Abstract
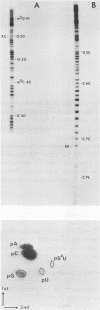
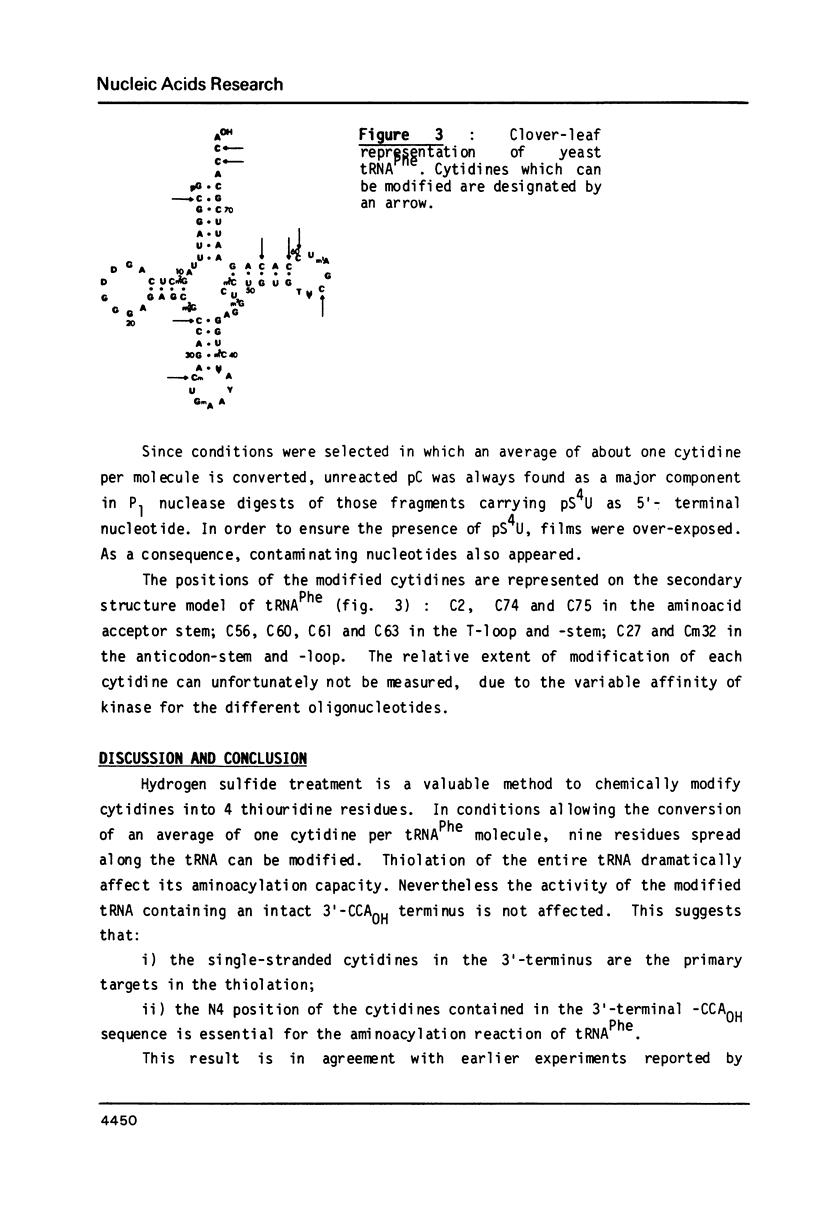
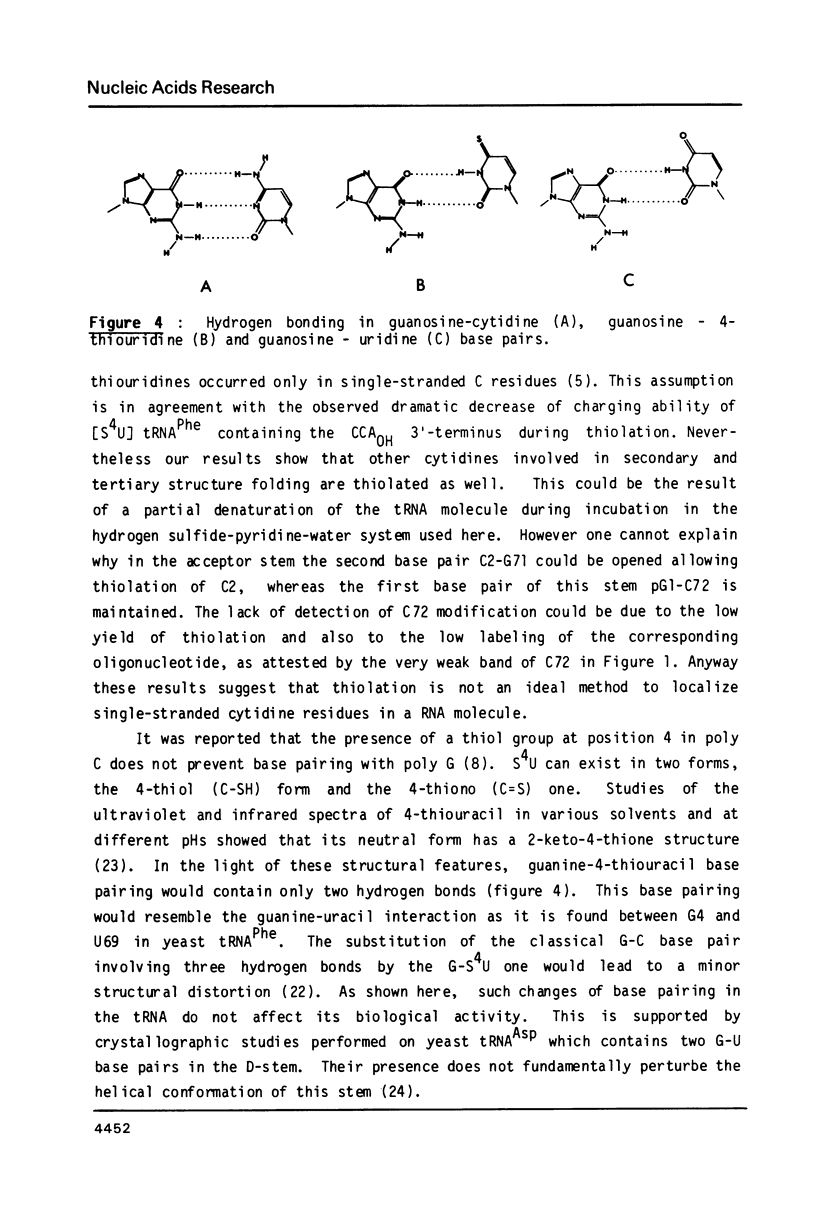
Treatment of yeast phenylalanine tRNA with pressurized hydrogen sulfide results in conversion of cytidine residues into 4-thiouridine residues. Under conditions leading to an average modification of one cytidine per tRNA molecule 9 positions are thiolated. The 4-thiouridine residues are distributed along the tRNA molecule. Four of the reactive cytidines are located in single-stranded regions: Cm32 , C60 , C74 and C75 . The five others are located in base pairs: C2, C27, C56 , C61 and C63 . Importance of replacement of an amino group by a thiol group on hydrogen bonding and on biological activity of the modified tRNA is discussed.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Dirheimer G., Ebel J. P. Fractionnement des tRNA de levure de bière par distribution en contre-courant. Bull Soc Chim Biol (Paris) 1967;49(12):1679–1687. [PubMed] [Google Scholar]
- Favre A., Fourrey J. L. Intramolecular cross-linking of single-stranded copolymers of 4-thiouridine and cytidine. Biochem Biophys Res Commun. 1974 May 20;58(2):507–515. doi: 10.1016/0006-291x(74)90394-5. [DOI] [PubMed] [Google Scholar]
- Hassur S. M., Whitlock H. W., Jr UV shadowing--a new and convenient method for the location of ultraviolet-absorbing species in polyacrylamide gels. Anal Biochem. 1974 May;59(1):162–164. doi: 10.1016/0003-2697(74)90020-7. [DOI] [PubMed] [Google Scholar]
- Holbrook S. R., Kim S. H. Correlation between chemical modification and surface accessibility in yeast phenylalanine transfer RNA. Biopolymers. 1983 Apr;22(4):1145–1166. doi: 10.1002/bip.360220410. [DOI] [PubMed] [Google Scholar]
- Kuchino Y., Watanabe S., Harada F., Nishimura S. Primary structure of AUA-specific isoleucine transfer ribonucleic acid from Escherichia coli. Biochemistry. 1980 May 13;19(10):2085–2089. doi: 10.1021/bi00551a013. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Iwano T., Tsuda S., Ueda T., Harada F., Kato N. Chemical modification of cytosine residues of tRNAVa1 with hydrogen sulfide (nucleosides and nucleotides. XL). Chem Pharm Bull (Tokyo) 1982 Nov;30(11):4126–4133. doi: 10.1248/cpb.30.4126. [DOI] [PubMed] [Google Scholar]
- Miura K., Shiga M., Ueda T. Nucleosides and nucleotides. VI. Preparation of diribonucleoside monophosphates containing 4-thiouridine. J Biochem. 1973 Jun;73(6):1279–1284. doi: 10.1093/oxfordjournals.jbchem.a130201. [DOI] [PubMed] [Google Scholar]
- Miura K., Tsuda S., Harada F., Ueda T. Chemical modification of cytosine residues of U6 snRNA with hydrogen sulfide (nucleosides and nucleotides. Part 49 [1]). Nucleic Acids Res. 1983 Sep 10;11(17):5893–5901. doi: 10.1093/nar/11.17.5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Tsuda S., Iwano T., Ueda T., Harada F., Kato N. Chemical modification of cytosine residues of mouse 5 S ribosomal RNA with hydrogen sulfide. (Nucleosides and nucleotides 43). Biochim Biophys Acta. 1983 Mar 10;739(2):181–189. doi: 10.1016/0167-4781(83)90028-3. [DOI] [PubMed] [Google Scholar]
- Moras D., Comarmond M. B., Fischer J., Weiss R., Thierry J. C., Ebel J. P., Giegé R. Crystal structure of yeast tRNAAsp. Nature. 1980 Dec 25;288(5792):669–674. doi: 10.1038/288669a0. [DOI] [PubMed] [Google Scholar]
- Quigley G. J., Rich A. Structural domains of transfer RNA molecules. Science. 1976 Nov 19;194(4267):796–806. doi: 10.1126/science.790568. [DOI] [PubMed] [Google Scholar]
- Renaud M., Ehrlich R., Bonnet J., Remy P. Lack of correlation between affinity of the tRNA for the aminoacyl-tRNA synthetase and aminoacylation capacity as studied with modified tRNAPhe. Eur J Biochem. 1979 Oct;100(1):157–164. doi: 10.1111/j.1432-1033.1979.tb02044.x. [DOI] [PubMed] [Google Scholar]
- Rether B., Bonnet J., Ebel J. P. Studies on tRNA nucleotidyltransferase from baker's yeast. 1. Purification of the enzyme. Protection against thermal inactivation and inhibition by several substrates. Eur J Biochem. 1974 Dec 16;50(1):281–288. doi: 10.1111/j.1432-1033.1974.tb03896.x. [DOI] [PubMed] [Google Scholar]
- Riehl N., Remy P., Ebel J. P., Ehresmann B. Crosslinking of N-acetyl-phenylalanyl [s4U]tRNAPhe to protein S10 in the ribosomal P site. Eur J Biochem. 1982 Nov 15;128(2-3):427–433. doi: 10.1111/j.1432-1033.1982.tb06982.x. [DOI] [PubMed] [Google Scholar]
- Schulman L. H., Goddard J. P. Loss of methionine acceptor activity resulting from a base change in the anticodon of Escherichia coli formylmethionine transfer ribonucleic acid. J Biol Chem. 1973 Feb 25;248(4):1341–1345. [PubMed] [Google Scholar]
- Silberklang M., Gillum A. M., RajBhandary U. L. The use of nuclease P1 in sequence analysis of end group labeled RNA. Nucleic Acids Res. 1977 Dec;4(12):4091–4108. doi: 10.1093/nar/4.12.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley J., Vassilenko S. A different approach to RNA sequencing. Nature. 1978 Jul 6;274(5666):87–89. doi: 10.1038/274087a0. [DOI] [PubMed] [Google Scholar]
- Uziel M., Khym J. X. Sequential degradation of nucleic acids. Degradation of Escherichia coli B phenylalanine transfer ribonucleic acid. Biochemistry. 1969 Aug;8(8):3254–3260. doi: 10.1021/bi00836a018. [DOI] [PubMed] [Google Scholar]
- Vlassov V. V., Giegé R., Ebel J. P. Tertiary structure of tRNAs in solution monitored by phosphodiester modification with ethylnitrosourea. Eur J Biochem. 1981 Sep;119(1):51–59. doi: 10.1111/j.1432-1033.1981.tb05575.x. [DOI] [PubMed] [Google Scholar]