimA (major) fimbriae; Mfa1 (minor) fimbriae | P. gingivalis | Host cells (αvβ3/α5β1 integrins); fibronectin; type I collagen; co-aggregation (S. gordonii) | |
| Flp1 | A. actinomycetemcomitans | Epithelial cells; salivary pellicle | |
| FimP/Q (type I)/ FimA/B (type 2) | A. oris | Salivary pellicle; host cells; (Gal-GalNAc receptors); co-aggregation (Streptococcus spp.) | |
| PilA, B, C | S. sanguinis | Fibronectin; epithelial cells | |
| Outer membrane proteins | FomA | F. nucleatum | Porin protein; salivary pellicle (statherin); co-aggregation (P. gingivalis) |
| HBP35 | P. gingivalis | Co-aggregation (A. naeslundii; S. gordonii; S. mutans) | |
| Msp | T. denticola | Epithelial cells; fibronectin, fibrinogen, laminin, type I collagen; co-aggregation (F. nucleatum; P. gingivalis) | |
| Auto-transporters | Aae/ApiA/EmaA | A. actinomycetemcomitans | Epithelial cells; type I collagen |
| RadD | F. nucleatum | Co-aggregation (Streptococcus spp.) |
Fibrillar proteins also serve as important adhesins for Gram-negative bacteria. The periodontal pathogen Porphyromonas gingivalis expresses at least two types of fimbriae: longer major fimbriae (from 0.3 to 1.6 µm), and shorter (from 80 to 120 nm) minor fimbriae (Amano, 2010). The predominant protein components of these structures are FimA and Mfa1, respectively. Major fimbriae are the principal mediators of initial P. gingivalis attachment to gingival epithelial cells (GECs) through recognition of αvβ3 and α5β1 integrin receptors on the epithelial cell surface (Yilmaz et al., 2002) (Fig. 1). At least 6 allelic forms of FimA have been discovered to date that bind integrin receptors with differing affinities, and this correlates with P. gingivalis strain pathogenicity (Amano et al., 2000). Ancillary subunits FimCDE of major fimbriae have also been shown to form a functional complex that mediates binding to ECM proteins fibronectin and type I collagen (Nishiyama et al., 2007; Pierce et al., 2009). The periodontal pathogen Aggregatibacter actinomycetemcomitans produces long fibrils of bundled pili resembling type IV pili (Kachlany et al., 2001). These appear to mediate adherence to a wide range of surfaces (Fig. 1), including salivary pellicle, suggesting that A. actinomycetemcomitans might in some aspects be considered an early colonizer (Fine et al., 2010).
The FadA fimbrial protein of F. nucleatum has received attention recently because of its novel structure and contribution to systemic complications associated with this oral bacterium. FadA exists in two forms: non-secreted pre-FadA (129-amino-acid residues) and secreted, mature FadA (mFadA, 111-amino-acid residues). The latter has recently been crystallized (Nithianantham et al., 2009), and an assembly model has been suggested in which mFadA subunits link in a head-to-tail pattern via a novel ‘leucine chain’ structural motif to form elongated filaments. These filaments are stabilized by intermolecular hydrophobic interactions between leucine residues, and may then intertwine or bundle to form thicker structures that support adhesion. FadA has been shown to promote attachment and invasion of F. nucleatum to epithelial and to endothelial cells (Han et al., 2005; Ikegami et al., 2009) (Fig. 1), but these functions are dependent upon the presence of both the pre-FadA and mFadA forms (M Xu et al., 2007). The role of pre-FadA in the overall structure of FadA filaments is not yet fully understood. The ability of FadA to promote endothelial cell interactions is one of the mechanisms by which F. nucleatum is posited to cause adverse pregnancy outcomes, enabling placental colonization to occur (Ikegami et al., 2009).
Outer Membrane Proteins – Msp
Oral spirochetes such as Treponema denticola are strongly associated with the progression of periodontal disease. One of the predominant adhesins of T. denticola is the major surface protein (Msp). Its precise localization within the cell is still under debate, but the N-terminal region appears to be responsible for many of the adhesive properties. These include binding ECM proteins fibronectin, fibrinogen, laminin, and type I collagen (Edwards et al., 2005), and epithelial or gingival fibroblast cells (Mathers et al., 1996; Fenno et al., 1998).
Auto-transporter Adhesins – EmaA
The other major class of non-fimbrial surface proteins found in Gram-negative bacteria is the auto-transporters (Henderson et al., 2004). They show rather diverse functions, such as the ability to condense host cell actin and to modulate apoptosis, but many remain uncharacterized. These proteins contain all the information required for their self-translocation to the bacterial cell surface and are divided into 3 domains: an N-terminal signal sequence, a central passenger domain, and a C-terminal translocation unit. The signal sequence directs passage of the protein into the periplasm via the Sec system. The translocation unit then inserts into the outer membrane to form a β-barrel pore, through which the passenger domain passes. This leads to presentation of the passenger domain at the cell surface, with the translocation unit serving as the membrane anchor.
The periodontopathogen A. actinomycetemcomitans expresses at least 3 auto-transporter adhesins, designated Aae, ApiA, and EmaA (Table 1). The Aae and ApiA proteins promote attachment of A. actinomycetemcomitans to human buccal cells and GECs (Asakawa et al., 2003; Rose et al., 2003; Fine et al., 2005), while EmaA binds type I collagen, a mechanism also associated with the initiation of infective endocarditis (Tang et al., 2008) (Fig. 1). EmaA is structurally distinct from the other two auto-transporters, with coiled-coil, glycosylated monomers trimerizing to form antenna-like protrusions on the bacterial cell surface. These comprise a stalk of approximately 150 nm that terminates in an ellipsoidal cap, within which the collagen-binding domain is located (Ruiz et al., 2006; Yu et al., 2008; Tang and Mintz, 2010). Specific amino acid residues within the stalk have been shown to introduce bends within the structure, conferring flexibility to ensure optimal adhesion to collagen (Yu et al., 2009).
Co-Aggregation and Co-adhesion
The initial attachment of primary colonizers to oral surfaces presents new receptors for the subsequent adhesion of other bacteria. The binding of bacterial cells to pre-adherent cells on a surface is thought to be important for the recruitment of secondary colonizers to the oral biofilm. The adhesion of different bacteria to one another in suspension is termed ‘co-aggregation’ (Cisar et al., 1979), and is readily observable in vitro. Over 1000 pair-wise co-aggregation interactions have been demonstrated between and among different strains of oral bacteria (Kolenbrander et al., 2006). In many cases, co-aggregation involves the recognition of carbohydrate structures on one organism by lectin-like protein adhesins on the compatible partner. The identification of the key protein adhesins is still very much a work in progress.
Antigen I/II Family Polypeptides
The streptococcal AgI/II polypeptides have diverse roles in adhesion to host substrates (see above), but are also important co-aggregation adhesins. The SspA and SspB polypeptides, produced by S. gordonii, interact directly with P. gingivalis. The SspB region containing the BAR (SspB Adhesion Region) domain is recognized by the short fimbriae of P. gingivalis. In the crystal structure of the C-terminal region of SspB, the BAR domain protrudes like a handle (Forsgren et al., 2010). Binding to the BAR domain may play an important role in the recruitment of P. gingivalis to pre-attached S. gordonii in dental plaque.
The S. gordonii AgI/II polypeptides also function in interactions with Actinomyces spp. and with Candida albicans. The SspB protein mediates co-aggregation with A. oris T14V (Jakubovics et al., 2005). By contrast, SspA interacts with Actinomyces sp. PK606 and is not involved in co-aggregation with A. oris T14V. The Actinomyces receptors for AgI/II are currently unknown. In contrast, interaction of S. gordonii SspB with the surfaces of C. albicans hyphal cells involves binding of SspB to glycoprotein receptor Als3p (Silverman et al., 2010). Als3p is a hyphal-specific cell wall protein and mediates a wide range of adherence functions in C. albicans (Hoyer et al., 2008). Several other glycosylphosphatidylinositol (GPI)-anchored proteins in C. albicans, including Hwp1, Eap1, and Rbt1, have been implicated in binding streptococci (Nobbs et al., 2010), thus enabling C. albicans to readily colonize surfaces already coated with streptococci.
Pili/Fibrillar Proteins
An AgI/II-independent interaction between A. oris T14V and Streptococcus oralis or S. sanguinis involves the recognition of streptococcal receptor polysaccharides (RPS) by A. oris type 2 fimbriae. In this interaction, the fimbrial shaft protein FimA acts as a lectin-like adhesin (Mishra et al., 2010). Antibodies against type 2 fimbriae and streptococcal receptor polysaccharides were co-localized in biofilms formed in the mouths of volunteers, suggesting that this association occurs in vivo (Palmer et al., 2003).
The short fimbriae (Mfa1) of P. gingivalis, which are involved in co-adhesion with S. gordonii, are regulated in mixed-species communities. The extracellular arginine deiminase of Streptococcus cristatus or Streptococcus intermedius signals the down-regulation of mfa and prohibits biofilm formation by P. gingivalis (Christopher et al., 2010; Wu and Xie, 2010). The signaling function of arginine deiminase resides within the C-terminal region and is not dependent upon enzyme activity. The arginine deiminase of S. gordonii is expressed at much lower levels than the equivalent protein of S. cristatus and does not induce down-regulation of mfa. It may be that P. gingivalis senses specific streptococcal arginine deiminase proteins and uses the information to select the most suitable streptococcal partner for co-adhesion and biofilm formation.
Outer Membrane Proteins – RadD, FomA, HBP35, and Msp
F. nucleatum has an unusual promiscuity for co-aggregation interactions with both early- and late-colonizers. For this reason, F. nucleatum is sometimes thought of as a bridging bacterium in plaque, adhering to primary colonizers but also facilitating the integration of later colonizers, including pathogens such as P. gingivalis (Kolenbrander et al., 2006) (Fig. 2). Fluorescence in situ hybridization has demonstrated that F. nucleatum colonizes the middle layer of subgingival dental plaque, supporting a bridging role for this organism (Zijnge et al., 2010). Adhesion of F. nucleatum to oral streptococci has been attributed to an arginine-sensitive protein adhesin on the surface of F. nucleatum. A spontaneous F. nucleatum ATCC 10953 mutant was isolated that was defective for co-aggregation with S. cristatus, S. gordonii, and S. sanguinis, but not with P. gingivalis (Edwards et al., 2007). The mutant lacked an approximately 360-kDa protein that was identified as a member of the auto-transporter family, RadD, confirmed as an arginine-inhibitable adhesin responsible for co-aggregation with streptococci (Kaplan et al., 2009). Dual-species biofilm formation with S. sanguinis was impaired in a radD mutant. In addition, RadD was shown to mediate arginine-sensitive interactions with secretory IgA and with human lymphocytes (Edwards et al., 2007; Kaplan et al., 2009).
Co-aggregation between F. nucleatum and P. gingivalis is sensitive to lactose but not to arginine, and has been attributed to a 42-kDa lectin-like adhesin FomA on the surface of F. nucleatum (Kinder and Holt, 1993). Co-inoculation of biofilm models with P. gingivalis and F. nucleatum resulted in increased biofilm formation compared with that achieved with either species on its own (Periasamy and Kolenbrander, 2009; Liu et al., 2010). Antibodies against FomA abrogated this mutualistic interaction (Liu et al., 2010). Co-aggregation between F. nucleatum and P. gingivalis may be important in periodontal disease. In a mouse model of periodontitis, gingival inflammation was enhanced in dual-species cultures compared with F. nucleatum or P. gingivalis monocultures. Antibodies against FomA, or vaccination targeting FomA, overcame this increased inflammation, indicating that FomA-mediated co-aggregation was important in vivo.
Non-fimbrial proteins of P. gingivalis, including Arg-gingipain, Lys-gingipain, and hemagglutinin (Hag), function in co-aggregation interactions. P. gingivalis cells also release vesicles that are capable of agglutinating a wide variety of micro-organisms. There is evidence that vesicles are important in biofilm formation (Kulp and Kuehn, 2010). Antibodies against the outer membrane heme-binding protein HBP35, which is found in vesicles, inhibit co-aggregation between P. gingivalis vesicles and A. naeslundii (Abiko et al., 1997). Mutants in hbp35 produce vesicles with reduced ability to agglutinate S. gordonii, S. mutans, or A. naeslundii (Hiratsuka et al., 2008). However, other potential co-aggregation-mediating adhesins, including hemagglutinins and fimbriae, were reduced in the hbp35 mutant, indicating that the role of HBP35 in co-aggregation may be indirect.
P. gingivalis also co-aggregates with T. denticola, and these two organisms form a mutualistic partnership for biofilm growth (Yamada et al., 2005). Co-aggregation depends upon T. denticola Msp (Rosen et al., 2008), and purified Msp binds P. gingivalis. Likewise, Msp mediates interactions between T. denticola and F. nucleatum (Fig. 1). However, differences in the mechanisms of co-aggregation were noted: Binding of T. denticola to F. nucleatum was inhibited by galactose, whereas co-aggregation with P. gingivalis was not (Rosen et al., 2008). De-glycosylation of Msp with the enzyme N-glycosidase F inhibited co-aggregation of T. denticola with F. nucleatum but not with P. gingivalis. Analysis of these data suggests that carbohydrate moieties on Msp interact with F. nucleatum, while the polypeptide backbone mediates co-aggregation with P. gingivalis.
Summary
New Approaches to the Identification of Adhesins
The identification and characterization of key adhesins that stabilize oral biofilm communities are important goals, since adhesins are potentially excellent targets for measures to control the microbial population in the mouth (see below). Large-scale genomic sequencing endeavors such as the human microbiome project (Nelson et al., 2010) are providing a wealth of data that can be mined to search for new adhesin genes (Table 2). Attempts to make sense of the vast amounts of data will benefit greatly from concerted efforts to organize the genome sequences and to make them accessible to scientists who do not have specialized bioinformatics training. Several databases for oral bacterial genome analysis are already in place—for example, the Human Oral Microbiome Database (Dewhirst et al., 2010). Genome sequences can provide useful information for the identification of potential adhesins, but the confirmation of adhesin roles ultimately depends upon biological experimentation (Table 2).
Table 2.
Some Approaches to the Identification of Microbial Adhesins
| Methodology | Example | Reference |
|---|---|---|
| Genome mining | Specialized software to identify potential adhesions, e.g., SPAAN | Sachdeva et al., 2005 |
| Functional genomics | High-throughput assays, e.g., receptors within salivary proteome recognized by Helicobacter pylori and vice versa | Walz et al., 2009 |
| Gene knockout | Mutation of srtA encoding sortase releases cell-wall-anchored adhesins in Gram-positive bacteria | Davies et al., 2009 |
| Protein expression | Recombinant proteins from H. pylori fixed to nickel-adsorbed beads and screened for adherence to epithelial cells | Rubinsztein- Dunlop et al., 2005 |
| Receptor tagging | Biotin-tagged receptor with linked photoreactive group that, when activated, biotinylates the cognate adhesin | Ilver et al., 1998 |
| Genome display library | Infection-associated antigens identified by screening pneumococcal genome display libraries with sera from infected individuals | Papasergi et al., 2010 |
Applications of Adhesin Identification
Dental caries and periodontal disease are chronic infectious diseases that are becoming increasingly prevalent throughout the global population at huge economic cost. Current chemoprophylactic therapies can be rendered ineffective by microbial antibiotic resistance, host toxicity, or problems with maintaining a functional dose within the oral cavity. The development of vaccines or other control regimens is therefore an attractive alternative strategy.
The principle of oral disease vaccines is to induce antigen-specific antibodies against the causative agent or agents that inhibit the infection process by one of 3 main mechanisms: antibody-mediated clearance, blocking of essential receptors required for microbial attachment, and inhibition of enzymes required for metabolism or colonization. For mutans streptococci, 3 proteins are promising vaccine candidates: AgI/II family polypeptide SpaP, GTF, and GbpB (Taubman and Nash, 2006). However, dental caries is not associated just with S. mutans; therefore, vaccines targeting only these organisms may not be 100% effective.
No clinical trials have been performed to date against periodontitis. Nonetheless, animal models have shown that vaccines against fimbriae, gingipains, or OMPs can successfully interfere with P. gingivalis colonization and reduce periodontal bone resorption (Persson, 2005; Beevi et al., 2009). Molecular information about adherence mechanisms can also be exploited in the design of replacement therapies, attachment-blocking peptides, and biomimetics or signaling inhibitors to disrupt microbial community development (Nobbs et al., 2009; Chen and Wang, 2010). Some of these approaches already show much promise. It is clear that members of the oral microbiota utilize an astounding repertoire of mechanisms, involving a multitude of surface components, to ensure their attachment, persistence, and pathogenicity within the oral cavity. Paradoxically, however, it is these very strengths that we hope may reveal their weaknesses, enabling the development of novel strategies to control colonization and infection.
Acknowledgments
We acknowledge the tremendous contributions that Paul Kolenbrander has made, not only to this field of research, but also to the development of our careers: He has been an inspiration to each of us.
Footnotes
Work in the authors’ laboratories is funded by the NIH (NIDCR) R01 DE016690 and the Wellcome Trust (#081855 and #084979), and by a SfAM New Lecturer Grant.
References
- Abiko Y, Ogura N, Matsuda U, Yanagi K, Takiguchi H. (1997). A human monoclonal antibody which inhibits the coaggregation activity of Porphyromonas gingivalis. Infect Immun 65:3966-3969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano A. (2010). Bacterial adhesins to host components in periodontitis. Periodontol 2000 52:12-37 [DOI] [PubMed] [Google Scholar]
- Amano A, Kuboniwa M, Nakagawa I, Akiyama S, Morisaki I, Hamada S. (2000). Prevalence of specific genotypes of Porphyromonas gingivalis fimA and periodontal health status. J Dent Res 79:1664-1668 [DOI] [PubMed] [Google Scholar]
- Asakawa R, Komatsuzawa H, Kawai T, Yamada S, Goncalves RB, Izumi S, et al. (2003). Outer membrane protein 100, a versatile virulence factor of Actinobacillus actinomycetemcomitans. Mol Microbiol 50:1125-1139 [DOI] [PubMed] [Google Scholar]
- Beevi L, Hedge S, Kashyap R, Kumar A. (2009). Vaccines and periodontal diseases—an insight. Dent Update 36:635-638 [DOI] [PubMed] [Google Scholar]
- Brady LJ, Maddocks SE, Larson MR, Forsgren N, Persson K, Deivanayagam CC, et al. (2010). The changing faces of Streptococcus antigen I/II polypeptide family adhesins. Mol Microbiol 77:276-286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Wang D. (2010). Novel technologies for the prevention and treatment of dental caries: a patent survey. Expert Opin Ther Pat 20:681-694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopher AB, Arndt A, Cugini C, Davey ME. (2010). A streptococcal effector protein that inhibits Porphyromonas gingivalis biofilm development. Microbiology 156:3469-3477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisar JO, Kolenbrander PE, McIntire FC. (1979). Specificity of coaggregation reactions between human oral streptococci and strains of Actinomyces viscosus or Actinomyces naeslundii. Infect Immun 24:742-752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JR, Svensater G, Herzberg MC. (2009). Identification of novel LPXTG-linked surface proteins from Streptococcus gordonii. Microbiology 155:1977-1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, et al. (2010). The human oral microbiome. J Bacteriol 192:5002-5017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AM, Jenkinson HF, Woodward MJ, Dymock D. (2005). Binding properties and adhesion-mediating regions of the major sheath protein of Treponema denticola ATCC 35405. Infect Immun 73:2891-2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AM, Grossman TJ, Rudney JD. (2007). Association of a high-molecular weight arginine-binding protein of Fusobacterium nucleatum ATCC 10953 with adhesion to secretory immunoglobulin A and coaggregation with Streptococcus cristatus. Oral Microbiol Immunol 22:217-224 [DOI] [PubMed] [Google Scholar]
- Eriksson C, Frängsmyr L, Danielsson Niemi L, Loimaranta V, Holmskov U, Bergman T, et al. (2007). Variant size- and glycoforms of the scavenger receptor cysteine-rich protein gp-340 with differential bacterial aggregation. Glycoconj J 24:131-142 [DOI] [PubMed] [Google Scholar]
- Fenno JC, Hannam PM, Leung WK, Tamura M, Uitto VJ, McBride BC. (1998). Cytopathic effects of the major surface protein and the chymotrypsin-like protease of Treponema denticola. Infect Immun 66:1869-1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine DH, Velliyagounder K, Furgang D, Kaplan JB. (2005). The Actinobacillus actinomycetemcomitans autotransporter adhesin Aae exhibits specificity for buccal epithelial cells from humans and old world primates. Infect Immun 73:1947-1953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine DH, Markowitz K, Furgang D, Velliyagounder K. (2010). Aggregatibacter actinomycetemcomitans as an early colonizer of oral tissues: epithelium as a reservoir? J Clin Microbiol 48:4464-4473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsgren N, Lamont RJ, Persson K. (2009). Crystal structure of the variable domain of the Streptococcus gordonii surface protein SspB. Protein Sci 18:1896-1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsgren N, Lamont RJ, Persson K. (2010). Two intramolecular isopeptide bonds are identified in the crystal structure of the Streptococcus gordonii SspB C-terminal domain. J Mol Biol 397:740-751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han YW, Ikegami A, Rajanna C, Kawsar HI, Zhou Y, Li M, et al. (2005). Identification and characterization of a novel adhesin unique to oral fusobacteria. J Bacteriol 187:5330-5340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Navarro-Garcia F, Desvaux M, Fernandez RC, Ala’Aldeen D. (2004). Type V protein secretion pathway: the autotransporter story. Microbiol Mol Biol Rev 68:692-744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsuka K, Hayakawa M, Kiyama-Kishikawa M, Sasaki Y, Hirai T, Abiko Y. (2008). Role of the hemin-binding protein 35 (HBP35) of Porphyromonas gingivalis in coaggregation. Microb Pathog 44:320-328 [DOI] [PubMed] [Google Scholar]
- Hoyer LL, Green CB, Oh SH, Zhao X. (2008). Discovering the secrets of the Candida albicans agglutinin-like sequence (ALS) gene family—a sticky pursuit. Med Mycol 46:1-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami A, Chung P, Han YW. (2009). Complementation of the fadA mutation in Fusobacterium nucleatum demonstrates that the surface-exposed adhesin promotes cellular invasion and placental colonization. Infect Immun 77:3075-3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilver D, Arnqvist A, Ogren J, Frick IM, Kersulyte D, Incecik ET, et al. (1998). Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science 279:373-377 [DOI] [PubMed] [Google Scholar]
- Jakubovics NS, Stromberg N, van Dolleweerd CJ, Kelly CG, Jenkinson HF. (2005). Differential binding specificities of oral streptococcal antigen I/II family adhesins for human or bacterial ligands. Mol Microbiol 55:1591-1605 [DOI] [PubMed] [Google Scholar]
- Jonasson A, Eriksson C, Jenkinson HF, Kallestal C, Johansson I, Stromberg N. (2007). Innate immunity glycoprotein gp-340 variants may modulate human susceptibility to dental caries. BMC Infect Dis 7:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachlany SC, Planet PJ, Desalle R, Fine DH, Figurski DH, Kaplan JB. (2001). flp-1, the first representative of a new pilin gene subfamily, is required for non-specific adherence of Actinobacillus actinomycetemcomitans. Mol Microbiol 40:542-554 [DOI] [PubMed] [Google Scholar]
- Kang HJ, Coulibaly F, Clow F, Proft T, Baker EN. (2007). Stabilizing isopeptide bonds revealed in Gram-positive bacterial pilus structure. Science 318:1625-1628 [DOI] [PubMed] [Google Scholar]
- Kaplan CW, Lux R, Haake SK, Shi W. (2009). The Fusobacterium nucleatum outer membrane protein RadD is an arginine-inhibitable adhesin required for inter-species adherence and the structured architecture of multispecies biofilm. Mol Microbiol 71:35-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinder SA, Holt SC. (1993). Localization of the Fusobacterium nucleatum T18 adhesin activity mediating coaggregation with Porphyromonas gingivalis T22. J Bacteriol 175:840-850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander PE, Palmer RJ, Jr, Rickard AH, Jakubovics NS, Chalmers NI, Diaz PI. (2006). Bacterial interactions and successions during plaque development. Periodontol 2000 42:47-79 [DOI] [PubMed] [Google Scholar]
- Koo H, Xiao J, Klein MI, Jeon JG. (2010). Exopolysaccharides produced by Streptococcus mutans glucosyltransferases modulate the establishment of microcolonies within multispecies biofilms. J Bacteriol 192:3024-3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulp A, Kuehn MJ. (2010). Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol 64:163-184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson MR, Rajashankar KR, Patel MH, Robinette RA, Crowley PJ, Michalek S, et al. (2010). Elongated fibrillar structure of a streptococcal adhesin assembled by the high-affinity association of alpha- and PPII-helices. Proc Natl Acad Sci USA 107:5983-5988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PF, Shi W, Zhu W, Smith JW, Hsieh SL, Gallo RL, et al. (2010). Vaccination targeting surface FomA of Fusobacterium nucleatum against bacterial co-aggregation: implication for treatment of periodontal infection and halitosis. Vaccine 28:3496-3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathers DA, Leung WK, Fenno JC, Hong Y, McBride BC. (1996). The major surface protein complex of Treponema denticola depolarizes and induces ion channels in HeLa cell membranes. Infect Immun 64:2904-2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra A, Das A, Cisar JO, Ton-That H. (2007). Sortase-catalyzed assembly of distinct heteromeric fimbriae in Actinomyces naeslundii. J Bacteriol 189:3156-3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra A, Wu C, Yang J, Cisar JO, Das A, Ton-That H. (2010). The Actinomyces oris type 2 fimbrial shaft FimA mediates co-aggregation with oral streptococci, adherence to red blood cells and biofilm development. Mol Microbiol 77:841-854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagaki H, Sekine S, Terao Y, Toe M, Tanaka M, Ito HO, et al. (2010). Fusobacterium nucleatum envelope protein FomA is immunogenic and binds to the salivary statherin-derived peptide. Infect Immun 78:1185-1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson KE, Weinstock GM, Highlander SK, Worley KC, Creasy HH, Wortman JR, et al. (2010). A catalog of reference genomes from the human microbiome. Science 328:994-999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama S, Murakami Y, Nagata H, Shizukuishi S, Kawagishi I, Yoshimura F. (2007). Involvement of minor components associated with the FimA fimbriae of Porphyromonas gingivalis in adhesive functions. Microbiology 153:1916-1925 [DOI] [PubMed] [Google Scholar]
- Nithianantham S, Xu M, Yamada M, Ikegami A, Shoham M, Han YW. (2009). Crystal structure of FadA adhesin from Fusobacterium nucleatum reveals a novel oligomerization motif, the leucine chain. J Biol Chem 284:3865-3872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobbs AH, Lamont RJ, Jenkinson HF. (2009). Streptococcus adherence and colonization. Microbiol Mol Biol Rev 73:407-450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobbs AH, Vickerman MM, Jenkinson HF. (2010). Heterologous expression of Candida albicans cell wall-associated adhesins in Saccharomyces cerevisiae reveals differential specificities in adherence and biofilm formation and in binding oral Streptococcus gordonii. Eukaryot Cell 9:1622-1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okahashi N, Nakata M, Sakurai A, Terao Y, Hoshino T, Yamaguchi M, et al. (2010). Pili of oral Streptococcus sanguinis bind to fibronectin and contribute to cell adhesion. Biochem Biophys Res Commun 391:1192-1196 [DOI] [PubMed] [Google Scholar]
- Palmer RJ, Jr, Gordon SM, Cisar JO, Kolenbrander PE. (2003). Coaggregation-mediated interactions of streptococci and actinomyces detected in initial human dental plaque. J Bacteriol 185:3400-3409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papasergi S, Garibaldi M, Tuscano G, Signorino G, Ricci S, Peppoloni S, et al. (2010). Plasminogen- and fibronectin-binding protein B is involved in the adherence of Streptococcus pneumoniae to human epithelial cells. J Biol Chem 285:7517-7524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Periasamy S, Kolenbrander PE. (2009). Mutualistic biofilm communities develop with Porphyromonas gingivalis and initial, early, and late colonizers of enamel. J Bacteriol 191:6804-6811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson GR. (2005). Immune responses and vaccination against periodontal infections. J Clin Periodontol 32(Suppl 6):39-53 [DOI] [PubMed] [Google Scholar]
- Pierce DL, Nishiyama S, Liang S, Wang M, Triantafilou M, Triantafilou K, et al. (2009). Host adhesive activities and virulence of novel fimbrial proteins of Porphyromonas gingivalis. Infect Immun 77:3294-3301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puntervoll P, Ruud M, Bruseth LJ, Kleivdal H, Hogh BT, Benz R, et al. (2002). Structural characterization of the fusobacterial non-specific porin FomA suggests a 14-stranded topology, unlike the classical porins. Microbiology 148:3395-3403 [DOI] [PubMed] [Google Scholar]
- Ramboarina S, Garnett JA, Zhou M, Li Y, Peng Z, Taylor JD, et al. (2010). Structural insights into serine-rich fimbriae from Gram-positive bacteria. J Biol Chem 285:32446-32457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JE, Meyer DH, Fives-Taylor PM. (2003). Aae, an autotransporter involved in adhesion of Actinobacillus actinomycetemcomitans to epithelial cells. Infect Immun 71:2384-2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen G, Genzler T, Sela MN. (2008). Coaggregation of Treponema denticola with Porphyromonas gingivalis and Fusobacterium nucleatum is mediated by the major outer sheath protein of Treponema denticola. FEMS Microbiol Lett 289:59-66 [DOI] [PubMed] [Google Scholar]
- Rubinsztein-Dunlop S, Guy B, Lissolo L, Fischer H. (2005). Identification of two new Helicobacter pylori surface proteins involved in attachment to epithelial cell lines. J Med Microbiol 54:427-434 [DOI] [PubMed] [Google Scholar]
- Ruiz T, Lenox C, Radermacher M, Mintz KP. (2006). Novel surface structures are associated with the adhesion of Actinobacillus actinomycetemcomitans to collagen. Infect Immun 74:6163-6170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdeva G, Kumar K, Jain P, Ramachandran S. (2005). SPAAN: a software program for prediction of adhesins and adhesin-like proteins using neural networks. Bioinformatics 21:483-491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine S, Kataoka K, Tanaka M, Nagata H, Kawakami T, Akaji K, et al. (2004). Active domains of salivary statherin on apatitic surfaces for binding to Fusobacterium nucleatum cells. Microbiology 150:2373-2379 [DOI] [PubMed] [Google Scholar]
- Silverman RJ, Nobbs AH, Vickerman MM, Barbour ME, Jenkinson HF. (2010). Interaction of Candida albicans cell wall Als3 protein with Streptococcus gordonii SspB adhesin promotes development of mixed-species communities. Infect Immun 78:4644-4652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamatsu D, Bensing BA, Prakobphol A, Fisher SJ, Sullam PM. (2006). Binding of the streptococcal surface glycoproteins GspB and Hsa to human salivary proteins. Infect Immun 74:1933-1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Mintz KP. (2010). Glycosylation of the collagen adhesin EmaA of Aggregatibacter actinomycetemcomitans is dependent upon the lipopolysaccharide biosynthetic pathway. J Bacteriol 192:1395-1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Kitten T, Munro CL, Wellman GC, Mintz KP. (2008). EmaA, a potential virulence determinant of Aggregatibacter actinomycetemcomitans in infective endocarditis. Infect Immun 76:2316-2324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubman MA, Nash DA. (2006). The scientific and public-health imperative for a vaccine against dental caries. Nat Rev Immunol 6:555-563 [DOI] [PubMed] [Google Scholar]
- Walz A, Odenbreit S, Stuhler K, Wattenberg A, Meyer HE, Mahdavi J, et al. (2009). Identification of glycoprotein receptors within the human salivary proteome for the lectin-like BabA and SabA adhesins of Helicobacter pylori by fluorescence-based 2-D bacterial overlay. Proteomics 9:1582-1592 [DOI] [PubMed] [Google Scholar]
- Wu H, Mintz KP, Ladha M, Fives-Taylor PM. (1998). Isolation and characterization of Fap1, a fimbriae-associated adhesin of Streptococcus parasanguis FW213. Mol Microbiol 28:487-500 [DOI] [PubMed] [Google Scholar]
- Wu H, Zeng M, Fives-Taylor P. (2007). The glycan moieties and the N-terminal polypeptide backbone of a fimbria-associated adhesin, Fap1, play distinct roles in the biofilm development of Streptococcus parasanguinis. Infect Immun 75:2181-2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Xie H. (2010). Role of arginine deiminase of Streptococcus cristatus in Porphyromonas gingivalis colonization. Antimicrob Agents Chemother 54:4694-4698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu CP, van de Belt-Gritter B, Busscher HJ, van der Mei HC, Norde W. (2007a). Calorimetric comparison of the interactions between salivary proteins and Streptococcus mutans with and without antigen I/II. Colloids Surf B Biointerfaces 54:193-199 [DOI] [PubMed] [Google Scholar]
- Xu CP, van de Belt-Gritter B, Dijkstra RJ, Norde W, van der Mei HC, Busscher HJ. (2007b). Interaction forces between salivary proteins and Streptococcus mutans with and without antigen I/II. Langmuir 23:9423-9428 [DOI] [PubMed] [Google Scholar]
- Xu M, Yamada M, Li M, Liu H, Chen SG, Han YW. (2007). FadA from Fusobacterium nucleatum utilizes both secreted and nonsecreted forms for functional oligomerization for attachment and invasion of host cells. J Biol Chem 282:25000-25009 [DOI] [PubMed] [Google Scholar]
- Yamada M, Ikegami A, Kuramitsu HK. (2005). Synergistic biofilm formation by Treponema denticola and Porphyromonas gingivalis. FEMS Microbiol Lett 250:271-277 [DOI] [PubMed] [Google Scholar]
- Yilmaz O, Watanabe K, Lamont RJ. (2002). Involvement of integrins in fimbriae-mediated binding and invasion by Porphyromonas gingivalis. Cell Microbiol 4:305-314 [DOI] [PubMed] [Google Scholar]
- Yu C, Ruiz T, Lenox C, Mintz KP. (2008). Functional mapping of an oligomeric autotransporter adhesin of Aggregatibacter actinomycetemcomitans. J Bacteriol 190:3098-3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Mintz KP, Ruiz T. (2009). Investigation of the three-dimensional architecture of the collagen adhesin EmaA of Aggregatibacter actinomycetemcomitans by electron tomography. J Bacteriol 191:6253-6261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zijnge V, van Leeuwen MB, Degener JE, Abbas F, Thurnheer T, Gmür R, et al. (2010). Oral biofilm architecture on natural teeth. PLoS One 5:e9321. [DOI] [PMC free article] [PubMed] [Google Scholar]


