Abstract
Bacterial infection of plants often begins with colonization of the plant surface, followed by entry into the plant through wounds and natural openings (such as stomata), multiplication in the intercellular space (apoplast) of the infected tissues, and dissemination of bacteria to other plants. Historically, most studies assess bacterial infection based on final outcomes of disease and/or pathogen growth using whole infected tissues; few studies have genetically distinguished the contribution of different host cell types in response to an infection. The phytotoxin coronatine (COR) is produced by several pathovars of Pseudomonas syringae. COR-deficient mutants of P. s. tomato (Pst) DC3000 are severely compromised in virulence, especially when inoculated onto the plant surface. We report here a genetic screen to identify Arabidopsis mutants that could rescue the virulence of COR-deficient mutant bacteria. Among the susceptible to coronatine-deficient Pst DC3000 (scord) mutants were two that were defective in stomatal closure response, two that were defective in apoplast defense, and four that were defective in both stomatal and apoplast defense. Isolation of these three classes of mutants suggests that stomatal and apoplastic defenses are integrated in plants, but are genetically separable, and that COR is important for Pst DC3000 to overcome both stomatal guard cell- and apoplastic mesophyll cell-based defenses. Of the six mutants defective in bacterium-triggered stomatal closure, three are defective in salicylic acid (SA)-induced stomatal closure, but exhibit normal stomatal closure in response to abscisic acid (ABA), and scord7 is compromised in both SA- and ABA-induced stomatal closure. We have cloned SCORD3, which is required for salicylic acid (SA) biosynthesis, and SCORD5, which encodes an ATP-binding cassette (ABC) protein, AtGCN20/AtABCF3, predicted to be involved in stress-associated protein translation control. Identification of SCORD5 begins to implicate an important role of stress-associated protein translation in stomatal guard cell signaling in response to microbe-associated molecular patterns and bacterial infection.
Author Summary
Pathogen entry into host tissue is a critical first step in causing infection. For foliar bacterial plant pathogens, natural surface openings, such as stomata, are important entry sites into the leaf apoplast (internal intercellular spaces). Recent studies have shown that plants respond to surface-inoculated bacterial pathogens by reducing stomatal aperture as part of the innate immune response to restrict bacterial invasion. Once inside plant tissue, bacteria encounter defenses in the apoplast. To counter host defenses during invasion and in the apoplast, bacterial pathogens produce a variety of virulence factors, such as the polyketide toxin coronatine produced by Pseudomonas syringae pv. tomato (Pst) DC3000. Coronatine-deficient Pst DC3000 mutants are compromised in virulence, especially when inoculated onto the plant surface. In this study, we conducted a random genetic screen to identify Arabidopsis mutants that could rescue the virulence of coronatine-deficient mutant bacteria and obtained three classes of Arabidopsis mutants: those that are defective in stomatal closure only, those defective in apoplastic defense only, and those compromised in both stomatal closure and apoplastic defenses. The isolation of these host mutants highlight the important role of COR, a molecular mimic of the plant hormone jasmonate, in overcoming both stomatal and apoplastic defenses during Pst DC3000 infection.
Introduction
Pseudomonas syringae pv. tomato (Pst) strain DC3000 is a Gram-negative bacterium that infects tomato and Arabidopsis and is a model pathogen used to investigate molecular mechanisms underlying plant-pathogen interactions [1]–[3]. In nature, P. syringae strains often exhibit an epiphytic phase (i.e., surviving and/or multiplying on the plant surface) before entering the intercellular space of the host through wounds or natural openings such as stomata. Once inside the apoplastic space of a susceptible host plant, P. syringae can multiply to high levels within days, a process aided by many virulence factors including secreted phytotoxins and effector proteins delivered into the host cells through the type III secretion system (T3SS). Successful colonization of P. syringae in the apoplastic space eventually results in the development of disease symptoms, which usually include localized tissue necrosis and discoloration.
Coronatine (COR) is a non-host-specific phytotoxin produced by several pathovars of P. syringae, including Pst DC3000 [4]–[8]. COR has been implicated in the inhibition of stomatal closure to facilitate bacterial invasion, promotion of bacterial multiplication and persistence in planta, induction of disease symptoms, and enhancement of disease susceptibility in uninfected parts of the plant [7], [9]–[19]. COR shows remarkable structural similarity to the active form of the plant hormone jasmonoyl isoleucine (JA-Ile), and targets the JA receptor complex directly [20]–[22]. Indeed, COR serves as a potent inducer of JA-associated responses and expression of JA-responsive genes in plants [7], [12], [23], [24].
The molecular mechanism(s) by which COR-mediated activation of JA signaling results in various virulence roles of COR throughout the bacterial infection cycle remains to be determined. COR-deficient mutants of Pst DC3000 are able to multiply to high levels in salicylic acid (SA)-deficient Arabidopsis plants nahG and sid2, suggesting that COR may be required to overcome SA-mediated defenses [12], [14], [19]. Indeed, SA-dependent expression of pathogenesis-related (PR) genes is suppressed in a COR-dependent manner during Pst DC3000 infection in tomato plants [15], [25]. However, such COR-dependent suppression of PR gene expression has not yet been observed in Pst DC3000 infection of Arabidopsis plants [12]. Interestingly, recent studies show that COR can suppress plant defense responses triggered by microbe-associated molecular patterns (MAMPs). For example, COR inhibits MAMP- and bacterium-triggered stomatal closure, which is dependent on SA signaling [14], [19], and COR and JA suppress MAMP-induced callose deposition in leaf mesophyll cells and root cells [26], [27] in Arabidopsis. Taken together, these results collectively indicate that a major function of COR is to suppress interconnected MAMP- and SA-activated defense responses during bacterial infection.
Thus far, attempts to elucidate the role of COR in virulence have relied heavily on known plant defense signaling mutants. We reasoned that an unbiased genetic screen for plant mutants that affect the function of COR in the context of bacterial infection would be useful. Accordingly, we conducted a genetic screen aimed at isolating Arabidopsis mutants that would rescue the virulence of COR-deficient mutant bacterium Pst DC3118. This screen allowed us to identify eight susceptible to coronatine-deficient Pst DC3000 (scord) mutants. Six of them are defective in stomatal closure responses to Pst DC3118; the other two have a normal stomatal closure response to bacteria, but show weakened apoplastic resistance to both Pst DC3000 and Pst DC3118. Further characterization of the six stomatal closure mutants revealed defects in salicylic acid (SA) accumulation, SA-triggered stomatal closure, and/or abscisic acid (ABA)-induced stomatal closure. We cloned SCORD3, which is required for SA biosynthesis, and SCORD5, which encodes an ABC-transporter-family protein (AtGCN20/AtABCF3), predicted to be involved in stress-associated protein translation control. The identification of SCORD5 revealed potentially a new layer of regulation of MAMP/bacterium-triggered stomatal closure in Arabidopsis.
Results
Experimental setup of the genetic screen
When inoculated onto leaves of wild-type Col-0 plants by dipping at 1×108 CFU/ml, COR-deficient mutants of Pst DC3000, such as Pst DC3118, do not cause significant disease symptoms or multiply to a high level [14], [19]. Pst DC3118 carries a Tn5 insertion in the cfa6 gene (Figure S1), which is essential for the biosynthesis of COR [5], [7] (Figure S1). However, Pst DC3118 exhibits aggressive growth and causes severe disease symptoms in Arabidopsis mutants that are defective in MAMP signaling, or in biosynthesis or signaling of ABA and SA [12], [14], [19]. For example, the ost1-2 mutant, defective in a guard cell/ABA signaling kinase, OST1 [28], [29], allows Pst DC3118 to multiply to high levels and cause prominent disease symptoms when surface-inoculated [14]. On the other hand, ost1-2 plants remained as resistant as wild-type Ler plants to the nonpathogenic hrcC mutant (defective in the T3SS) of Pst DC3000 (Figure S2), indicating that the greatly enhanced susceptibility of ost1-2 plants was specific to Pst DC3118.
For large-scale mutant screens we needed to grow plants in high density (approximately 20 to 30 plants per pot and 15 pots per flat). Accordingly, we tested whether ost1-2 mutant plants would still exhibit enhanced susceptibility to Pst DC3118 under this growth condition. Indeed, when dip-inoculated with Pst DC3118, ost1-2 plants allowed much higher bacterial multiplication and exhibited much more severe disease symptoms than wild-type Ler plants (Figure S2). In subsequent mutant screening, ost1-2 plants were included in each flat as a positive control.
Identification and characterization of scord mutants
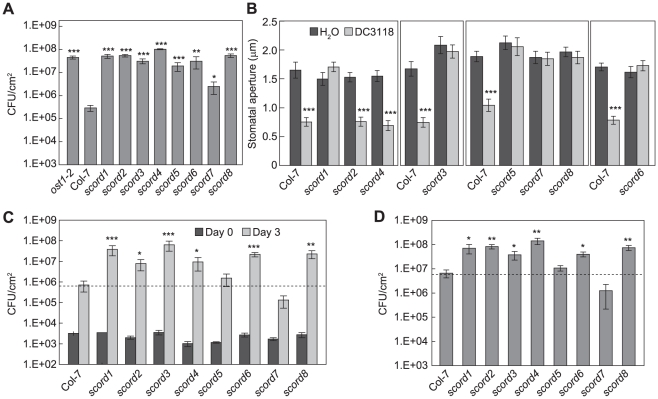
About 14,000 activation-tagged T-DNA lines were screened by dip-inoculation with 1×108 CFU/ml of Pst DC3118. Plants that showed disease symptoms similar to ost1-2 plants were selected as putative mutants and allowed to set seeds. The progeny of each putative mutant were screened at least four more times in the following two generations. Here we report eight mutants that reproducibly showed enhanced disease symptoms, and are named susceptible to coronatine-deficient Pst DC3000 (scord). Pst DC3118 populations in these mutants were almost 100 times higher than in wild-type plants when dip-inoculated, except for the scord7 mutant, in which Pst DC3118 population was about 10 times higher (Figure 1A, Table 1).
Figure 1. Characterization of scord mutants.
(A) Pst DC3118 population at 3 dpi after dip-inoculation at 1×108 CFU/ml. (B) Stomata apertures (µm) from leaf peels incubated with Pst DC3118 (1×108 CFU/ml) for 1 h. (C) Pst DC3118 populations when inoculated by infiltration at 1×106 CFU/ml. (D) Pst DC3000 population at 3 dpi after dip-inoculation at 1×107 CFU/ml. Differences were detected by comparing each scord mutant with Col-7 for (A), (C) and (D) Statistical analyses for this and following figures are described in MATERIALS and METHODS.
Table 1. Phenotype summary of scord mutants.
| Col-7 | scord1 | scord2 | scord3 | scord4 | scord5 | scord6 | scord7 | scord8 | |
| stomatal response to DC3118 | closure | open | closure | open | closure | open | open | open | open |
| log(cfu/cm2), 3 dpi, DC3118 dipping (1E+8 cfu/ml) | 5.36±0.21 | 7.68±0.09 | 7.72±0.06 | 7.45±0.11 | 8.00±0.02 | 7.13±0.19 | 7.09±0.55 | 6.01±0.35 | 7.72±0.09 |
| log(cfu/cm2), 3 dpi, DC3118 infiltration (1E+6 cfu/ml) | 5.70±0.20 | 7.45±0.18 | 6.69±0.24 | 7.64±0.22 | 6.71±0.26 | 5.97±0.24 | 7.29±0.13 | 4.84±0.31 | 7.24±0.19 |
| log(cfu/cm2), 3 dpi, DC3000 dipping (1E+7 cfu/ml) | 6.68±0.23 | 7.74±0.18 | 7.89±0.09 | 7.47±0.18 | 8.10±0.12 | 6.98±0.12 | 7.57±0.09 | 5.65±0.33 | 7.84±0.09 |
| endogenous SA a | ++++ | + | ++++ | − | ++++ | ++++ | ++ | ++++ | + |
| DC3000-induced SA a (12 hpi, infiltration at 1E+8 cfu/ml) | ++++ | − | ++++ | − | ++++ | ++++ | +++ | ++++ | +++ |
| stomatal response to SA | closure | closure | closure | closure | closure | closure | open | open | open |
| stomatal response to ABA | closure | closure | closure | closure | closure | closure | closure | open | closure |
| rosette size | ++++ | ++++ | +++++ | +++++ | ++++ | +++ | ++++ | ++++ | +++ |
| leaf morphology | normal | normal | normal | normal | serrated edge | pale green | smooth edge | thin | serrated edge |
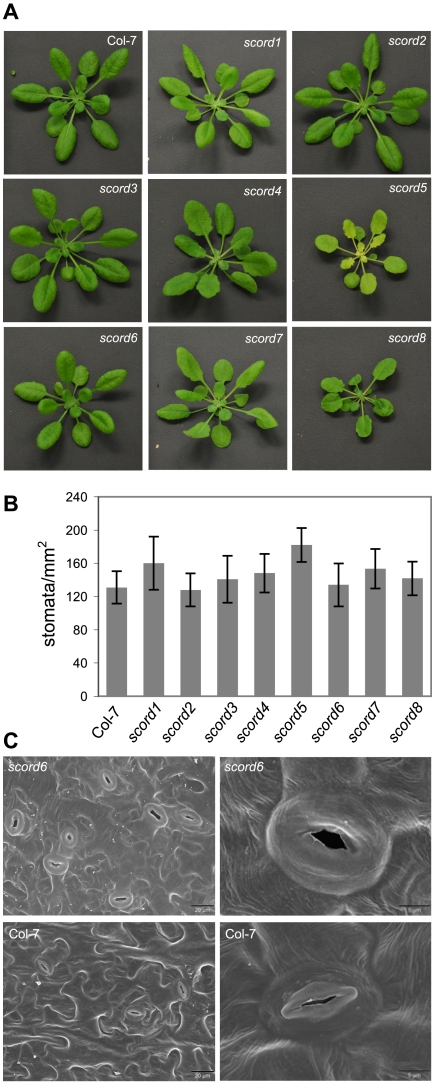
| stomatal density | normal | normal | normal | normal | normal | normal | normal | normal | normal |
| stomatal morphology | normal | normal | normal | normal | normal | normal | large and missing central ridges | normal | normal |
| gene/chromosome | At4g39030/IV | At1g64550/I | 18.84 Mb–19.03 Mb/III | 1.9 Mb–2.14 Mb/V |
SA levels from scord mutants were compared to those from Col-7, based on Figure 2.
As the scord mutants may be affected in different aspects of COR action during Pst DC3000 infection, we first determined whether some of them are defective in the bacterium-induced stomatal closure response. When subjected to leaf peel assays with suspensions of Pst DC3118 [14], [19], most stomata of wild-type Col-7 plants closed and resulted in a significant reduction of the mean aperture (Figure 1B). However, the stomata of six scord mutants—scord1, -3, -5, -6, -7, and -8—did not show significant closure, indicating loss of normal stomatal closure response to Pst DC3118 (Figure 1B). The other two mutants, scord2 and scord4, maintained normal stomatal closure responses to bacteria, at least within the time frame of our assays (i.e., 1 to 1.5 h after incubation) (Figure 1B).
Since increased plant susceptibility in dip-inoculation experiments could also be caused by a deficiency in apoplastic defense after bacterial invasion, we next tested scord mutants by infiltration inoculation with Pst DC3118 (Figure 1C). Of the eight scord mutants, scord1, -3, -6, and -8 plants showed a much higher apoplastic susceptibility to Pst DC3118, and contained 50 to 100 times more bacteria 3 days after inoculation, compared to Col-7 plants. scord2 and scord4 also showed a higher susceptibility to Pst DC3118 in infiltration experiments, harboring about 10 times more bacteria than Col-7 plants at day 3 (Figure 1C). In contrast, scord5 and scord7 did not show significantly higher susceptibility to Pst DC3118 in infiltration experiments compared to Col-7 plants (Figure 1C). Although statistically (Student's t-test) not significant, we have consistently observed in all repeats that Pst DC3118 growth is actually less in scord7 plants than in Col-7 plants. Overall, these results suggest that scord1, -3, -6, and -8 are deficient not only in stomatal defense but also in apoplastic defense. On the other hand, scord5 and scord7 are deficient in stomatal defense, and scord2 and scord4 are affected in apoplastic defense.
Next, we tested the susceptibility of the scord mutants to wild type Pst DC3000 by dipping inoculation. We found that scord1, -2, -3, -4, -6, and -8 mutants were also more susceptible to Pst DC3000 than Col-7 plants (Figure 1D). In contrast, scord5 and scord7 were not hyper-susceptible to Pst DC3000 compared to Col-7 plants. Again, although statistically not significant, Pst DC3000 growth was consistently less in scord7 plants than in Col-7 plants.
Finally, we examined the susceptibility of these eight scord mutants to the nonpathogenic Pst DC3000 hrcC mutant by dipping inoculation. None of the eight scord mutants allowed significant growth of Pst DC3000 hrcC (Figure S3). Thus, scord mutants maintained normal resistance to nonpathogenic bacteria.
Effects of scord mutations on SA accumulation and/or SA-induced stomatal closure
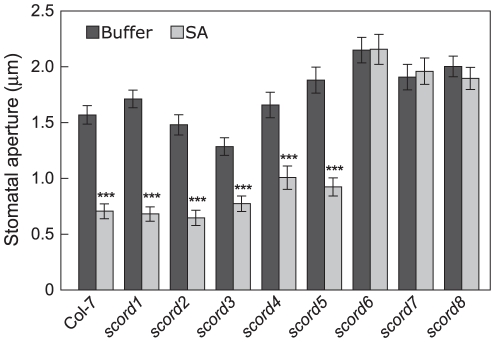
We have previously shown that SA plays a critical role in bacterium-induced stomatal closure response [14], [19]. To position the scord mutants in relation to SA, we tested SA-induced stomatal closure in the scord mutants (Figure 2). In leaf peel assays, we found that scord6, -7 and -8 mutants are defective in SA-induced stomatal closure, but scord1, -2, -3, -4 and -5 had normal stomatal closure response, similar to wild type Col-7 plants.
Figure 2. Stomatal closure responses of scord mutants to SA.
Stomatal apertures (µm) were measured from leaf peels incubated with SA (100 µM) for 1 h.
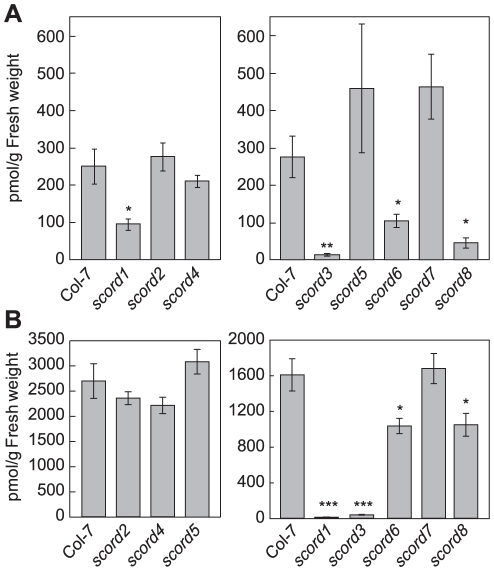
We conducted further experiments to examine whether any of the scord mutants were affected in SA accumulation. We found that scord1, -3, -6, and -8 plants all had significantly lower basal levels of SA, containing only 10–30% of what was detected in wild-type Col-7 plants (Figure 3A). To the contrary, SA levels in scord2, -4, -5, and -7 plants were similar to those in wild-type Col-7 plants (Figure 3A).
Figure 3. Salicylic acid (SA) levels in leaves of Col-7 and scord mutants.
Numbers represent means and standard errors of values from 4 different plants. Similar results were obtained from two different experiments. (A) Plants without treatment. (B) Plants were infiltrated with Pst DC3000 at 1×108 CFU/ml. Leaves were collected at 12 hpi.
We also examined scord mutants for SA levels 12 hours after infiltration with Pst DC3000 at 1×108 CFU/cm (Figure 3B). Similar to what was observed without bacterium challenge, the SA levels in scord1, -3, -6, and -8 plants were reduced, whereas those of scord2, -4, -5, and -7 plant were similar to those in Col-7 plants (Figure 3B).
Effects of scord mutations on ABA-induced stomatal closure
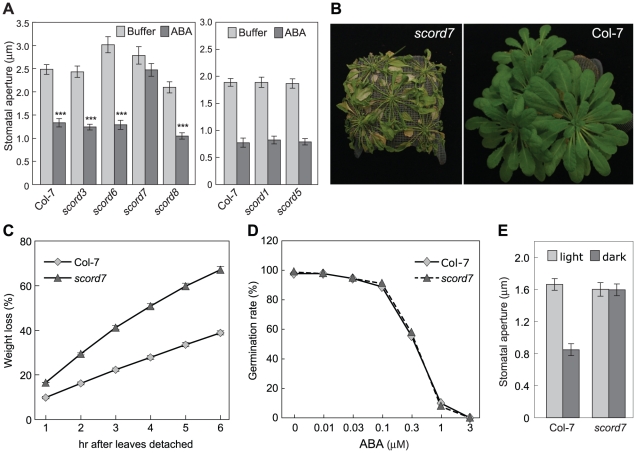
Besides SA, we have previously shown that ABA biosynthesis and signaling are also critical for the stomatal closure response to bacteria or MAMPs [14], [19]. To position the six stomata-closure-defective scord mutations in relation to ABA, we examined the stomatal response of scord1, -3, -5, -6, -7, and -8 mutants to ABA. We found that only scord7 stomata failed to close in response to ABA (Figure 4A), whereas stomata of Col-7 and the other five scord mutants (scord1, -3, -5, -6, and -8) showed normal closure responses to ABA (Figure 4A), suggesting that the scord7 mutant might be affected in ABA response or a general step in stomatal closure that is common to multiple stomatal closure pathways. ABA response mutants often show hypersensitivity to drought stress. Indeed, when subjected to water withholding, scord7 plants wilted faster than wild-type Col-7 plants (Figure 4B). When detached leaves were tested for water loss, leaves from scord7 plants lost water at a much faster rate than those from Col-7 wild type plants (Figure 4C). The scord7 mutant, however, showed a similar ABA inhibition response as wild type Col-7 in seed germination assay (Figure 4D). We also tested scord7 plants for dark-induced stomatal response, which is thought to be independent of ABA signaling [30]. The scord7 plants were defective in dark-induced stomatal closure (Figure 4E). Thus, SCORD7 is involved either in a guard cell-specific ABA response pathway or in a later step shared by SA-, ABA- and dark signaling pathways leading to stomatal closure.
Figure 4. Responses of scord mutants to ABA.
(A) Stomatal apertures of Col-7 and scord1, -3, -5, -6, -7 and -8 in response to ABA (10 µM) in leaf peel assay. (B) Appearance of 5-week-old scord7 and Col-7 plants after being withheld water for 10 days. (C) Water loss of detached leaves. Four plants and 4 to 5 leaves from each plant were sampled for Col-7 or scord7. Values are means with standard errors displayed. (D) Germination rates of Col-7 and scord7 seeds on MS plates supplemented with ABA. Values are means with standard errors. (E) Stomata apertures of Col-7 and scord7 in response to dark. Leaf peels were incubated in buffer under light or in the dark for 3 h before stomatal aperture measurements.
Morphological phenotypes of scord mutants
In addition to the phenotypes characterized above, we also noticed morphological phenotypes for some of the scord mutants. Among the eight mutants, the appearances of scord1, -2, -3, and -7 plants are similar to wild type Col-7 plants (Figure 5; Table 1). On the other hand, compared to Col-7 plants, scord5 and 8 plants have slightly smaller rosettes, scord5 plants have pale green leaves, scord4 and 8 plants have more serrated leaf edges, and scord6 has smoother leaf edges (Figure 5A). We also examined the stomata density and stomata morphology in these mutants, and did not notice any dramatic difference on stomata density (Figure 5B), except that scord6 stomata appear to be larger and lack the central ridges (Figure 5C). At present, it is not known whether these morphological phenotypes are caused by the same mutations that lead to defects in stomatal closure and/or bacterial susceptibility phenotypes.
Figure 5. Morphological phenotypes of scord mutants of 4 to 5 weeks old.
(A) Rosettes. (B) Stomata densities of leaf abaxial surfaces. (C) Scanning electron microscopy pictures of leaf abaxial surfaces of Col-7 and scord6 plants. Note that raised central ridges in stomata are missing in the scord6 mutant.
The scord3 and scord5 mutants carry a T-DNA insertion in the EDS5 gene and AtGCN20/AtABCF3 gene, respectively
We attempted plasmid rescue experiments with all scord mutants. However, we were able to correlate a T-DNA insertion with mutant phenotypes in only the scord3 and scord5 mutants. In the scord3 mutant, the T-DNA insertion is located in the first intron of At4g39030 (Figure S4), which encodes EDS5, a putative transporter protein required for SA biosynthesis [31]. Similar to the scord3 mutant, the eds5-1 mutant showed a defect in SA accumulation and stomatal response (Figure S5). Furthermore, allelism tests showed that scord3 and eds5-1 have mutations in the same gene (Figure S6). In the literature, three alleles of eds5 have been characterized [31]. We therefore have named scord3 eds5-4.
As described above, in addition to the scord3/eds5-4 mutant, the scord1 mutant also contained little SA before or after bacteria challenge (Figure 2). Like the scord3/eds5-4 mutant, the scord1mutant maintained a normal SA-induced stomatal closure response, suggesting that the scord1 mutant is deficient in the synthesis of SA, but is normal in SA signaling. We therefore examined whether the scord1 mutant also contains a mutation in the EDS5 gene. We amplified and sequenced the genomic region of EDS5 from scord1 plants. However, we did not find any mutation in EDS5 (data not shown). Therefore, the scord1 mutant likely has a mutation in a gene, other than EDS5, that is involved in SA biosynthesis.
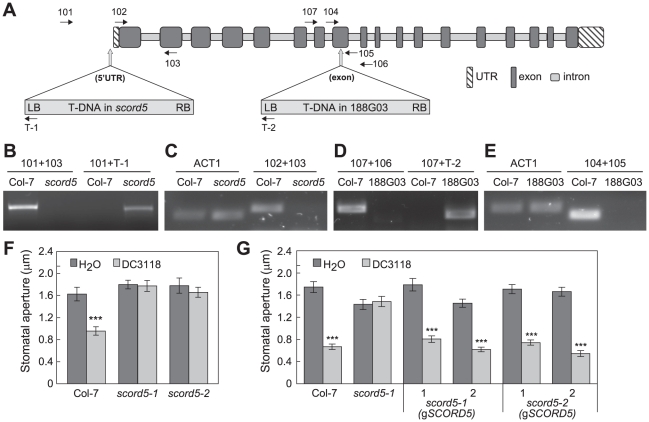
The T-DNA insertion site in the scord5 mutant lies in the 5′UTR region of At1g64550 (108 bps upstream of the start codon ATG; Figure 6A). The existence of this T-DNA insertion in the scord5 genome was confirmed by PCR using At1g64550-specific and T-DNA-specific primers (Figure 6B). Furthermore, the transcript of At1g64550 was not detectable in scord5 plants (Figure 6C). To confirm that the phenotypes observed in scord5 plants were due to the T-DNA insertion in At1g64550, we identified an independent T-DNA insertion line, 188G03, in the GABI-Kat collection [32]. Genomic PCR confirmed the presence of a T-DNA insertion in the eighth exon of At1g64550 (Figure 6A, D). RT-PCR using primers near the insertion site showed that 188G03 is also a knockout line for At1g64550 (Figure 6E). Like the scord5 mutant, 188G03 was also impaired in stomatal closure response to Pst DC3118 (Figure 6F). Based on these molecular and phenotypic analyses, we have designated the GABI-Kat line 188G03 as scord5-2 and the original scord5 line as scord5-1.
Figure 6. Characterization of SCORD5 (At1g64550).
(A) The intron/exon structure of At1g64550 and T-DNA insertion sites in scord5 (scord5-1) and 188G03 (scord5-2) mutants. Locations of primers used in this study are indicated. (B) and (D) PCR reactions using genomic DNA from Col-7 and scord5 (scord5-1) (B) or 188G03 (scord5-2) (D) as template. (C) and (E) RT-PCR reactions using total RNA from Col-7 and scord5 (scord5-1) (C) or 188G03 (scord5-2) (E) as template. The RT-PCR product of ACT1 (Arabidopsis actin 1; At2g37620) was used as a loading control. (F) Stomata apertures of Col-0, scord5-1, and scord5-2 leaf peels incubated with H2O or Pst DC3118 (1×108 CFU/ml) for 1 h. (G) Stomata apertures of Col-7, scord5-1 and scord5 mutant plants complemented with a 5.4-kb-genomic region of At1g64550, in response to Pst DC3118 (1×108 CFU/ml) for 1 h.
To complement the phenotypes of scord5 mutants, a 5.4-kb genomic fragment that includes a 480-bp region upstream of the start codon and a 400-bp region downstream of the stop codon of SCORD5 (At1g64550) was amplified using Col-7 genomic DNA. This SCORD5 fragment was cloned into the plant transformation vector pCAMBIA1300 and introduced into both scord5-1 and scord5-2 mutant plants via Agrobacterium-mediated transformation. Examination of four randomly chosen, independent T1 transformants showed that stomatal closure response to Pst DC3118 was restored in both scord5-1 (gSCORD5) and scord5-2 (gSCORD5) lines (Figure 6G), providing further evidence that SCORD5 is required for bacterium-triggered stomatal closure.
Sequence analysis showed that SCORD5 encodes a gene that has been annotated as AtGCN20/AtABCF3, which belongs to the F subfamily of ATP-binding cassette (ABC) proteins. AtGCN20/AtABCF3 is a 4.8-kb gene with 17 introns, and encodes a protein of 715 amino acids. AtGCN20/AtABCF3 (SCORD5 hereinafter) is predicted to have only nucleotide-binding domains (NBDs), lacking transmembrane domains that are characteristic of the AtABCA, -B, -C, -D, and -G protein subfamilies [33]–[35].
Molecular characterizations of other scord mutations
Because T-DNA insertions in other scord mutants (i.e., other than scord 3 and scord5) either could not be amplified, or were amplified but not linked to Pst DC3118-hypersusceptibility phenotypes (data not shown), we have initiated physical mapping of two of these scord mutations: scord6 and scord7. To this end, we have narrowed the scord6 mutation to a 190-kb region on chromosome III, and the scord7 mutation to a 240-kb region on chromosome V (Table 1). Neither region contains genes that are known to be involved in MAMP, SA or ABA signaling or in stomatal closure response. Therefore, SCORD6 and SCORD7 likely encode new regulators of stomatal closure response.
In summary, our molecular and phenotype characterization of the eight scord mutants suggests that scord3, -5, -6, and -7 represent mutations in four different genes (i.e., EDS5, AtGCN20/AtABCF3, SCORD6, and SCORD7). Although we cannot ascertain the genetic relationships among the other four scord mutations (scord1, -2, -4, and -8), the distinct phenotypes (stomatal closure response, apoplast defense, SA levels, and morphology; Table 1) exhibited in the corresponding mutants suggest that at least some of them would be affected in additional genes.
The scord5 mutant is affected in MAMP-induced stomatal closure, but not other MAMP-induced responses in the leaves
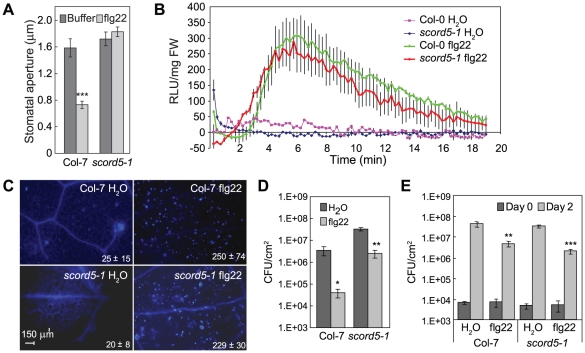
Because SCORD5 has never been implicated in stomatal closure or plant defense response, we focused on this gene for further analysis. We recently showed that the signaling cascade for bacterium/MAMP-induced stomata closure involves MAMP signaling, followed by SA and ABA signaling [19], [36]. As stomata of the scord5 mutant responded normally to SA and ABA (Figures 2, 4), we investigated a possible defect in MAMP response using flg22 (a bioactive 22-aa peptide from bacterial flagellin). We found flg22 triggered stomatal closure in Col-7 plants, but not in scord5 plants (Figure 7A). Similarly, elf18 (a bioactive 18-aa peptide from E. coli EF-Tu) induced stomatal closure in Col-7 plants, but not in scord5 plants (Figure S7). These results are consistent with the observed defect in stomatal closure response to Pst DC3118 bacteria (Figure 1B), which are expected to produce flagellin and possibly other MAMPs during infection [19].
Figure 7. scord5 mutant plants have normal flg22-induced responses other than stomatal closure response.
(A) Stomata apertures of Col-7 and scord5-1 leaf peels treated with flg22 (10 µM). (B) H2O2-dependent luminescence of luminol induced by flg22 (100 nM) in Col-7 and scord5-1seedlings. (C) Callose deposition induced by flg22 (10 nM) in Col-7 and scord5-1 leaves. (D) and (E) Populations of Pst DC3000 at 2 dpi after inoculation with dipping (D; 1×108 CFU/ml) or infiltration (E; 1×106 CFU/ml) following spray with H2O or flg22 (3 µM) a day earlier. Differences were detected by comparing H2O and flg22 treatment. Experiments in (B) and (C) were repeated two and three times, respectively.
Stomatal closure is only one of several functional outputs induced by MAMPs. To determine whether other responses induced by MAMPs are also affected in the scord5 mutant, we first examined the production of reactive oxygen species and deposition of callose in scord5 leaves in response to flg22. The production of H2O2 after flg22 treatment (100 nM) was similar in scord5-1 and Col-7 plants (Figure 7B). Similarly, the amounts of callose deposition induced by infiltration of flg22 (10 nM) were comparable in scord5-1 and Col-7 leaves (Figure 7C). Next, we studied the effect of flg22 pretreatment on the induced resistance against Pst DC3000 (i.e., flg22 protection assay) in scord5-1 and Col-7 plants. One day after spraying scord5-1 and Col-7 plants with 3 µM flg22, we inoculated plants with Pst DC3000 by dipping at 1×108CFU/ml (Figure 7D). We noticed that scord5 plants were more susceptible than Col-7 to Pst DC3000 in the H2O treatment control, and allowed more Pst DC3000 multiplication, compared to Col-7 after flg22 pretreatment (Figure 7D). Nonetheless, the flg22 treatment still induced resistance in scord5 plants, albeit to a smaller degree compared to that in Col-7 plants. To find out whether the higher susceptibility of the scord5 mutant shown in the flg22 protection assay in dipping inoculation was caused by reduced apoplastic defense, we repeated the flg22 protection experiment by infiltrating Pst DC3000 (1×106 CFU/ml) directly into the apoplast. We found that the multiplication of Pst DC3000 was suppressed to similar levels in scord5-1 and Col-7 leaves (Figure 7E). These experiments suggest that flg22-induced apoplastic defense is normal in the scord5 mutant and that the partial loss of flg22-induced resistance in scord5 plants is likely caused by a defect in bacterium-triggered stomatal closure.
ILITYHIA (ILA) is also required for bacterium-triggered stomatal closure response
We conducted further experiments to increase our understanding of the involvement of the SCORD5-associated pathway in regulating bacterium-triggered stomatal closure. SCORD5 shares significant sequence similarities with yeast (46% identity and 66% similarity) and mammalian (41% identity and 61% similarity) GCN20 proteins (EDN59158 and NP_038880, respectively), which, together with GCN1, regulates stress-associated protein translation [37]–[39]. Arabidopsis has a putative GCN1 orthologue named ILITYHIA (ILA; At1g64790), which was shown recently to be involved in defense against Pst DC3000 [40]. Arabidopsis ILA shares 35% identity and 54% similarity with rat GCN1 (NP_001162135) and 34% identity and 54% similarity with yeast GCN1 (EEU04663), but shares no sequence similarity with SCORD5 (AtGCN20/AtABCF3). We examined the possibility that ILA may also be required for bacterium-triggered stomatal closure. Indeed, similar to the scord5 mutant, stomata of ila-3 mutant were completely unresponsive to Pst DC3118 (Figure 8A). This result provides further evidence that bacterium-triggered stomatal closure is likely regulated by an authentic GCN1/GCN20-associated protein translation control mechanism in plants.
Figure 8. Stomatal closure response in the ila-3 mutant.
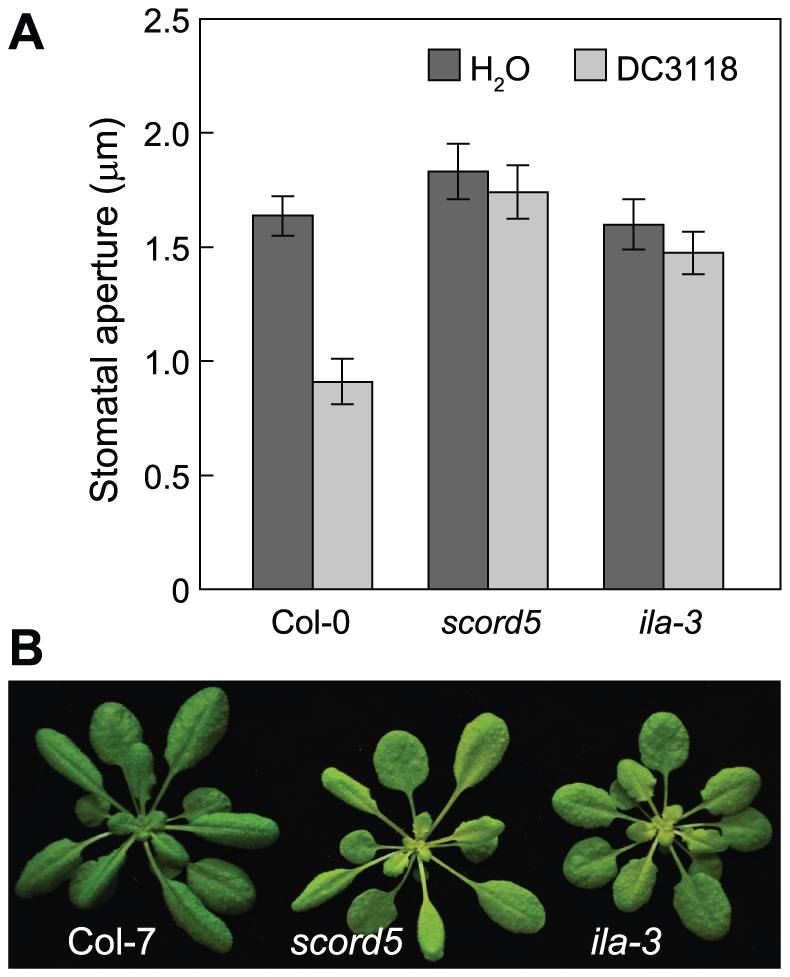
(A) Stomata apertures of Col-7, scord5 and ila-3 in response to Pst DC3118 (1×108 CFU/ml) at 1 hpi. (B) Four-week-old rosettes.
Discussion
In the Arabidopsis-Pst DC3000 interaction, COR has been implicated in several aspects of pathogenesis. To date, however, a random genetic screen for host mutants that rescue the virulence of COR-deficient Pst mutants has not been reported. In principle, such a genetic screen not only could identify new alleles of host genes known to be involved in COR-dependent interactions during infection, but also has the potential to uncover novel components in the host that are required for the various virulence functions of COR. Indeed, from an initial screen of ∼14,000 T-DNA insertion lines, we were able to isolate eight scord mutants. Further detailed characterization of these mutants uncovered both known (e.g., EDS5) and novel (e.g., AtGCN20/AtABCF3, and possibly SCORD6 and SCORD7) regulators of stomatal and/or apoplastic defenses, illustrating the exciting prospect of future large-scale Pst DC3118-based genetic screens in the identification of cell-type-specific defense mutants in plants.
Stomatal closure response has been shown to be an integral part of Arabidopsis resistance to COR-deficient mutants of Pst DC3000, and SA is necessary for this response [14], [19]. Indeed, of the eight scord mutants isolated in this study, six are defective in the stomatal closure response to Pst DC3118 (Figure 1B). Among them, both basal and bacterium-induced levels of SA are significantly lower in scord1, -3, -6 and -8 plants than those in wild-type plants (Figure 3), indicating that these four mutants have defects in the accumulation of SA to various degrees. The scord7 mutant is defective in stomatal closure response to SA, ABA and dark, and is hypersensitive to drought (Figure 4). These results point to a possible deficiency in a step common to multiple signaling pathways leading to stomatal closure in the scord7 mutant plant.
SA is also known to be essential for apoplast defense, which can be revealed using infiltration inoculation. In our screen, SA-compromised scord1,-3, -6 and -8 mutants allowed more bacterial growth, compared with wild-type plants, when Pst DC3118 was infiltrated into the apoplast directly (Figure 1C), supporting the importance of SA in mediating the apoplastic defense against Pst DC3118. In contrast, although the scord7 mutant is more susceptible than Col-7 plants to surface-inoculated DC3118, this mutant showed slightly more resistance to Pst DC3118 in infiltration experiments (Figure 1C). This is a particularly interesting result because ABA mutants have been observed to be more resistant to Pst DC3000 in infiltration experiments [41]. The enhanced apoplastic resistance of the scord7 mutant to Pst DC3118 therefore could be related to the inability of scord7 stomata to respond to ABA. However, considering that scord7 stomata are not responsive to SA and dark, we cannot rule out the possibility that the apoplast of the scord7 mutant has elevated resistance due to an ABA signaling-independent mechanism. Regardless, the overall higher level of susceptibility of the scord7 mutant to surface-inoculated Pst DC3118 can now be explained by a combined effect of compromised stomatal defense and enhanced apoplastic defense, with the stomatal closure defect overpowering the enhanced apoplastic defense in this mutant. We noted that the overall enhancement of Pst DC3118 growth in the scord7 mutant when inoculated by dipping was less than in other scord mutants (Figure 1A), which is consistent with the observation that the apoplast of other scord mutants was not more resistant to Pst DC3118, as revealed in the infiltration experiments (Figure 1C).
It is interesting that, although the scord5 and scord7 mutants are much more susceptible to surface-inoculated Pst DC3118, compared with Col-7 plants, they are not more susceptible to surface-inoculated Pst DC3000. Thus, the disease hyper-susceptibility of the scord5 and scord7 mutants is dependent on COR production in bacteria. This finding is consistent with the observations that stomatal defense in wild-type Col-7 plants presents a significant barrier to infection by Pst DC3118 and that the scord5 and scord7 mutants are defective in stomatal defense, thereby allowing Pst DC3118 to infect from the leaf surface. On the other hand, Pst DC3000 can infect not only scord5 and scord7 mutants, but also wild-type Col-7 plants because of its ability to produce COR, which counteracts stomatal closure in wild-type Col-7 plants. COR is not necessary for Pst DC3000 to infect scord5 and scord7 mutants, in which stomatal closure is already compromised.
In this study, we also identified two mutants, scord2 and scord4, that showed an apparently normal stomatal closure response, but compromised apoplastic defense (Figure 1). Interestingly, in contrast to scord5 and scord7 mutants, scord2 and scord4 mutants were hyper-susceptible to both Pst DC3118 and Pst DC3000, suggesting that these two mutants have a general defect in the apoplast (defense or nutrition) and that the hyper-susceptibility phenotype is not dependent on COR production in bacteria. The ability of these mutants to rescue the virulence of Pst DC3118 suggests that a hyper-susceptible apoplast can compensate, at least genetically, for the reduced ability of Pst DC3118 to overcome stomatal defense. We hypothesize that these apoplast defense mutants are likely affected in cellular processes other than the biosynthesis or signaling of SA and ABA, because defects in SA or ABA biosynthesis or signaling cause altered stomatal closure in response to bacteria [14], [19] and would have been detected in this study. Accordingly, further characterization of the scord2 and scord4 mutants has potential to yield novel genes/pathways that control apoplast defense (or nutrition) in Arabidopsis.
We were successful in cloning SCORD3 and SCORD5. Whereas SCORD3 turns out to be EDS5, which is known to be involved in plant defense, SCORD5 is a new component required for bacterium-triggered stomata closure response. The scord5 mutation affected mainly MAMP-induced stomatal closure, but not SA- or ABA-induced stomatal closure, suggesting that SCORD5 likely acts early in the stomatal closure response pathway. Although we could not detect a significant defect in apoplastic defense in the scord5 mutant, the ila-3 mutant has been shown to be more susceptible to Pst DC3000 compared with Col-0 plants by infiltration inoculation, suggesting a defect in apoplastic defense [40]. We noticed that the ila-3 plants are slightly smaller than scord5 plants (Figure 8B). Because SCORD5 belongs to a five-member gene family (Figure S8), whereas there is only a single ILA gene in Arabidopsis, we speculate that some of the SCORD5-family genes may have partially redundant functions and that one or more of SCORD5-like genes may compensate for the loss of SCORD5 in the mesophyll cells. Future research should determine whether higher-order mutants of the SCORD5 family genes would phenocopy the pleiotropic defects in both stomatal and apoplastic defenses in the ila-3 mutant.
Overall, the genetic screen reported in this paper illustrates an unbiased and useful approach in the understanding of Arabidopsis signaling pathways that govern cell-type-specific (i.e., stomatal and mesophyll) responses to Pst DC3000 infection, as well as the molecular action of COR during various stages of Pst DC3000 infection of Arabidopsis.
Materials and Methods
Unless specified, all experiments presented were repeated three times and statistical differences were detected with a two-tailed T-test (*, P<0.05; **, P<0.01; ***, P<0.001).
Plant materials and growth conditions
Arabidopsis plants were grown in soil under a 12/12 h photoperiod at 110 µmol m−2 s−1 at 22°C. Seeds of the activation-tagged T-DNA lines (CS21995, CS23153) were purchased from the Arabidopsis Biological Resource Center with Col-7 accession as the wild-type parent [42]. For ABA inhibition of germination, seeds are sterilized in 30% bleach for 15 min and washed in sterilized ddH2O for 5 times before put on MS (1% sucrose) plates. Ninety seeds were used for each treatment. Plates were left at 4°C for two days before placed in growth chambers, and the numbers of seeds germinated were recorded two days later. For stomata counting, three leaves were chosen from 5-week old plants. A middle section of each leaf was cut out and mounted in distilled water, and visualized with bright field microscopy. From each section, three areas of 0.45 µm2 were chosen for observation. The chosen areas were scanned to a depth of 40–60 µm with a Zeiss 510 Meta ConfoCor3 laser scanning confocal microscope (Carl Zeiss MicroImaging, Thornwood, NY) to generate Z-scans. Zeiss LSM software was used to assemble the z-series into stacked images representing the leaf surface for stomata counting.
Stomata assays
Stomatal assays followed procedures described in a previous study [19]. Leaf peels from the abaxial side were collected from mature leaves of 5-week-old plants and placed in 250–300 µl of ddH2O or bacteria resuspended in ddH2O (1×108 CFU/ml), buffer (25 mM MES, 10 mM KCl, pH 6.15) or buffer containing SA or ABA (Sigma, St. Louis, MO, USA), flg22 or elf18 (EZBiolab, Westfield, IN, USA) on glass slides in square Petri dishes with lids on. To preserve the stomatal aperture status in plants used for both stomatal and bacterial pathogenesis assays, we did not further treat leaf peels in any stomatal opening buffer. The Petri dishes were left for an hour in the growth chamber in which plants were grown before being viewed under the microscope. Leaf peels on slides were observed under a light microscope and images were randomly taken to ensure that at least 30 stomata were recorded for each sample treatment. Images were opened in Adobe Photoshop for measurement of stomatal apertures. Bacteria were used at 1×108 CFU/ml (OD600 = 0.2). Stomatal apertures shown in results are means and standard errors of 30–60 measurements for each experiment.
Bacterial infection assays
A 10-ml low-salt LB liquid culture was started using bacteria (Pst DC3118) scraped from a plate that had been stored at −4°C for less than two weeks. After 12 h at 28°C, a larger subculture was started using 1∶100 dilution at 28°C. Bacterial cells were harvested when the OD600 of the culture reached 0.8 to 1.0. Bacteria were resuspended in ddH2O to an OD600 of 0.2 (1×108 CFU/ml). Silwet L-77 was added to a final concentration of 0.02 to 0.05%. For DC3118 dipping screening, four-week-old plants grown in meshed pots, at a density of about 20 seedlings per pot, were dipped upside down in the bacterial solution for a few seconds to coat all leaves uniformly with bacterial suspension. Plants were returned to a growth room in a tray covered with a plastic lid. At day 3 post-inoculation (3 dpi), plants showing no disease symptoms were removed from the pot. Diseased plants were allowed to set seeds. Seeds from the putative mutant plants were sown and the screening process was repeated. For other bacterial infection assays via dipping or infiltration inoculation, disease symptoms were recorded by camera and bacterial populations were monitored by serial-dilution assays according to Katagiri et al. [3]. All bacteria growth results are recorded as means of four different leaves from four different plants, with standard errors indicated.
flg22 protection assay
Five-week-old Arabidopsis plants were sprayed with H2O or 3 µM flg22 (EZBiolab, Carmel, IN) 24 h before bacterial inoculation. The flg22-treated plants were inoculated with Pst DC3000 by dipping at 1×108 CFU/ml or infiltration at 1×106 CFU/ml. Two days after bacterium inoculation, leaf samples were collected for bacterial enumeration.
Plasmid rescue cloning of T-DNA flanking fragments from the scord3 mutant
The plasmid rescue procedure was adapted from Weigel et al. [42]. Genomic DNA was prepared from 1 g (fresh weight) leaf tissue of 5-week-old plants using the Nucleon Phytopure Plant DNA Extraction Kit (Amersham Biosciences, Piscataway, NJ). One to five µg of purified genomic DNA was digested in a 120-µl reaction mixture overnight with restriction enzymes BamHI, SpeI, and NotI for the left border and KpnI, EcoRI, and HindIII for the right border of the T-DNA [42]. After digestion, ddH2O was added in the reaction mixture to bring the volume to 500 µl, followed with phenol-chloroform extraction and then chloroform extraction. Digested DNA was precipitated with 2 volumes of ethanol and 1/10 volume of sodium acetate (3 M, pH 5.2), and washed twice with 70% ethanol. Dried DNA was resuspended in 100 µl ddH2O, then used in a ligation reaction of 120 µl with 20–40 U of T4 DNA ligase (NEB, Beverly, MA) overnight at 16°C. DNA from the ligation mixture was precipitated in the same way as before and resuspended in 10 µl ddH2O. One to two µl of the ligation mixture was used for transformation into the SURE Electroporation-Competent Cells (Stratagene, La Jolla, CA). Plasmids from the Apr colonies were digested with the same restriction enzymes used for plasmid rescue reactions, and the inserts were subjected to sequencing using primers T3 (5′-ATTAACCCTCACTAAAGGGA-3′), T7 (5′-TAATACGACTCACTATAGGG-3′), T-1 (5′-AAGTGCAGGTCAAACCTTGAC-3′), T-3 (5′-GGTAATTACTCTTTCTTTTCCTC-3′), or sfEcoRI (5′-AGCCTTGCTTCCTATTATATCT-3′). Primers used to confirm the T-DNA insertion in scord3/eds5-4 plants also include 108 (5′-AAGGAAGCTCCATCGAACT-3′) and 109 (5′-TGTTTGGCAAGAGAAGTAGCA-3′).
iPCR cloning of T-DNA flanking fragments from the scord5 mutant
Genomic DNA was prepared the same way as described for plasmid rescue. One µg of purified genomic DNA was digested in a 100-µl reaction mixture overnight with the restriction enzyme EcoRV, HindIII, DraI, or EcoRI. After digestion, ddH2O was added to the reaction mixture to bring the volume to 500 µl, and the DNA mixtures were purified using a QIAquick PCR purification kit (Qiagen, Valencia, CA) and eluted in 50 µl ddH2O. Ten µl of the purified DNA mixture was incubated overnight at 16°C in a ligation reaction of 100 µl with 10 to 30 U of T4 DNA ligase (NEB, Beverly, MA). Five µl of the ligation mixture was used in each PCR reaction, along with the following primers specific to the T-DNA [42]: iPCR1 (5′-TGGATCTCAACAGCGGTAAGA-3′), iPCR2 (5′-TTCGACGTGTCTACATTCACG-3′), iPCR3 (5′-TCGGTGTGTCGTAGATACTAG-3′), iPCR4 (5′-TGGTTGACGATGGTGCAGAC-3′), iPCR1g (5′-TGACCATCATACTCATTGCTGATCC-3′), iPCR2g (5′-CGATCCGTCGTATTTATAGGCGAAAG-3′), T-1 (5′-AAGTGCAGGTCAAACCTTGAC-3′), T-3 (5′-GGTAATTACTCTTTCTTTTCCTC-3′), and T7 (5′-TAATACGACTCACTATAGGG-3′). PCR products obtained were purified from agarose gels after electrophoresis and sequenced. The T-DNA flanking sequence was identified from a PCR fragment amplified using primers T-3 and iPCR3 and DraI-digested scord5 genomic DNA as template.
Gene cloning
The primers used for PCR to confirm the T-DNA insertions in scord5-1 and scord5-2 plants, and for RT-PCR to confirm lack of expression of SCORD5 in the mutant plants were as follows: 101 (5′-CAAGTTATAGATAATATGAGTTTGT-3′), 102 (5′-GAATCTCGTCGCTTCTGTGTT-3′), 103 (5′-AACTTATCCGACAGTTTGCTAC-3′), 104 (5′-GTGGTCACGGACATCATCCAT-3′), 105 (5′-CTGCATATGTGACCGTGATCG-3′), 106 (5′-CAAACCAGAGACGAGTTGGAAACAG-3′), 107 (5′-CAGGTTTCTGACATTCTTTTCCTC-3′), T-1 (5′-AAGTGCAGGTCAAACCTTGAC-3′), T-2 (5′-ATATTGACCATCATACTCATTGC-3′), ACT1 (5′-GGTCGTACTACCGGTATTGTGCT-3′; 5′-TGACAATTTCACGCTCTGCTGTG-3′).
The 5.4-kb genomic fragment of SCORD5/At1g64550 was amplified using the genomic DNA of Col-7 as template. This fragment contains 480 bps upstream of the start codon ATG with an NheI restriction site attached (5′-CTGCTAGCCAATCTAAGCTCTCTTC-3′) and 400 bps downstream of the stop codon TAA with a SalI restriction site attached (5′-TGGGTCGACGGTGCTCGATTCGAA-3′). The fragment was fully sequenced before being cloned into pCambia1300 at the XbaI and SalI sites. The clone was transferred into GV3101 (pMP90) for plant transformation using the flower-dipping protocol. Independent T1 plants were identified based on hygromycin resistance and analyzed for complementation of the scord5-1 and 188G03 (scord5-2) mutations.
Identification of the Tn5 insertion in the Pst DC3118 genome
The site of the Tn5 insertion in Pst DC3118 was identified using iPCR performed according to Huang et al. [43]. Single colonies of Pst DC3000 and Pst DC3118 were grown overnight in 4 ml of low-salt LB culture media with appropriate antibiotics (Pst DC3000: Rif 100 µg/ml; Pst DC3118: Rif 100 µg/ml, Km 25 µg/ml). Overnight cultures were centrifuged (5 min at 2000×g) and cell pellets were resuspended in 400 µl of TES lysis solution (50 mM Tris-HCl, pH 7.5, 10 mM NaCl, 10 mM EDTA) supplemented with Sarcosyl (final concentration of 1%) and proteinase K (100 µg/ml). Lysates became clear after 45 min, whereupon 225 µl of ice cold NH4OAc (7.5 M) was added. Genomic DNA was extracted from lysates and purified using a conventional phenol chloroform extraction protocol [44], resuspended in 10 mM Tris-HCl (pH 8.0) and stored at −20°C. One µg of purified genomic DNA was digested using 20 U of restriction enzyme BamHI (New England Biolabs) for 16 h at 37°C and concentrated by ethanol precipitation before self-ligation using T4 DNA ligase (Promega) at 16°C overnight. The ligation product was used as the template for PCR with primers BR/IR0 and BL/IR0 according to manufacturer's directions (PfuUltra II Fusion HS DNA Polymerase, Agilent Technologies) and was isolated by gel electrophoresis (Figure S1). The band was purified using the QIAquick Gel Extraction Kit (Qiagen) and sequenced using the primers IR0 and BL. Amplicon sequencing indicated that the genomic region flanking the Tn5 insertion corresponds to cfa6. We confirmed the Tn5 insertion locus to be in cfa6 by PCR amplification of Pst DC3118 gDNA using Tn5- and cfa6-specific primer sets (Figure S1) and amplicon sequencing. The sequences of primers used here are: IR0 (5′-GCCGAAGAGAACACAGATTTAGC-3′), BL (5′-GGGGACCTTGCACAGATAGC-3′), BR (5′-CATTCCTGTAGCGGATGGAGATC-3′), cfa6F (5′-AGTCATGGACGGACAGGTTC-3′), and cfa6R (5′-CCAAGCTCTACGATTCCGAG-3′).
Callose deposition
Leaves from 4-week-old plants were infiltrated with water or 10 nM flg22 using a needleless syringe. After 24 h, about eight leaves from at least five independent plants were cleared and dehydrated with 100% ethanol. Leaves were fixed in an acetic acid∶ethanol (1∶3) solution for 2 h, sequentially incubated for 15 min in 75% ethanol, 50% ethanol, and 150 mM phosphate buffer, pH 8.0, and then stained overnight at 4°C in 150 mM phosphate buffer (pH 8.0) containing 0.01% (w/v) aniline blue supplemented with carbinicillin (100 µg/ml). After staining, leaves were mounted in 50% glycerol and examined by UV epifluorescence using an Axio Image M1 microscope (Zeiss). Callose quantification was performed using Image J software.
Oxidative burst in Arabidopsis seedlings
Active oxygen species released by seedlings were assayed by H2O2-dependent luminescence of luminol [45]. Two-week-old seedlings were transferred from liquid culture into 96-well plates and incubated overnight in 200 µl H2O supplemented with carbinicillin (100 µg/ml) at room temperature. The next morning, 100 µl H2O containing 20 µM luminol and 1 µg horseradish peroxidase (Sigma) was added and luminescence was measured for 20 min after the addition of the test solutions (100 nM flg22). Luminescence was detected using a Spectra Max L plate reader (Molecular Devices, Sunnyvale, CA). Seedlings were weighed to normalize results.
Quantification of SA by liquid chromatography-tandem mass spectrometry (LC/MS)
Approximately 100–300 mg (fresh weight) of leaf tissues were frozen in liquid nitrogen, ground, and extracted with 1 ml methanol∶water (1∶1 v/v) containing 0.1% formic acid and 0.1 g L−1 butylated hydroxytoluene (BHT) at 4°C for 24 h. Homogenates were mixed and centrifuged at 12,000×g for 10 min at 4°C. Supernatants were filtered through 0.2 µm PTFE membrane (Millipore, Bedford, MA) and transferred to autosampler vials. Injections of plant extracts (10 µl per injection) were separated on a fused core Ascentis Express C18 column (2.1×50 mm, 2.7 µm; Supelco, Bellefonte, PA) installed in the column heater of an Acquity Ultra Performance Liquid Chromatography (UPLC) system (Waters Corporation, Milford, MA). A gradient of 0.15% aqueous formic acid (solvent A) and methanol (solvent B) was applied in a 5-min program with a mobile phase flow rate of 0.4 ml/minute. The separation consisted of a linear increase from A∶B (9∶1, v/v) to 100% B. The column, which was maintained at 50°C, was interfaced to a Quattro Premier XE tandem quadrupole mass spectrometer (Waters Corporation, Milford, MA) equipped with electrospray ionization and operated in negative ion mode. The capillary voltage, cone voltage, and extractor voltage were set at 3 kV, 30 V, and 3V, respectively. The flow rates of cone gas and desolvation gas were 100 and 800 L h−1, respectively. The source temperature was 120°C and the desolvation temperature was 350°C. Propyl 4-hydroxybenzoate was added as the internal standard for quantification of SA. Transitions from deprotonated molecules to characteristic product ions were monitored for SA (m/z 137>93) and propyl 4-hydroxybenzoate (179>93). Collision energies and source cone potentials were optimized for each transition using QuanOptimize software. Peak areas were integrated, and the analytes were quantified based on standard curves generated from peak area ratios of analytes related to the corresponding internal standard. Data acquisition and processing were performed using Masslynx 4.1 software (Waters, Milford, MA).
TAIR accession numbers
Sequence data for genes and proteins described in this article can be found in The Arabidopsis Information Resource (TAIR, http://www.arabidopsis.org/) under the following ID numbers: ACT1 (Arabidopsis actin 1): At2g37620; OST1: At4g33950; SCORD3/EDS5: At4g39030; SCORD5/AtGCN20/AtABCF3: At1g64550.
Supporting Information
Pst DC3118 carries a Tn 5 insertion in the cfa6 gene and does not produce coronatine in planta. (A) A schematic diagram of gene structures surrounding the Tn5 insertion in the Pst DC3118 genome. B, BamHI. (B) PCR reactions showing that the Tn5 insertion is in the genome of Pst DC3118 but not that of wild-type Pst DC3000. Primers used were specific to cfa6 and Tn5, as indicated in (A). (C) Detection of coronatine in Col-0 plants infiltrated with Pst DC3000 or Pst DC3118 bacteria (1×108 CFU/ml) at various time points after infiltration.
(EPS)
Responses of L er and ost1-2 plants to Pst DC3118 (COR-deficient) and Pst DC3000 hrcC mutant (T3SS-deficient) when dip-inoculated at 1×108 CFU/ml. (A) Leaf appearance at 3 dpi. (B) Bacterial populations at 3 dpi. Statistical analyses for this and following figures are described in MATERIALS and METHODS.
(TIF)
scord mutants are not susceptible to Pst DC3000 hrcC mutant. Plants were dip-inoculated at 1×108 CFU/ml and assayed for bacteria populations at 3 dpi. No significant difference was detected between Col-7 and each of the scord mutants in Student's t-tests.
(EPS)
A T-DNA insertion is present in the first intron of EDS5 ( At4g39030 ) in the scord3 mutant. (A) The exon-intron structure of EDS5, showing positions of the T-DNA insertion and EDS5- (108 and 109) and T-DNA-specific (T-1 and T-3) primers. (B) PCR products generated with indicated primer sets and genomic DNA templates. (C) RT-PCR products using ACT1 (Arabidopsis actin 1; At2g37620)- or EDS5-specific primers (108 and 109) and cDNA from Col-7 or scord3 as template.
(EPS)
Similar phenotypes of eds5-1 and scord3 plants. (A) SA and SAG levels in leaves 12 h after infiltration with H2O or Pst DC3000 at 1×108 CFU/ml. Numbers represent means and standard errors (n = 4). (B) Populations of Pst DC3118 at day 3 after dip-inoculation at 1×108 CFU/ml. (C) and (D) Stomata apertures in response to Pst DC3118 at 1×108 CFU/ml (C) or ABA at 10 µM (D).
(EPS)
Phenotypes of F1 plants from scord3 and eds5-1 crossings. (A) Eight different F1 plants (designated 1, 2, 3, 5, 6, 7, 8, 9) were selected from five different crosses (designated I, II, III, IV, V) between scord3 and eds5-1. The Col-0 plant was designated as plant 4 in a blind assignment of numbers, and used as a control. (B) PCR products amplified with primers T-3 and 109 (see Figure S4) and genomic DNA from the nine plants selected in (A) as template. (C) Stomata apertures in leaf peels of the nine plants selected in (A) in response to Pst DC3118 at 1×108 CFU/ml. (D) SA and SAG levels in the leaves of the nine plants selected in (A).
(EPS)
Stomatal response to elf18 in scord5 plants. Stomatal apertures (µm) were measured after leaf peels had been incubated with buffer or elf18 (100 µM) for 1 h. One hundred µM of elf18 was used to ascertain that elf18-induced stomatal response of scord5 plants is lost even at this high concentration.
(EPS)
A phylogenetic tree of the SCORD5 (At1g64550/AtGCN20/AtABCF3) family of proteins. Sequences were retrieved from The Arabidopsis Information Resource (http://www.arabidopsis.org/), aligned with ClustalX (http://www.clustal.org) and viewed by Dendroscope (http://ab.inf.uni-tuebingen.de/software/dendroscope/).
(EPS)
Acknowledgments
We thank Dr. Xin Li (University of British Columbia, Vancouver, Canada) for ila-3 seeds. We also thank Karen Bird for editing the manuscript, Marlene Cameron for formatting figures, and members of the SYH lab for their comments on the manuscript.
Footnotes
The authors have declared that no competing interests exist.
This work was supported by funding from NIH (AI068718) and the Chemical Sciences, Geosciences and Biosciences Division, Office of Basic Energy Sciences, Office of Science, U.S. Department of Energy (DE-FG02-91ER20021) to SYH. The Quattro Premier mass spectrometer was purchased with funds from NSF (DBI-0619489) to ADJ. XG received support from a Michigan State University Plant Science Fellowship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Whalen M, Innes R, Bent A, Staskawicz B. Identification of Pseudomonas syringae pathogens of Arabidopsis thaliana and a bacterial gene determining avirulence on both Arabidopsis and soybean. Plant Cell. 1991;3:49–59. doi: 10.1105/tpc.3.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Preston GM. Pseudomonas syringae pv. tomato: the right pathogen, of the right plant, at the right time. Mol Plant Pathol. 2000;1:263–275. doi: 10.1046/j.1364-3703.2000.00036.x. [DOI] [PubMed] [Google Scholar]
- 3.Katagiri F, Thilmony R, He SY. The Arabidopsis thaliana-Pseudomonas syringae interaction. In: Somerville CR, Meyerowitz EM, editors. The Arabidopsis Book. Rockville: American Society of Plant Biologists; 2002. pp. 1–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitchell RE. Coronatine production by some phytopathogenic Pseudomonads. Physiol Plant Pathol. 1982;20:83–89. [Google Scholar]
- 5.Ma SW, Morris VL, Cuppels DA. Characterization of a DNA region required for production of the phytotoxin coronatine by Pseudomonas syringae pv. tomato. Mol Plant Microbe Interact. 1991;4:69–77. [Google Scholar]
- 6.Mittal S, Davis KR. Role of the phytotoxin coronatine in the infection of Arabidopsis thaliana by Pseudomonas syringae pv. tomato. Mol Plant Microbe Interact. 1995;8:165–171. doi: 10.1094/mpmi-8-0165. [DOI] [PubMed] [Google Scholar]
- 7.Bender CL, Alarcón-Chaidez F, Gross DC. Pseudomonas syringae phytotoxins: mode of action, regulation and biosynthesis by peptide and polyketide synthetases. Microbiol Mol Biol Rev. 1999;63:266–292. doi: 10.1128/mmbr.63.2.266-292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cintas NA, Koike ST, Bull CT. A new pathovar, Pseudomonas syringae pv. alisalensis, proposed for the causal agent of bacterial blight of broccoli and broccoli raab. Plant Dis. 2002;86:992–998. doi: 10.1094/PDIS.2002.86.9.992. [DOI] [PubMed] [Google Scholar]
- 9.Bender CL, Stone HE, Sims JJ, Cooksey DA. Reduced pathogen fitness of Pseudomonas syringae pv. tomato Tn5 mutants defective in coronatine production. Physiol Mol Plant Pathol. 1987;30:273–283. [Google Scholar]
- 10.Peñaloza-Vázquez A, Preston GM, Collmer A, Bender CL. Regulatory interactions between the hrp Type III protein secretion system and coronatine biosynthesis in Pseudomonas syringae pv. tomato DC3000. Microbiology. 2000;146:2447–2456. doi: 10.1099/00221287-146-10-2447. [DOI] [PubMed] [Google Scholar]
- 11.Brooks DM, Hernández-Guzmán G, Kloek AP, Alarcón-Chaidez F, Sreedharan A, et al. Identification and characterization of a well-defined series of coronatine biosynthetic mutants of Pseudomonas syringae pv. tomato DC3000. Mol Plant Microbe Interact. 2004;17:162–174. doi: 10.1094/MPMI.2004.17.2.162. [DOI] [PubMed] [Google Scholar]
- 12.Brooks DM, Bender CL, Kunkel BN. The Pseudomonas syringae phytotoxin coronatine promotes virulence by overcoming salicylic acid-dependent defenses in Arabidopsis thaliana. Mol Plant Pathol. 2005;6:629–639. doi: 10.1111/j.1364-3703.2005.00311.x. [DOI] [PubMed] [Google Scholar]
- 13.Cui J, Bahrami AK, Pringle EG, Hernandez-Guzman G, Bender CL, et al. Pseudomonas syringae manipulates systemic plant defenses against pathogens and herbivores. Proc Natl Acad Sci U S A. 2005;102:1791–1796. doi: 10.1073/pnas.0409450102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Melotto M, Underwood W, Koczan J, Nomura K, He SY. Plant stomata function in innate immunity against bacterial invasion. Cell. 2006;126:969–980. doi: 10.1016/j.cell.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 15.Uppalapati SR, Ishiga Y, Wangdi T, Kunkel BN, Anand A, et al. The phytotoxin coronatine contributes to pathogen fitness and is required for suppression of salicylic acid accumulation in tomato inoculated with Pseudomonas syringae pv. tomato DC3000. Mol Plant Microbe Interact. 2007;20:955–965. doi: 10.1094/MPMI-20-8-0955. [DOI] [PubMed] [Google Scholar]
- 16.Uppalapati SR, Ishiga Y, Wangdi T, Urbanczyk-Wochniak E, Ishiga T, et al. Pathogenicity of Pseudomonas syringae pv. tomato on tomato seedlings: phenotypic and gene expression analyses of the virulence function of coronatine. Mol Plant Microbe Interact. 2008;21:383–395. doi: 10.1094/MPMI-21-4-0383. [DOI] [PubMed] [Google Scholar]
- 17.Ishiga Y, Uppalapati SR, Ishiga T, Elavarthi S, Martin B, et al. The phytotoxin coronatine induces light-dependent reactive oxygen species in tomato seedlings. New Phytol. 2009;181:147–160. doi: 10.1111/j.1469-8137.2008.02639.x. [DOI] [PubMed] [Google Scholar]
- 18.Freeman BC, Beattle GA. Bacterial growth restriction during host resistance to Pseudomonas syringae is associated with leaf water loss and localized cessation of vascular activity in Arabidopsis thaliana. Mol Plant Microbe Interact. 2009;22:857–867. doi: 10.1094/MPMI-22-7-0857. [DOI] [PubMed] [Google Scholar]
- 19.Zeng W, He SY. A prominent role of the flagellin receptor FLAGELLIN-SENSING2 in mediating stomatal response to Pseudomonas syringae pv tomato DC3000 in Arabidopsis. Plant Physiol. 2010;153:1188–1198. doi: 10.1104/pp.110.157016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc Natl Acad Sci U S A. 2008;105:7100–7105. doi: 10.1073/pnas.0802332105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fonseca S, Chini A, Hamberg M, Adie B, Porzel A, et al. (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat Chem Biol. 2009;5:344–350. doi: 10.1038/nchembio.161. [DOI] [PubMed] [Google Scholar]
- 22.Sheard LB, Tan X, Mao H, Withers J, Nissan GB, et al. Mechanism of jasmonate recognition by an inositol phosphate-potentiated COI1-JAZ co-receptor. Nature. 2010;468:400–405. doi: 10.1038/nature09430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uppalapati SR, Ayoubi P, Weng H, Palmer DA, Mitchell RE, et al. The phytotoxin coronatine and methyl jasmonate impact multiple phytohormone pathways in tomato. Plant J. 2005;42:201–217. doi: 10.1111/j.1365-313X.2005.02366.x. [DOI] [PubMed] [Google Scholar]
- 24.Thilmony R, Underwood W, He SY. Genome-wide transcriptional analysis of the Arabidopsis thaliana interaction with the plant pathogen Pseudomonas syringae pv. tomato DC3000 and the human pathogen Escherichia coli O157:H7. Plant J. 2006;46:34–53. doi: 10.1111/j.1365-313X.2006.02725.x. [DOI] [PubMed] [Google Scholar]
- 25.Zhao Y, Thilmony R, Bender CL, Schaller A, He SY, et al. Virulence systems of Pseudomonas syringae pv. tomato promote bacterial speck disease in tomato by targeting the jasmonate signaling pathway. Plant J. 2003;36:485–499. doi: 10.1046/j.1365-313x.2003.01895.x. [DOI] [PubMed] [Google Scholar]
- 26.Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM. Glucosinolate metabolites required for an Arabidopsis innate immune response. Science. 2009;323:95–101. doi: 10.1126/science.1164627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Millet YA, Danna CH, Clay NK, Songnuan W, Simon MD, et al. Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns. Plant Cell. 2010;22:973–990. doi: 10.1105/tpc.109.069658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mustilli A-C, Merlot S, Vavasseur A, Fenzi F, Giraudat J. Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell. 2002;14:3089–3099. doi: 10.1105/tpc.007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie X, Wang Y, Williamson L, Holroyd GH, Tagliavia C, et al. The identification of genes involved in the stomatal response to reduced atmospheric relative humidity. Curr Biol. 2006;16:882–887. doi: 10.1016/j.cub.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 30.Desikan R, Cheung M-K, Clarke A, Golding S, Sagi M, et al. Hydrogen peroxide is a common signal for darkness- and ABA-induced stomatal closure in Pisum sativum. Funct Plant Biol. 2004;31:913–920. doi: 10.1071/FP04035. [DOI] [PubMed] [Google Scholar]
- 31.Nawrath C, Heck S, Parinthawong N, Métraux JP. EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell. 2002;14:275–286. doi: 10.1105/tpc.010376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosso MG, Li Y, Strizhov N, Riess B, Dekker K, et al. An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol. 2003;53:247–259. doi: 10.1023/B:PLAN.0000009297.37235.4a. [DOI] [PubMed] [Google Scholar]
- 33.Sánchez-Fernández R, Davies TGE, Coleman JOD, Rea PA. The Arabidopsis thaliana ABC protein superfamily, a complete inventory. J Biol Chem. 2001;276:30231–30244. doi: 10.1074/jbc.M103104200. [DOI] [PubMed] [Google Scholar]
- 34.Garcia O, Bouige P, Forestier C, Dassa E. Inventory and comparative analysis of rice and Arabidopsis ATP-binding cassette (ABC) systems. J Mol Biol. 2004;343:249–265. doi: 10.1016/j.jmb.2004.07.093. [DOI] [PubMed] [Google Scholar]
- 35.Verrier PJ, Bird D, Burla B, Dassa E, Forestier C, et al. Plant ABC proteins - a unified nomenclature and updated inventory. Trends Plant Sci. 2008;13:151–159. doi: 10.1016/j.tplants.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 36.Zeng W, Melotto M, He SY. Plant stomata: a checkpoint of host immunity and pathogen virulence. Curr Opin Biotechnol. 2010;21:599–603. doi: 10.1016/j.copbio.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Aldana CRV, Marton MJ, Hinnebusch AG. GCN20, a novel ATP binding cassette protein, and GCN1 reside in a complex that mediates activation of the eIF-2 alpha kinase GCN2 in amino acid-starved cells. EMBO J. 1995;14:3184–3199. doi: 10.1002/j.1460-2075.1995.tb07321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marton MJ, de Aldana CRV, Qiu H, Chakraburtty K, Hinnebusch AG. Evidence that GCN1 and GCN20, translational regulators of GCN4, function on elongating ribosomes in activation of eIF2α kinase GCN2. Mol Cell Biol. 1997;17:4474–4489. doi: 10.1128/mcb.17.8.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 40.Monaghan J, Li X. The HEAT repeat protein ILITYHIA is required for plant immunity. Plant Cell Physiol. 2010;51:742–753. doi: 10.1093/pcp/pcq038. [DOI] [PubMed] [Google Scholar]
- 41.de Torres-Zabala M, Truman W, Bennett MH, Lafforgue G, Mansfield JW, et al. Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signaling pathway to cause disease. EMBO J. 2007;26:1434–1443. doi: 10.1038/sj.emboj.7601575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weigel D, Ahn JH, Blazquez MA, Borevitz JO, Christensen SK, et al. Activation tagging in Arabidopsis. Plant Physiol. 2000;122:1003–1013. doi: 10.1104/pp.122.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang G, Zhang L, Birch RG. Rapid amplification and cloning of Tn5 flanking fragments by inverse PCR. Lett Appl Microbiol. 2000;31:149–153. doi: 10.1046/j.1365-2672.2000.00781.x. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook J, MacCallum P, Russell DW. Molecular Cloning: A Laboratory Manual, 3rd edition. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 45.Keppler LD, Baker CJ, Atkinson MM. Active oxygen production during a bacteria-induced hypersensitive reaction in tobacco suspension cells. Phytopathology. 1989;79:974–978. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pst DC3118 carries a Tn 5 insertion in the cfa6 gene and does not produce coronatine in planta. (A) A schematic diagram of gene structures surrounding the Tn5 insertion in the Pst DC3118 genome. B, BamHI. (B) PCR reactions showing that the Tn5 insertion is in the genome of Pst DC3118 but not that of wild-type Pst DC3000. Primers used were specific to cfa6 and Tn5, as indicated in (A). (C) Detection of coronatine in Col-0 plants infiltrated with Pst DC3000 or Pst DC3118 bacteria (1×108 CFU/ml) at various time points after infiltration.
(EPS)
Responses of L er and ost1-2 plants to Pst DC3118 (COR-deficient) and Pst DC3000 hrcC mutant (T3SS-deficient) when dip-inoculated at 1×108 CFU/ml. (A) Leaf appearance at 3 dpi. (B) Bacterial populations at 3 dpi. Statistical analyses for this and following figures are described in MATERIALS and METHODS.
(TIF)
scord mutants are not susceptible to Pst DC3000 hrcC mutant. Plants were dip-inoculated at 1×108 CFU/ml and assayed for bacteria populations at 3 dpi. No significant difference was detected between Col-7 and each of the scord mutants in Student's t-tests.
(EPS)
A T-DNA insertion is present in the first intron of EDS5 ( At4g39030 ) in the scord3 mutant. (A) The exon-intron structure of EDS5, showing positions of the T-DNA insertion and EDS5- (108 and 109) and T-DNA-specific (T-1 and T-3) primers. (B) PCR products generated with indicated primer sets and genomic DNA templates. (C) RT-PCR products using ACT1 (Arabidopsis actin 1; At2g37620)- or EDS5-specific primers (108 and 109) and cDNA from Col-7 or scord3 as template.
(EPS)
Similar phenotypes of eds5-1 and scord3 plants. (A) SA and SAG levels in leaves 12 h after infiltration with H2O or Pst DC3000 at 1×108 CFU/ml. Numbers represent means and standard errors (n = 4). (B) Populations of Pst DC3118 at day 3 after dip-inoculation at 1×108 CFU/ml. (C) and (D) Stomata apertures in response to Pst DC3118 at 1×108 CFU/ml (C) or ABA at 10 µM (D).
(EPS)
Phenotypes of F1 plants from scord3 and eds5-1 crossings. (A) Eight different F1 plants (designated 1, 2, 3, 5, 6, 7, 8, 9) were selected from five different crosses (designated I, II, III, IV, V) between scord3 and eds5-1. The Col-0 plant was designated as plant 4 in a blind assignment of numbers, and used as a control. (B) PCR products amplified with primers T-3 and 109 (see Figure S4) and genomic DNA from the nine plants selected in (A) as template. (C) Stomata apertures in leaf peels of the nine plants selected in (A) in response to Pst DC3118 at 1×108 CFU/ml. (D) SA and SAG levels in the leaves of the nine plants selected in (A).
(EPS)
Stomatal response to elf18 in scord5 plants. Stomatal apertures (µm) were measured after leaf peels had been incubated with buffer or elf18 (100 µM) for 1 h. One hundred µM of elf18 was used to ascertain that elf18-induced stomatal response of scord5 plants is lost even at this high concentration.
(EPS)
A phylogenetic tree of the SCORD5 (At1g64550/AtGCN20/AtABCF3) family of proteins. Sequences were retrieved from The Arabidopsis Information Resource (http://www.arabidopsis.org/), aligned with ClustalX (http://www.clustal.org) and viewed by Dendroscope (http://ab.inf.uni-tuebingen.de/software/dendroscope/).
(EPS)