Abstract
In asymmetrically dividing cells, a failure to coordinate cell polarity with the site of cell division can lead to cell fate transformations and tumorigenesis. Cell polarity in C. elegans embryos is defined by PAR proteins, which occupy reciprocal halves of the cell cortex. During asymmetric division, the boundary between the anterior and posterior PAR domains precisely matches the site of cell division, ensuring exclusive segregation of cell fate. The PAR domains determine the site of cell division by positioning the mitotic spindle, suggesting one means by which cell polarity and cell division might be coordinated. Here, we report that cell polarity and cell division are coordinated through an additional mechanism: the site of cell division repositions the PAR-2 boundary. Gα-mediated microtubule-cortex interactions appear to direct cortical flows of PAR-2 and myosin toward the site of cell division, which acts as a PAR-2 and myosin sink. Embryos with defects in PAR-2 boundary correction undergo mis-segregation of cortical polarity and cytoplasmic determinants, suggesting that PAR domain correction might help prevent cell fate transformation.
Keywords: PAR polarity, Asymmetric cell division, Cytokinesis, C. elegans
INTRODUCTION
Asymmetric cell divisions play essential roles throughout development in all organisms. They provide the basis of cellular differentiation and thus underlie numerous cell fate determination events. Three basic processes ensure that the resulting daughter cells have different fates (Gonczy and Hyman, 1996; Nelson, 2003). First, a polarity axis is established in response to a cue; the formation of a cortical domain marks the polarity axis. Second, fate determinants segregate according to the cortical polarity axis. Third, the mitotic spindle aligns parallel to the polarity axis and positions the cytokinesis furrow. The cortical polarity domain and fate determinants are restricted to one daughter cell.
The importance of coordinating cortical polarity domains with the site of cell division is emphasized by the finding that spindle misorientation relative to the polarity axis can prove tumorigenic in Drosophila (for a review, see Gonzalez, 2007; Knoblich, 2008; Zhong and Chia, 2008). Mutations in the tumor suppressors lethal giant larvae (lgl), scribble (scrib) and discs large (dlg) and regulators of centrosome duplication and spindle orientation [overexpressed Polo kinase and partner of inscuteable (pins) mutants] exhibit defects in asymmetric cell division and undergo overproliferation of stem cell lineages in the brain, leading to massive tumors that can continue to divide indefinitely (Basto et al., 2008; Caussinus and Gonzalez, 2005; Gonzalez, 2007; Knoblich, 2008). In wild-type neuroblasts, a crescent of the fate determinant Numb and its adaptors Pon and Miranda is generated at the basal cortex upon the onset of mitosis through activation of the conserved Bazooka/Par3–Par6–aPKC complex. The cortical polarity defined by the PAR complex is also responsible for spindle orientation, ensuring parallel alignment of the spindle and polarity axes. In the mutant neuroblasts, the spindle axis is randomized relative to the cell polarity axis and cell polarity is distributed randomly during division. In some cases, both daughter cells inherit polarity that would normally be restricted to one self-renewing daughter cell. The downstream effect of this mis-segregation of polarity is continued cell division in both of the daughters and their progeny.
Spindle misorientation does not always lead to symmetric proliferative divisions and tumors. Drosophila PAR polarity mutants such as bazooka (baz or Par3), aPKC and inscuteable (insc) use a correction process known as telophase rescue to position cortical polarity according to the spindle (Cai et al., 2003; Siegrist and Doe, 2007). At the onset of mitosis in these mutant neuroblasts, spindle orientation is random and the Numb crescent is absent. In late mitosis, Numb is recruited to the cortex overlying one of the spindle poles through interactions between the astral microtubules and the cortex. The kinesin motor protein Khc73 and the microtubule-binding protein Mud facilitate formation of a cortical domain of Gαi and its regulator Pins, thereby establishing cell polarity (for a review, see Knoblich, 2008; Siegrist and Doe, 2005). After telophase rescue, the spindle and polarity axes lie in parallel and the mutant neuroblasts can divide asymmetrically. It remains unclear why some polarity defects are corrected by telophase rescue but others are not.
We sought to address, first, if polarity correction mechanisms exist in other asymmetrically dividing cells; second, where the positional information for correction comes from and how it is transduced; and finally, what circumstances eliminate the ability to correct cortical polarity during cell division. Previous work proposed that C. elegans spn-4 mutant embryos coordinate the cell polarity axis with spindle orientation even when spindle orientation is defective (Gomes et al., 2001) but the mechanism remained unexamined. C. elegans embryos provide several advantages for investigating how polarity domains are corrected during asymmetric cell division, offering high spatial and temporal resolution as well as the possibility of both genetic and physical manipulations of the cell division machinery.
C. elegans embryos undergo a series of asymmetric cell divisions to establish the founding cells of five developmental lineages (for a review, see Gonczy and Rose, 2005). One-cell embryos establish a polarity axis shortly after fertilization (for a review, see Cowan and Hyman, 2007), thereby defining the anterior and posterior of the embryo. One-cell embryos divide: the anterior blastomere of the two-cell embryo is already restricted in its developmental potential, whereas the posterior blastomere is not. The posterior blastomere divides three more times in a series of stem cell-like divisions (reviewed by Strome, 2005).
These asymmetric divisions depend on cell polarity defined by PAR proteins (reviewed by Cowan and Hyman, 2004b; Gonczy and Rose, 2005; Schneider and Bowerman, 2003). PAR-3, PAR-6 and aPKC (PKC-3) localize to the anterior half of the one-cell embryo and PAR-2 and PAR-1 localize to the posterior half. The anterior and posterior PAR domains are mutually exclusive. The PAR proteins control the segregation of fate determinants and position the spindle. During division, the anterior PAR proteins are inherited by the anterior daughter cell and the posterior PAR proteins are inherited by the posterior daughter cell. This exclusive segregation of polarity requires that the position of the boundary between the two PAR domains is coordinated with the site of cell division. It has been assumed that because the PAR domains position the spindle, they ensure their own exclusive segregation, but this has not been demonstrated.
We addressed the question of how polarity domains are segregated during asymmetric division by examining the behavior of the PAR domains in response to different genetic or mechanical perturbations. Using mutants with either large or small posterior domains, we have found that a domain correction process requiring Gα and its regulators GPR-1/2 and LIN-5 acts during division to reposition the boundary between the PAR domains to match the cytokinesis furrow. The Gα pathway regulates microtubule-cortex interactions and thereby large-scale cortical reorganization that moves PAR-2 toward the furrow. In cases of extreme spindle positioning defects, cortical polarity is mis-segregated and cells divide symmetrically, suggesting the corrective function can be distorted. Correction of cortical polarity domains is likely to be a general mechanism in asymmetrically dividing cells and might help differentiate between symmetric and asymmetric cell divisions in more complex systems.
MATERIALS AND METHODS
C. elegans strains
All C. elegans strains were maintained at 16°C on NGM (nematode growth media) with E. coli OP50 as a food source. Strain genotypes and details of their construction can be found in Table 1.
Table 1.
Worm strains used in this study
RNAi and mutant analysis
RNAi was performed either by injection or feeding as detailed in Table S1 in the supplementary material. Our previous work showed that approximately half of cye-1(RNAi) and cdk-2(RNAi) embryos exhibit no cortical PAR-2 domain throughout the entire cell cycle in one-cell embryos (Cowan and Hyman, 2006). Only cye-1(RNAi)/cdk-2(RNAi) embryos that exhibited cortical PAR-2 prior to anaphase onset were included in the analysis. For analysis of temperature-sensitive zyg-8(b235ts) and spd-1(oj5ts) alleles, worms were shifted to 25°C at the fourth larval (L4) stage and incubated at the non-permissive temperature for at least 24 hours prior to recording. For generating partial centrosome duplication phenotypes, mixed-stage zyg-1(b1ts) males and hermaphrodites were shifted to 25°C for 16-24 hours, at which point, L4 and young adult males were selected and mated to GFP::PAR-2;GFP::SPD-2 hermaphrodites for at least 20 hours at 25°C. A homozygous deletion mutant of F26H9.2 is available, but the mutant does not reproducibly exhibit the large posterior domain phenotype (C.R.C., unpublished observations). We have not characterized the mutant in any further detail.
Mechanical manipulations
Displacement of the spindle and cytokinesis furrow using a needle was performed as described (Bringmann et al., 2007). Briefly, using a motorized micromanipulator, a microinjection capillary needle was laid on the free surface of the egg, roughly perpendicular to the long axis of the embryo. The needle was gradually pressed onto the embryo until the eggshell surface was deformed and normal spindle movement was prevented. To generate posterior spindles and furrows, the needle was applied at the time of pronuclear meeting. To generate anterior spindles and furrows, the needle was applied at the maximum anterior position of the centrosome-pronuclei complex, during centration.
The laser ablation experiments were performed on a custom-built setup (developed by Katrin Heinze, IMP) based on an inverted epifluorescence microscope (Zeiss Axiovert 200M) and a UV-VIS-IR 63× C-Apochromat 1.25 NA lens (Zeiss), and equipped with a cooled CCD camera (CoolSnap HQ, Photometrics) and LED illumination (465 nm and 565 nm, CoolLED, Precise Excite). The stage was controlled by an XYZ driver (ASI Inc.). The ablation laser (365 nm, 50 mW, Teem Photonics) was focused through the rear slider port. An external pulse generator (1000 Hz, Pc_Lab2000SE, Velleman Components) was used to control the laser, and ablation was regulated by an external shutter (Thor Labs) placed immediately before the laser path entered the microscope and controlled via the computer. Continuous pulses were used to cut the anterior kinetochore microtubules during early anaphase (determined by posterior spindle pole movement toward the cortex) and then destroy the anterior centrosome to generate maximum anterior displacement of the astral microtubule furrow (Bringmann and Hyman, 2005).
Time-lapse microscopy, drug treatments and image display
Worms were dissected in 0.1 M NaCl 4% sucrose solution on coverslips to release early embryos and mounted directly on 2% agarose (0.1 M NaCl 4% sucrose) pads on standard microscope slides. For nocodazole treatments, worms were cut open at the vulva in a solution of 7.5 μg/ml nocodazole 0.1 M NaCl 4% sucrose on a coverslip. The cut worms were incubated in a humid chamber for 10-20 minutes and then further dissected to release embryos and mounted as described. All images were acquired on Zeiss wide-field fluorescence microscopes with 40× Plan NeoFluor 1.3 NA objectives equipped with a cooled CCD camera (ORCA-ER, Hamamatsu or CoolSnap HQ, Photometrics) and appropriate filters. All components and image acquisition were controlled by MetaMorph (Molecular Devices) software. Image intensity scaling, rotation, cropping and QuickTime movie assembly were performed either directly in MetaMorph, imported as .stk files into ImageJ64 (1.41o, NIH) or imported as TIFF files into GraphicConverter (LemkeSoft).
Phenotype quantification and data analysis
Measurements and point tracking were performed directly in MetaMorph or imported into ImageJ64 (.stk format). Data was exported to Octave (v. 2.9.17, www.gnu.org/software/octave) for further analysis. Graphs were generated in Octave and AquaTerm directly or with the graph tool in Adobe Illustrator using the exported data. Values are given as mean ± standard deviation. Times given relative to anaphase onset were determined by spindle pole movement, chromosome splitting and/or time elapsed relative to nuclear envelope breakdown, as appropriate to the GFP strain and phenotype. Owing to an apparent change in the time of cortical reorganization in nocodazole-treated embryos relative to controls, times are indicated relative to nuclear envelope breakdown, judged by the change in nuclear GFP intensity.
Kymographs were generated from points picked manually and represent the distance from the posterior pole to the point projected onto the anterior-posterior axis (depicting the linear extension of PAR-2, for example, but not the absolute size of the PAR-2 domain). Kymograph data is from a single representative embryo, unless otherwise stated. For clarity, only one PAR-2 boundary line is shown. The PAR-2 domain was assigned based on the change in GFP signal intensity along the anterior-posterior cortex: intensity values 20% or more above the anterior intensity were considered to be the PAR-2 domain. The boundary was defined as the anterior-most position in the continuous domain.
The linear placement of the PAR-2 domain boundary, centrosomes and cytokinesis furrow in partial centriole duplication embryos [zyg-1(b1ts)-paternal] were calculated at the onset of furrow ingression as the distance from the posterior pole to the point projected onto the anterior-posterior axis, as for kymographs. Centrosome size was estimated from the amount of GFP::SPD-2 fluorescence.
The size of the PAR-2 domain relative to embryo or P1 cell circumference was calculated as the length of PAR-2 around the cortex as a fraction of the total length of the cortex at the completion of furrow ingression, roughly 1:30 after anaphase onset. The distance of the PAR-2 domain boundary to the cytokinesis furrow was calculated as the linear distance along the cortex as a fraction of the length of embryo long axis approximately one minute after the onset of furrow ingression. A lower threshold of 75% and a higher threshold of 1% were applied to the images and the presence of PAR-2 was scored as regions in which the remaining signal was (1) at the cortex (judged from corresponding brightfield or DIC images) and (2) of higher intensity than the neighboring internal cytoplasm. Corresponding DIC images were used to confirm the furrow position.
Cortical PAR-2 and PAR-6 intensities were measured as the maximum values of a 10-pixel-wide line at the completion of cytokinesis. The line included the invagination where the furrow bisected the embryo but did not include the furrow itself. The intensity values were standardized to cortex length and to the maximum intensity along the line.
Kymographs of cortical PAR-2 and NMY-2 intensities were generated by tracking the maximum intensity along a 20-pixel-wide line drawn along the anterior-posterior cortex (mid-embryo focal plane) at each time point. The data for individual embryos was averaged along the cortex length to give 200 bins. Data from n embryos was combined and averaged over time intervals of 10 seconds.
RESULTS
The site of cell division repositions the PAR domain boundary
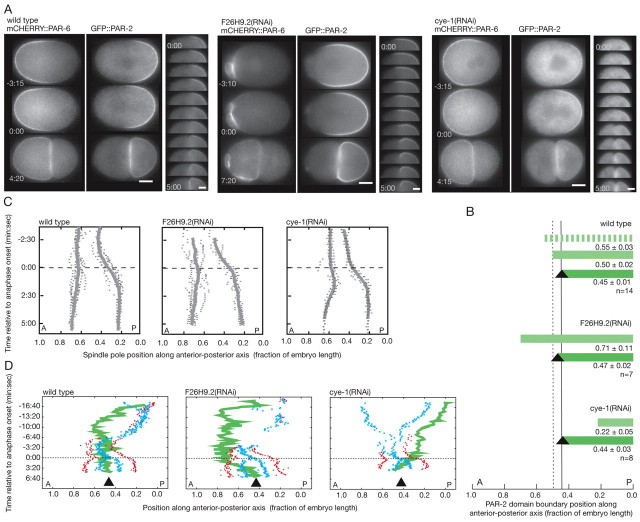
How are the PAR domains segregated in C. elegans embryos so that one cell inherits the anterior PAR domain and the other cell the posterior PAR domain? In one-cell embryos, the boundary between the anterior and posterior PAR domains is located mid-way along the embryo axis but cell division is asymmetric, displaced toward the posterior (Fig. 1A,B). To investigate how cell polarity and cell division are coordinated, we looked through RNAi-based screen data (Gunsalus et al., 2004; Piano et al., 2000; Sonnichsen et al., 2005; Zipperlen et al., 2001) to find phenotypes in which the PAR boundary is displaced from the middle of the embryo. The position of the PAR boundary is coincident with the position of the pseudocleavage furrow, a deep but transient cortical ingression that reaches its maximum in the middle of the embryo, roughly ten minutes after polarity is initiated (for a review, see Cowan and Hyman, 2004b). We identified several gene products that, when depleted, lead to either anterior or posterior displacement of the pseudocleavage furrow. Further analysis of these RNAi phenotypes using time-lapse microscopy of GFP::PAR-2, a component of the posterior PAR domain, and mCHERRY::PAR-6, a member of the anterior PAR complex, indicated that the PAR domain boundary was displaced away from the embryo middle (Fig. 1A; see Movie 1 in the supplementary material). After cell division in these domain-size mutants, however, PAR-2 occupied only the cortex of the posterior cell and PAR-6 was located only in the cortex of the anterior cell, as in wild type. Two possibilities could explain the exclusive inheritance of cell polarity in PAR domain-size mutants: the site of cell division moved to match the altered position of the PAR domain boundary or the PAR domain boundary moved to match the site of cell division.
Fig. 1.
Mis-sized PAR domains are corrected during cell division. Wild-type (left), F26H9.2(RNAi) big PAR-2 domain (middle) and cye-1(RNAi)/cdk-2(RNAi) small PAR-2 domain (right) embryos. (A) Time-lapse images of GFP::PAR-2 and mCHERRY::PAR-6, marking the posterior and anterior PAR domains, respectively. F26H9.2(RNAi) embryos exhibit increased cortical activity in the anterior domain as the domain size decreases. Higher temporal resolution time-lapse sequences (30 second intervals) of GFP::PAR-2 are shown to the right. (B) Posterior PAR-2 domain size in domain-size mutants. Striped green, maximum PAR-2 domain; light green, PAR-2 domain at nuclear envelope breakdown; dark green, PAR-2 domain after cell division; black triangles, cytokinesis furrow position; vertical dotted line, wild-type PAR boundary position at nuclear envelope breakdown; vertical solid line, wild-type cytokinesis furrow position. (C) Spindle pole position in domain-size mutants. Kymographs were generated by tracking centrosome position in embryos expressing GFP::PAR-2;GFP::γ-tubulin;GFP::histone or GFP::PAR-2;GFP::SPD-2. Gray x's, individual positions; gray lines, average position (wild type, n=7; F26H9.2(RNAi), n=5; cye-1(RNAi), n=6). (D) PAR-2 boundary position relative to the cell cycle. Green lines, PAR-2 boundary (the domain extends to the posterior pole); red dots, centrosomes; blue x's, pronuclei/chromosomes; black triangles, cytokinesis furrow position. In cye-1(RNAi) embryos, the PAR-2 domain continues to expand throughout the cell cycle but the rate of PAR-2 boundary movement increases significantly at anaphase onset. In A, C and D, times are indicated relative to anaphase onset. A, anterior; P, posterior; posterior pole, 0.0. Scale bars: 10 μm.
To determine how the PAR domain boundary and the site of cell division were coordinated, we first examined whether the change in the position of the PAR boundary resulting from mis-sized PAR domains affected the site of cell division. We measured anaphase spindle displacement and the position of the cytokinesis furrow from time-lapse movies of wild type and PAR domain-size mutant embryos. We selected one large domain mutant, F25H9.2(RNAi), and two indistinguishable small domain mutants, cye-1(RNAi) and cdk-2(RNAi) (Cowan and Hyman, 2006), for further analysis. Embryos depleted of the predicted actin and phosphatase regulator F26H9.2 (www.wormbase.org) exhibited large posterior domains extending to approximately 70% embryo length (Fig. 1A,B; see Movie 1 in the supplementary material). However, spindle displacement and the site of cytokinesis were not significantly affected in F26H9.2(RNAi) embryos compared with wild type (Fig. 1B,C). Similarly, in cyclin E- or CDK-2-depleted [cye-1(RNAi) or cdk-2(RNAi)] embryos that established polarity late in the cell cycle and thus had small posterior domains at the time of mitosis (Fig. 1A,B; see Movie 1 in the supplementary material), spindle displacement and the position of cytokinesis were similar to wild-type embryos (Fig. 1B,C). The position of the PAR domain boundary, as determined by domain size, had little influence on the site of cell division.
It seemed probable that the PAR domain boundary was repositioned to match the site of cell division. We analyzed the position of the PAR domain boundary relative to the site of cell division by time-lapse microscopy of GFP::PAR-2. In F26H9.2(RNAi) embryos, the PAR boundary was located in the anterior embryo half prior to cell division (Fig. 1). As the cell began to divide, the PAR boundary shifted posteriorly to match the site of cell division (Fig. 1A,D; see Movie 1 in the supplementary material). In cye-1(RNAi) or cdk-2(RNAi) embryos exhibiting small posterior domains, the PAR boundary was in the posterior embryo half prior to cell division (Fig. 1). The PAR-2 domain expanded during mitosis until the domain boundary matched the site of cell division (Fig. 1A,D; see Movie 1 in the supplementary material). Although the PAR-2 domain expanded throughout the cell cycle in cye-1(RNAi) and cdk-2(RNAi) embryos owing to delayed polarity establishment, the rate at which PAR-2 moved changed significantly during division (Fig. 1D). Similar PAR domain size correction can be observed in other mutants with enlarged posterior domains, such as rga-3/4(RNAi) embryos (Schmutz et al., 2007; Schonegg et al., 2007), in two-cell embryos with spindle orientation defects (Gomes et al., 2001) and in subsequent divisions of the germline lineage in wild-type C. elegans embryos (see Figs S1 and S2 in the supplementary material; see Movie 2 in the supplementary material). C. elegans embryos appear to match the PAR domain boundary to the site of cell division.
The PAR domain boundary responds to the position of the cytokinesis furrow
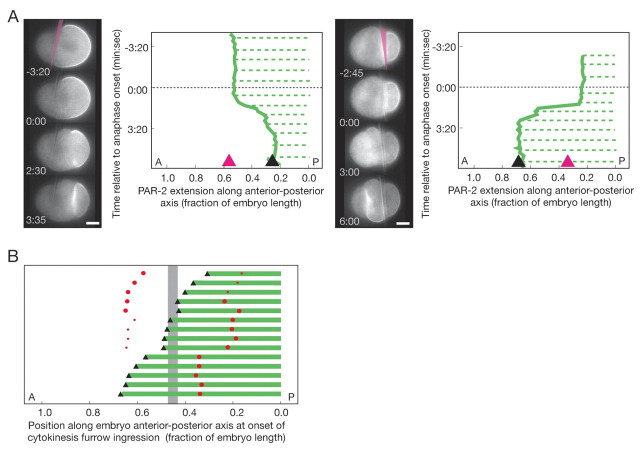
What is the positional information that corrects the PAR domain boundary during cell division? Two general mechanisms could provide a spatial signal: a cell division-independent landmark or a cell division-dependent cue, such as the spindle or cytokinesisfurrow. We tested the existence of a static cellular landmark by asking whether a misplaced site of cell division could reposition a correctly positioned PAR domain boundary. Because the posterior PAR-2 boundary is sharper than the PAR-6 boundary and thus easier to identify, we focused on the PAR-2 domain in subsequent experiments. First, we displaced the site of cell division toward either the anterior or posterior by pushing the spindle with a glass needle (see Materials and methods). The cleavage furrow formed in the anterior- or posterior-third of the embryo. After anaphase onset, regardless of the direction or extent of displacement, the PAR-2 domain boundary shifted to match the site of cell division (anterior spindle, n=4; posterior spindle, n=7; Fig. 2A; see Movie 3 in the supplementary material). Second, we generated spindles with varying spindle pole sizes by partially inhibiting centriole duplication through partial inactivation of ZYG-1, an essential regulator of this process (O'Connell et al., 2001), using a conditional mutant allele. The resulting asymmetric spindles led to a range of cleavage sites. Following more complete zyg-1 inactivation, centriole duplication did not occur, leaving only one centrosome, and the furrow was displaced to the anterior. In all cases, the PAR-2 domain boundary matched the site of cytokinesis furrow initiation (n=14; Fig. 2B; see Movie 4 in the supplementary material). The PAR-2 domain boundary can be repositioned to match the site of cell division, regardless of location within the cell.
Fig. 2.
The site of cell division repositions the PAR-2 domain boundary. (A) Time-lapse images and kymographs of the GFP::PAR-2 domain following manual spindle displacement using a glass needle (pink in the first images). The spindle was pushed toward the posterior (left panel) or the anterior (right panel). Green solid lines, PAR-2 boundary; green dotted lines, cortical PAR-2; black triangle, cytokinesis furrow position; pink triangle, needle position. Times are indicated relative to anaphase onset. (B) Linear position of the PAR-2 domain (green bar), cytokinesis furrow (black triangle) and centrosomes (red dots) in zyg-1(b1ts) embryos with asymmetric (partial loss-of-function) or monopolar (complete loss-of-function) spindles. Bars represent individual embryos. The size of centrosome dots indicates the relative size of GFP::SPD-2 centrosomes (normal versus small centrosome) but not absolute size. The vertical gray bar indicates the range of furrow positions in wild-type embryos. A, anterior; P, posterior; posterior pole, 0.0. Scale bars: 10 μm.
The PAR-2 domain boundary matched the furrow in embryos with a single centrosome [zyg-1(b1)], thus lacking a mitotic spindle. Similarly, embryos depleted of microtubules do not form a spindle but the posteriorly located centrosomes can still induce furrow ingression in the anterior (Werner et al., 2007). The PAR-2 boundary matched the furrow position in embryos depleted of microtubules (Fig. 3A), indicating that the cytokinesis furrow, rather than the spindle, is important for PAR domain correction. When cytokinesis furrow ingression was prevented by depletion of the actin nucleation factor CYK-1, the PAR-2 domain boundary fluctuated after anaphase onset and eventually shrank toward the posterior pole (Fig. 3A). Together, these experiments indicate that the cytokinesis furrow positions the PAR domain boundary during cell division.
Fig. 3.
The PAR-2 boundary and cytokinesis furrow respond to the same positional information. (A) Time-lapse images and kymographs of GFP::PAR-2 in wild-type, tba-4 + tbb-2(RNAi) and cyk-1(RNAi) embryos. Green solid lines, PAR-2 boundary; green dotted lines, cortical PAR-2; black triangle, cytokinesis furrow position. Only the anterior-most furrow in the tba-4 + tbb-2(RNAi) embryos is indicated. The pink asterisk marks the approximate position of the centrosomes. (B) Time-lapse images, kymograph and cortical intensity distribution of the GFP::PAR-2 domain following asymmetric spindle cutting. Only the upper cortex is indicted. Left panel: green solid lines, PAR-2 boundary; green dotted lines, cortical PAR-2; light gray line/arrowhead, aster-positioned cytokinesis furrow; dark gray line/arrowhead, midzone-positioned cytokinesis furrow. Right panel: green lines, PAR-2 intensity along anterior-posterior cortex; light gray line, aster-positioned cytokinesis furrow; dark gray line, midzone-positioned cytokinesis furrow. Spindle microtubules were cut between the metaphase chromosomes and the anterior centrosome, followed by ablation of the anterior centrosome. Times are indicated relative to anaphase onset. The gap in B corresponds to the ablation. A, anterior; P, posterior; posterior pole, 0.0. Scale bars: 10 μm.
In C. elegans embryos, the cytokinesis furrow is positioned by two signals: one from the spindle pole asters and one from the spindle midzone (Bringmann and Hyman, 2005; Dechant and Glotzer, 2003; Verbrugghe and White, 2004; Werner et al., 2007). Normally these two signals converge, generating one furrow. The positions of the two signals can be separated by asymmetrically cutting the spindle with a laser, leading to two spatially and temporally distinct furrows (Bringmann and Hyman, 2005). The first furrow forms in response to the asters and the second in response to the midzone. To see whether the PAR domain boundary distinguished between the aster and midzone furrows, we followed the PAR-2 boundary position relative to the two furrows following asymmetric spindle cutting (n=3; Fig. 3B; see Movie 6 in the supplementary material). The PAR-2 boundary shifted anteriorly to match the first furrow specified by the asters. When the second midzone-specified furrow ingressed, the PAR-2 boundary retracted slightly from the astral furrow but did not precisely match the midzone furrow. The astral cytokinesis furrow appears to dominate PAR-2 boundary positioning.
PAR domain correction depends on Gα, GPR-1/2 and LIN-5
What are the molecular requirements for PAR domain correction? The PAR-2 domain boundary appeared to respond to the astral cytokinesis furrow and so we asked whether molecules involved in positioning the furrow are required to position the PAR domain boundary. At the onset of anaphase in embryos depleted of two redundant Gα subunits [goa-1+gpa-16(RNAi)] required for the astral signal (Bringmann et al., 2007; Dechant and Glotzer, 2003), the PAR-2 domain boundary shrank toward the posterior pole of the embryo, and after division, the PAR-2 domain occupied only a subset of the posterior cell circumference (Fig. 4A,B; see Movie 7 in the supplementary material). A similar failure in domain correction was observed in embryos lacking the conserved Gα regulator GPR-1/2 (AGS3/Pins) (Gotta et al., 2003; Colombo et al., 2003; Srinivasan et al., 2003) and the Gα effector LIN-5 (NuMA/Mud) (Srinivasan et al., 2003) (see Movie 7 in the supplementary material). LET-99, a DEP-domain protein required for aster-dependent cytokinesis (Bringmann et al., 2007), also appears to affect PAR domain boundary repositioning (H.B. and C.R.C., unpublished observations), but the role of LET-99 in spindle orientation makes this difficult to assess accurately. By contrast, in embryos defective for the midzone signal components ZEN-4 [zen-4(RNAi)] or SPD-1 [spd-1(oj5)] (Raich et al., 1998; Verbrugghe and White, 2004), the PAR-2 domain boundary matched the site of the cytokinesis furrow (see Fig. S2 and Movie 8 in the supplementary material). These experiments indicate that PAR domain correction depends on Gα and two members of a conserved Gα signaling pathway, GPR-1/2 and LIN-5.
Fig. 4.

Gα is required for PAR-2 domain correction. (A) Time-lapse images and kymographs of the GFP::PAR-2 domain in wild-type embryos and embryos depleted of Gα [goa-1+gpa-16(RNAi)], and Gα-depleted embryos treated with the microtubule depolymerizing drug nocodazole. Green solid lines, PAR-2 boundary; green dotted lines, cortical PAR-2; black triangle, cytokinesis furrow position. The pink asterisk marks the approximate position of the centrosomes. The reduced cortical PAR-2 intensity following nocodazole treatment in Gα-depleted embryos is unlikely to be responsible for domain correction: gpr-1/2(RNAi) embryos showed reduced cortical PAR-2 but failed to correct PAR-2 (see Movie 8 in the supplementary material). (B) PAR-2 domain size following cell division in goa-1+gpa-16(RNAi) (red squares), gpr-1/2(RNAi) (pink squares) and lin-5(RNAi) (orange squares) embryos. The amount of PAR-2 was calculated as a percentage of the circumference of the P1 cell at the completion of furrow ingression. In wild-type embryos, PAR-2 occupies the entire cortex of the posterior cell following division, indicated by the black dotted line. (C) Time-lapse images of PIE-1::GFP in two-cell wild-type embryos and embryos lacking Gα [goa-1+gpa-16(RNAi)]. In Gα-depleted embryos, spindle orientation in the posterior cell of two-cell embryos is defective. In A and C, times are indicated relative to anaphase onset or, in the case of nocodazole-treated embryos, nuclear envelope breakdown. A, anterior; P, posterior; posterior pole, 0.0. Scale bars: 10 μm.
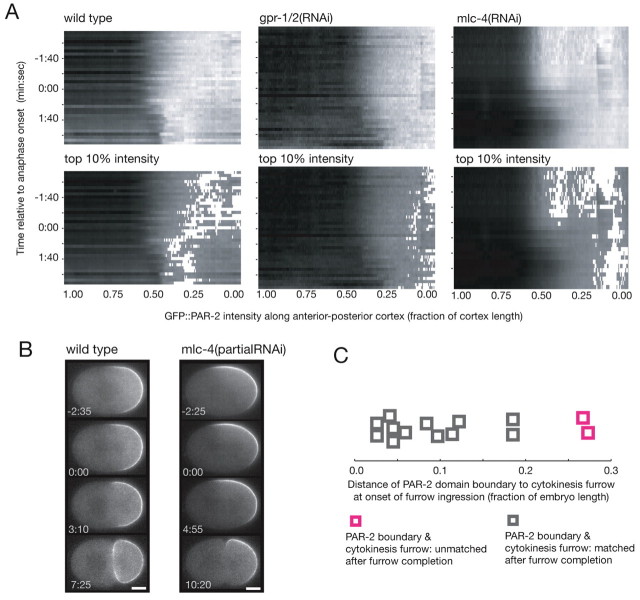
Gα-dependent cytokinesis furrow specification involves the reorganization of the cell cortex, including a domain of the Gα regulator GPR-1/2 (Bringmann et al., 2007). By tracking the cortical intensity of PAR proteins during domain correction, we found that the PAR-2 domain also undergoes reorganization: the maximum PAR-2 concentration shifted from the posterior pole to the nascent furrow position during anaphase (Fig. 5A; see Fig. S3 in the supplementary material). The morphology of the PAR-6 domain did not appear to change (see Fig. S3 in the supplementary material). At the same time, cortical myosin moved toward the nascent furrow, predominantly from the posterior (see Fig. S4 in the supplementary material). In goa-1+gpa-16(RNAi), gpr-1/2(RNAi) and lin-5(RNAi) embryos, the distribution of PAR-2 within the shrinking posterior domain did not change in anaphase: the maximum PAR-2 intensity remained near the posterior pole (Fig. 5A; see Fig. S3 in the supplementary material; data not shown). The PAR-6 domain extended into the posterior as the PAR-2 domain shrank (see Fig. S3 in the supplementary material). Cortical myosin did not exhibit furrow-directed movement in embryos depleted of Gα, GPR-1/2 or LIN-5 (see Fig. S4 in the supplementary material). These experiments suggest that during cortical domain correction, Gα is required to shift the distribution of PAR-2 toward the cytokinesis furrow. This cortical reorganization is accompanied by a coincident Gα-dependent myosin flow.
Fig. 5.
Domain correction involves myosin-dependent large-scale cortical reorganization. (A) Kymographs of cortical GFP::PAR-2 intensity in wild-type (n=6), gpr-1/2(RNAi) (n=6) and mlc-4(RNAi) (n=5) embryos. In the upper panels, gray values correspond to PAR-2 levels (black, low; white, high). In the bottom panels, the gray range was lowered and the top 10% intensity values are shown in white. Kymographs represent mean values from n embryos. (B) Time-lapse images of GPF::PAR-2 in wild-type embryos and embryos partially depleted of the myosin regulatory light chain MLC-4 [mlc-4(partialRNAi)]. In about one third of mlc-4(partialRNAi) embryos examined, the PAR-2 axis was tilted with respect to the long axis of the cell and subsequent division site, represented by the embryo shown. (C) Capacity for domain correction relative to the distance of the PAR-2 domain boundary to the furrow in embryos partially depleted of myosin contractility [mlc-4(partialRNAi)].
The coincidence of PAR-2 redistribution and furrow-directed myosin flows, both of which required Gα, suggested a role for cortical flow in domain correction. We therefore analyzed PAR-2 domain reorganization in embryos with reduced acto-myosin contractility, the mechanical force thought to drive cortical flow. The myosin regulatory light chain MLC-4 is required for cortical contractility, cortical flow and furrow ingression. Embryos strongly depleted of mlc-4 fail to establish polarity (Shelton et al., 1999). Embryos partially depleted of mlc-4 [mlc-4(partialRNAi)] could establish a normal PAR-2 cortical domain (Fig. 5B; see Fig. S2 in the supplementary material), although the polarity axis was sometimes tilted with respect to the long axis of the cell in contrast to wild-type embryos. During anaphase in mlc-4(partialRNAi) embryos, the PAR-2 domain did not exhibit an anterior shift in PAR-2 intensity (Fig. 5A). If the PAR-2 domain boundary was within 0.25 embryo length of the cytokinesis furrow, the boundary shifted to match the furrow position. If the PAR-2 domain boundary was further from the cytokinesis furrow, the boundary did not correct (Fig. 5B,C). The limited contractility remaining in mlc-4(partialRNAi) embryos might be sufficient for small shifts in the position of the PAR-2 domain boundary. Large-scale reorganization of the PAR-2 domain and bigger shifts in boundary position both appear to depend on myosin-driven cortical contractility, consistent with the finding that Gα-directed cortical flows are required for domain correction.
During cell division, Gα has an established role controlling spindle position through regulation of microtubule-cortex interactions. To see whether Gα-dependent cortical reorganization is controlled through Gα-dependent microtubule regulation as opposed to direct regulation of contractility, we asked whether we could rescue the lack of domain correction in Gα-depleted embryos by depolymerizing microtubules. We treated goa-1+gpa-16(RNAi) embryos with the microtubule poison nocodazole and assessed the position of the PAR-2 domain boundary and cortical myosin during cell division. Whereas Gα-depleted embryos do not undergo cortical reorganization during anaphase, Gα-depleted embryos treated with nocodazole underwent PAR-2 domain correction and furrow-directed myosin flow. Thus, Gα controls microtubule-cortex interactions that facilitate large-scale cortical reorganizations, the probable mechanical basis of PAR-2 domain boundary repositioning during cell division.
PAR domain size and spindle orientation might limit domain correction
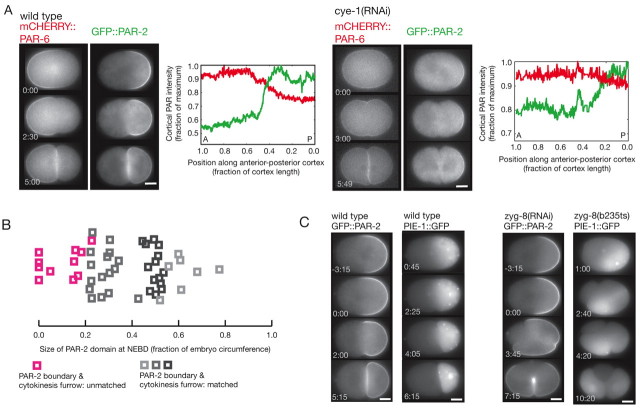
Not all embryos with altered domain sizes repositioned the PAR domain boundary to match the site of cell division. We found that cye-1(RNAi) and cdk-2(RNAi) embryos did not correct PAR domains if the PAR-2 domain occupied less than ~20% cell circumference (n=7 out of 20; Fig. 6A,B; see Movie 9 in the supplementary material). A similar size limit was seen in mex-5(RNAi) embryos, which often exhibited small PAR-2 domains (S. Reiter and C.R.C., unpublished observations). In embryos in which the PAR-2 domain extended into both nascent daughter cells owing to spindle misorientation [zyg-8(RNAi) or zyg-8(b235ts)], PAR-2 expanded around the cortex of both cells (Fig. 6C), but only when the PAR-2 domain occupied 30% or more of the cortex bounded by the cytokinesis furrow (n=25 out of 28 cortical segments). The mis-segregation of PAR-2 in zyg-8(−) embryos often correlated with mis-segregation of cell fate determinants, such that both daughter cells expressed the germline marker PIE-1 [n=4 out of 8 zyg-8(b235) embryos; Fig. 6C]. Equal segregation of PAR-2 also occurred if the cytokinesis furrow was induced to bisect the domain by physically manipulating spindle orientation in genetically wild-type embryos (n=2; see Fig. S5 and Movie 10 in the supplementary material). Thus, either a small PAR-2 domain or spindle misorientation relative to the PAR-2 domain reduced the ability of the embryo to undergo domain correction and exclusively segregate polarity. Although we cannot rule out a size-independent mechanism, these results are consistent with a model in which PAR-2 domain size influences whether or not the domain boundary will be corrected: if the domain size is above a threshold, it expands to match the furrow, and if it is below a threshold, it does not.
Fig. 6.
Small PAR-2 domains and severe spindle misorientation are associated with defects in domain correction. (A) Time-lapse images and intensity distribution of mCHERRY::PAR-6 and GFP::PAR-2 in wild-type and small PAR-2 domain [cye-1(RNAi)] embryos. The cortical intensities of PAR-2 (green) and PAR-6 (red) along one lateral half of the embryo at the completion of cytokinesis are shown. The PAR domain boundary did not match the furrow in the cye-1(RNAi) embryo. After division, the PAR-2 and PAR-6 domains overlapped. A, anterior; P, posterior; posterior pole, 0.0. (B) Size of the PAR-2 domain relative to the capacity for domain correction in one-cell embryos. The embryos analyzed include wild-type (dark gray), F26H9.2(RNAi) (light gray) and cye-1(RNAi)/cdk-2(RNAi) (medium gray and pink) embryos. (C) Time-lapse images of GFP::PAR-2 and PIE-1::GFP in wild-type and zyg-8(RNAi) or zyg-8(b235ts) embryos. In embryos defective for the microtubule regulator ZYG-8, the spindle moves into a transverse orientation in the posterior during anaphase (Gonczy et al., 2001) and the plane of cell division intersects the posterior cortical domain. In the zyg-8(−) embryos, three furrows are visible. The PAR-2 boundary responds to two furrows but the middle furrow bisects the PAR-2 domain. PIE-1 segregates to both the anterior and posterior cells. Times are indicated relative to anaphase onset. Scale bars: 10 μm.
Together, our experiments show that C. elegans embryos posses a cortical domain correction mechanism that ensures the PAR boundary matches the site of cell division. The PAR boundary responds to the position of the cytokinesis furrow by following cortical myosin flows into the furrow. Gα-mediated microtubule-cortex interactions facilitate cortical flow and thus domain correction. Embryos defective for Gα exhibit cell fate transformations in subsequent embryonic divisions (n=5 out of 8; Fig. 4C), suggesting that domain correction might be important to maintain cell fates. Domain correction fails either when the PAR-2 domain is small or when the spindle axis is misoriented relative to the polarity axis, indicating that the corrective capacity is limited.
DISCUSSION
Our data demonstrate that the boundary between PAR polarity domains is repositioned during anaphase to match the site of cell division. This correction of cortical polarity domains ensures that the two resulting cells inherit the appropriate complement of PAR proteins and thus the capacity for differentiation or for continued asymmetric division. Initially, the PAR domains determine the position of the spindle (Grill et al., 2001; Kemphues et al., 1988). Later, the spindle indirectly determines the position of the cytokinesis furrow. Finally, the cytokinesis furrow controls the position of the PAR domain boundary. This circular regulatory pathway ensures the proper segregation of PAR domains during cell division.
PAR domain correction entails large-scale reorganization of the cortex, such that cortical myosin and PAR-2 exhibit bulk movement toward the cytokinesis furrow. These flows of cortical myosin and PAR-2 never moved through the furrow but rather the furrow appeared to act as a myosin and PAR-2 sink. Cortical flows depended on Gα, GPR-1/2 and LIN-5, members of a conserved Gα signaling complex. This molecular requirement for cortical flow is largely shared by cytokinesis furrow specification in response to signals from the astral microtubules (Fig. 4) (Bringmann et al., 2007; Dechant and Glotzer, 2003; Werner et al., 2007), suggesting that PAR domain correction and aster-dependent cytokinesis might result from the same cortical reorganization. By contrast, midzone-specified furrowing is thought to involve local control of the cortex and thus might have a limited capacity to influence the PAR-2 domain.
Gα appeared to control cortical flows indirectly through its role in regulating microtubule-cortex interactions as depolymerization of microtubules could restore cortical flows in embryos lacking Gα. Stable microtubules generally suppress contractility (for a review, see Mandato et al., 2000). In the posterior, Gα might help promote microtubule instability, which could facilitate spindle movement and increase cortical contractility. The spatial asymmetry in contractility arising from the displacement of the spindle toward the posterior might be sufficient to induce cortical flow along the contractility gradient, away from the posterior pole (Bray and White, 1988) (reviewed by Cowan and Hyman, 2004b). It remains to be determined how PAR-2 localizes to the cell cortex and the mechanical basis by which PAR-2 is coupled to cortical myosin flows.
In C. elegans embryos and Drosophila neuroblasts, spindle orientation and polarity correction mechanisms both depend on heterotrimeric Gα, its regulator GPR-1/2/Pins and LIN-5/Mud (Siegrist and Doe, 2005). In vertebrates, spindle orientation is coordinated with cell polarity (Chang et al., 2007; Fleming et al., 2007; Konno et al., 2008; Lamprecht et al., 1986; Lechler and Fuchs, 2005; Yingling et al., 2008), as in C. elegans and Drosophila. Likewise, regulators of heterotrimeric G proteins have been shown to mediate spindle orientation during asymmetric divisions in birds and mammals (Morin et al., 2007; Sanada and Tsai, 2005). In both the epidermal and neural progenitor divisions in the mouse brain, the orientation of the mitotic spindle relative to polarity cues correlates with the fate of the daughter cells. Inhibition of G protein regulators interferes with correct spindle orientation and alters cell fates (Sanada and Tsai, 2005). The emphasis in these studies has been on the alignment of the spindle relative to the existing cell polarity. The contribution of the Gα pathway to correcting cell polarity remains unexplored in vertebrate systems.
Given that cell polarity can be corrected to match the site of cell division, why does mis-segregation of cell polarity occur? We have demonstrated that in C. elegans embryos, limitations in the correction process might contribute to the mis-segregation of cortical polarity in two ways: when correction cannot act or when correction is active in the inappropriate cell. In cases where the PAR-2 domain size was small or in embryos depleted of Gα, the PAR-2 domain boundary did not match the furrow. In cases where the spindle was misaligned with respect to the polarity axis, PAR-2 domains exceeded the apparent size threshold in both daughter cells and PAR-2 expanded around both to match the furrow. Either type of mis-segregation could alter cell fate segregation (Figs 4, 6). By analogy, Drosophila mutant neuroblasts that exhibit symmetric proliferative divisions might result when domain thresholds were surpassed in both nascent cells, either owing to spindle misorientation, changes in domain size, or both, leading to correction acting in both daughters.
Two distinct outcomes were observed in embryos that accumulated PAR-2 in the anterior cortex. In large PAR-2 domain [F26H9.2(RNAi)] embryos, PAR-2 disappeared from the anterior and was inherited by only one cell. In zyg-8(−) embryos, PAR-2 expanded around the anterior and was inherited by both daughter cells. In both cases, the PAR-2 domain boundary appeared to move toward the cytokinesis furrow, suggesting that correction was active. One difference between the two conditions was the presence of PAR-2 on the cortex overlying the spindle pole, raising the possibility that astral microtubule interactions with the PAR-2 cortex determine domain expansion. Gα-regulated microtubule-cortex interactions indeed appear to be essential for domain correction (Fig. 4) and PAR polarity is thought to direct Gα activity (Colombo et al., 2003; Gotta et al., 2003), suggesting a mechanistic connection between PAR-2 domain size and Gα-induced cortical flow. Alternatively, another domain property that we could not detect, such as protein amount, might be monitored. Further experiments will be necessary to understand what cortical properties determine domain correction and how those properties are sensed.
Cortical domain size thresholds provide a useful basis for considering how stem cells maintain the capacity for both proliferation through symmetric division and differentiation through asymmetric division (for a review, see Morrison and Kimble, 2006). The domain size perceived during division appears to be determined by the orientation of cell division relative to the polarity axis. Cells can ensure either symmetric or asymmetric division by controlling the site of cell division relative to polarity, for instance, by regulating spindle orientation. Such a domain size threshold-based mechanism might be relevant in the embryonic mouse brain, for example, where a precise distinction between spindle orientation during symmetric and asymmetric divisions has been difficult to determine (for a review, see Zhong and Chia, 2008).
Supplementary Material
Acknowledgements
Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, funded by the NIH National Center for Research Resources. The authors thank A. Schwager, H. Wada and A. Zinke for worm maintenance; J. Kong and S. Ernst for strain construction; A. Göppert for lin-5 and gpr-1/2 feeding constructs. C.R.C. thanks J. Knoblich and N. Goehring for discussions, suggestions and comments on the manuscript. Research in the laboratory of A.A.H. is funded by the Max Planck Society and the DFG. H.B. was funded in part by a BI Fonds student fellowship. Research at the IMP is supported by Boehringer Ingelheim.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.040436/-/DC1
References
- Basto R., Brunk K., Vinadogrova T., Peel N., Franz A., Khodjakov A., Raff J. W. (2008). Centrosome amplification can initiate tumorigenesis in flies. Cell 133, 1032-1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray D., White J. G. (1988). Cortical flows in animal cells. Science 239, 883-888 [DOI] [PubMed] [Google Scholar]
- Bringmann H., Hyman A. A. (2005). A cytokinesis furrow is positioned by two consecutive signals. Nature 436, 731-734 [DOI] [PubMed] [Google Scholar]
- Bringmann H., Cowan C. R., Kong J., Hyman A. A. (2007). LET-99, GOA-1/GPA-16, and GPR-1/2 are required for aster-positioned cytokinesis. Curr. Biol. 17, 185-191 [DOI] [PubMed] [Google Scholar]
- Cai Y., Yu F., Lin S., Chia W., Yang X. (2003). Apical complex genes control mitotic spindle geometry and relative size of daughter cells in Drosophila neuroblast and pI asymmetric divisions. Cell 112, 51-62 [DOI] [PubMed] [Google Scholar]
- Caussinus E., Gonzalez C. (2005). Induction of tumor growth by altered stem-cell asymmetric division in Drosophila melanogaster. Nat. Genet. 37, 1125-1129 [DOI] [PubMed] [Google Scholar]
- Chang J. T., Palanivel V. R., Kinjyo I., Schambach F., Intlekofer A. M., Banerjee A., Longworth S. A., Vinup K. E., Mrass P., Oliaro J., et al. (2007). Asymmetric T lymphocyte division in the initiation of adaptive immune responses. Science 315, 1687-1691 [DOI] [PubMed] [Google Scholar]
- Colombo K., Grill S. W., Kimple R. J., Willard F. S., Siderovski D. P., Gönczy P. (2003). Translation of polarity cues into asymmetric spindle positioning in Caenorhabditis elegans embryos. Science 300, 1957-1961 [DOI] [PubMed] [Google Scholar]
- Cowan C. R., Hyman A. A. (2004a). Centrosomes direct cell polarity independently of microtubule assembly in C. elegans embryos. Nature 431, 92-96 [DOI] [PubMed] [Google Scholar]
- Cowan C. R., Hyman A. A. (2004b). Asymmetric cell division in C. elegans: cortical polarity and spindle positioning. Annu. Rev. Cell Dev. Biol. 20, 427-453 [DOI] [PubMed] [Google Scholar]
- Cowan C. R., Hyman A. A. (2006). Cyclin E-Cdk2 temporally regulates centrosome assembly and establishment of polarity in Caenorhabditis elegans embryos. Nat. Cell Biol. 8, 1441-1447 [DOI] [PubMed] [Google Scholar]
- Cowan C. R., Hyman A. A. (2007). Acto-myosin reorganization and PAR polarity in C. elegans. Development 134, 1035-1043 [DOI] [PubMed] [Google Scholar]
- Dechant R., Glotzer M. (2003). Centrosome separation and central spindle assembly act in redundant pathways that regulate microtubule density and trigger cleavage furrow formation. Dev. Cell 4, 333-344 [DOI] [PubMed] [Google Scholar]
- Fleming E. S., Zajac M., Moschenross D. M., Montrose D. C., Rosenberg D. W., Cowan A. E., Tirnauer J. S. (2007). Planar spindle orientation and asymmetric cytokinesis in the mouse small intestine. J. Histochem. Cytochem. 55, 1173-1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes J. E., Encalada S. E., Swan K. A., Shelton C. A., Carter J. C., Bowerman B. (2001). The maternal gene spn-4 encodes a predicted RRM protein required for mitotic spindle orientation and cell fate patterning in early C. elegans embryos. Development 128, 4301-4314 [DOI] [PubMed] [Google Scholar]
- Gonczy P., Hyman A. A. (1996). Cortical domains and the mechanisms of asymmetric cell division. Trends Cell. Biol. 6, 382-387 [DOI] [PubMed] [Google Scholar]
- Gonczy P., Rose L. S. (2005). Asymmetric cell division and axis formation in the embryo. WormBook, 1-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonczy P., Bellanger J. M., Kirkham M., Pozniakowski A., Baumer K., Phillips J. B., Hyman A. A. (2001). zyg-8, a gene required for spindle positioning in C. elegans, encodes a doublecortin-related kinase that promotes microtubule assembly. Dev. Cell 1, 363-375 [DOI] [PubMed] [Google Scholar]
- Gonzalez C. (2007). Spindle orientation, asymmetric division and tumour suppression in Drosophila stem cells. Nat. Rev. Genet. 8, 462-472 [DOI] [PubMed] [Google Scholar]
- Gotta M., Dong Y., Peterson Y. K., Lanier S. M., Ahringer J. (2003). Asymmetrically distributed C. elegans homologs of AGS3/PINS control spindle position in the early embryo. Curr. Biol. 13, 1029-1037 [DOI] [PubMed] [Google Scholar]
- Grill S. W., Gonczy P., Stelzer E. H., Hyman A. A. (2001). Polarity controls forces governing asymmetric spindle positioning in the Caenorhabditis elegans embryo. Nature 409, 630-633 [DOI] [PubMed] [Google Scholar]
- Gunsalus K. C., Yueh W. C., MacMenamin P., Piano F. (2004). RNAiDB and PhenoBlast: web tools for genome-wide phenotypic mapping projects. Nucleic Acids Res. 32, D406-D410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemphues K. J., Priess J. R., Morton D. G., Cheng N. S. (1988). Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell 52, 311-320 [DOI] [PubMed] [Google Scholar]
- Knoblich J. A. (2008). Mechanisms of asymmetric stem cell division. Cell 132, 583-597 [DOI] [PubMed] [Google Scholar]
- Konno D., Shioi G., Shitamukai A., Mori A., Kiyonari H., Miyata T., Matsuzaki F. (2008). Neuroepithelial progenitors undergo LGN-dependent planar divisions to maintain self-renewability during mammalian neurogenesis. Nat. Cell Biol. 10, 93-101 [DOI] [PubMed] [Google Scholar]
- Lamprecht J., Zieba P., Strojny P. (1986). Spatial orientation of the mitotic apparatus and its stability in a polarized epithelial cell. A computer-assisted morphometric analysis. Anat. Embryol. 175, 129-135 [DOI] [PubMed] [Google Scholar]
- Lechler T., Fuchs E. (2005). Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature 437, 275-280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandato C. A., Benink H. A., Bement W. M. (2000). Microtubule-actomyosin interactions in cortical flow and cytokinesis. Cell Motil. Cytoskel. 45, 87-92 [DOI] [PubMed] [Google Scholar]
- Morin X., Jaouen F., Durbec P. (2007). Control of planar divisions by the G-protein regulator LGN maintains progenitors in the chick neuroepithelium. Nat. Neurosci. 10, 1440-1448 [DOI] [PubMed] [Google Scholar]
- Morrison S. J., Kimble J. (2006). Asymmetric and symmetric stem-cell divisions in development and cancer. Nature 441, 1068-1074 [DOI] [PubMed] [Google Scholar]
- Nelson W. J. (2003). Adaptation of core mechanisms to generate cell polarity. Nature 422, 766-774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell K. F., Caron C., Kopish K. R., Hurd D. D., Kemphues K. J., Li Y., White J. G. (2001). The C. elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell 105, 547-558 [DOI] [PubMed] [Google Scholar]
- Piano F., Schetter A. J., Mangone M., Stein L., Kemphues K. J. (2000). RNAi analysis of genes expressed in the ovary of Caenorhabditis elegans. Curr. Biol. 10, 1619-1622 [DOI] [PubMed] [Google Scholar]
- Raich W. B., Moran A. N., Rothman J. H., Hardin J. (1998). Cytokinesis and midzone microtubule organization in Caenorhabditis elegans require the kinesin-like protein ZEN-4. Mol. Biol. Cell 9, 2037-2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanada K., Tsai L. H. (2005). G protein betagamma subunits and AGS3 control spindle orientation and asymmetric cell fate of cerebral cortical progenitors. Cell 122, 119-131 [DOI] [PubMed] [Google Scholar]
- Schmutz C., Stevens J., Spang A. (2007). Functions of the novel RhoGAP proteins RGA-3 and RGA-4 in the germ line and in the early embryo of C. elegans. Development 134, 3495-3505 [DOI] [PubMed] [Google Scholar]
- Schneider S. Q., Bowerman B. (2003). Cell polarity and the cytoskeleton in the Caenorhabditis elegans zygote. Annu. Rev. Genet. 37, 221-249 [DOI] [PubMed] [Google Scholar]
- Schonegg S., Constantinescu A. T., Hoege C., Hyman A. A. (2007). The Rho GTPase-activating proteins RGA-3 and RGA-4 are required to set the initial size of PAR domains in Caenorhabditis elegans one-cell embryos. Proc. Natl. Acad. Sci. USA 104, 14976-14981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton C. A., Carter J. C., Ellis G. C., Bowerman B. (1999). The nonmuscle myosin regulatory light chain gene mlc-4 is required for cytokinesis, anterior-posterior polarity, and body morphology during Caenorhabditis elegans embryogenesis. J. Cell Biol. 146, 439-451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegrist S. E., Doe C. Q. (2005). Microtubule-induced Pins/Galphai cortical polarity in Drosophila neuroblasts. Cell 123, 1323-1335 [DOI] [PubMed] [Google Scholar]
- Siegrist S. E., Doe C. Q. (2007). Microtubule-induced cortical cell polarity. Genes Dev. 21, 483-496 [DOI] [PubMed] [Google Scholar]
- Sonnichsen B., Koski L. B., Walsh A., Marschall P., Neumann B., Brehm M., Alleaume A. M., Artelt J., Bettencourt P., Cassin E., et al. (2005). Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434, 462-469 [DOI] [PubMed] [Google Scholar]
- Srinivasan D. G., Fisk R. M., Xu H., van den Heuvel S. (2003). A complex of LIN-5 and GPR proteins regulates G protein signaling and spindle function in C. elegans. Genes Dev. 17, 1225-1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S. (2005). Specification of the germ line. WormBook, 1-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbrugghe K. J., White J. G. (2004). SPD-1 is required for the formation of the spindle midzone but is not essential for the completion of cytokinesis in C. elegans embryos. Curr. Biol. 14, 1755-1760 [DOI] [PubMed] [Google Scholar]
- Wallenfang M. R., Seydoux G. (2000). Polarization of the anterior-posterior axis of C. elegans is a microtubule-directed process. Nature 408, 89-92 [DOI] [PubMed] [Google Scholar]
- Werner M., Munro E., Glotzer M. (2007). Astral signals spatially bias cortical myosin recruitment to break symmetry and promote cytokinesis. Curr. Biol. 17, 1286-1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yingling J., Youn Y. H., Darling D., Toyo-Oka K., Pramparo T., Hirotsune S., Wynshaw-Boris A. (2008). Neuroepithelial stem cell proliferation requires LIS1 for precise spindle orientation and symmetric division. Cell 132, 474-486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W., Chia W. (2008). Neurogenesis and asymmetric cell division. Curr. Opin. Neurobiol. 18, 4-11 [DOI] [PubMed] [Google Scholar]
- Zipperlen P., Fraser A. G., Kamath R. S., Martinez-Campos M., Ahringer J. (2001). Roles for 147 embryonic lethal genes on C. elegans chromosome I identified by RNA interference and video microscopy. EMBO J. 20, 3984-3992 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.