Abstract
Schizophrenia is a serious brain disease of uncertain etiology. A role for retroviruses in the etiopathogenesis of some cases of schizophrenia has been postulated on the basis of clinical and epidemiological observations. We found sequences homologous to retroviral pol genes in the cell-free cerebrospinal fluids (CSFs) of 10 of 35 (29%) individuals with recent-onset schizophrenia or schizoaffective disorder. Retroviral sequences also were identified in the CSFs of 1 of 20 individuals with chronic schizophrenia. However, retroviral sequences were not identified in any of the CSFs obtained from 22 individuals with noninflammatory neurological diseases or from 30 individuals without evidence of neurological or psychiatric diseases (χ2 = 19.25, P < 0.001). The nucleotide sequences identified in the CSFs of the individuals with schizophrenia or schizoaffective disorder were related to those of the human endogenous retroviral (HERV)-W family of endogenous retroviruses and to other retroviruses in the murine leukemia virus genus. Transcription of RNA homologous to members of the HERV-W family of retroviruses also was found to be up-regulated differentially in the frontal cortex regions of brains obtained postmortem from individuals with schizophrenia, as compared with corresponding tissue from individuals without psychiatric diseases. The transcriptional activation of certain retroviral elements within the central nervous system may be associated with the development of schizophrenia in at least some individuals. The further characterization of retroviral elements within the central nervous system of individuals with schizophrenia might lead to improved methods for the diagnosis and management of this disorder.
Schizophrenia is a pervasive neuropsychiatric disease of uncertain etiology. In the United States, schizophrenia has a lifetime prevalence of greater than 1% and results in the annual expenditure of more than 65 billion dollars (1, 2). Family and adoption studies have demonstrated a significantly increased risk of schizophrenia in individuals who have a first-degree birth relative with this disease, indicating that there is a substantial genetic component in disease pathogenesis. Epidemiological studies have identified a number of environmental factors associated with the development of schizophrenia, including perinatal infections, winter–spring birth, household crowding, and upbringing in urban areas (3, 4). These genetic and environmental findings can be reconciled by the concept that the disease can result from infectious processes occurring in genetically susceptible individuals (5–8).
Retroviruses have been hypothesized as one of the infectious agents involved in the pathogenesis of schizophrenia (9–11). Humans can be infected with retroviruses such as strains of human immunodeficiency virus (HIV) and human T-cell leukemia virus. These exogenous retroviruses can replicate within the central nervous system and cause neurological and psychiatric symptoms in some infected individuals. The clinical response to infection with HIV is determined, to some extent, by the genetic susceptibility of the infected individual (12–14).
The human genome also contains many endogenous retroviral (HERV) elements, which have homology to known animal retroviruses. These elements probably arose from reverse transcriptase-mediated integration into the germ line of progenitors of Homo sapiens after exogenous retroviral infection. Many of the HERV sequences in the human genome are incomplete, although some full-length proviral genomic sequences have been identified. Some of these proviruses contain long open reading frames capable of encoding complete viral proteins and engendering viral particles (15–20). The tissue-specific expression of HERVs has been associated with a number of chronic human diseases, including multiple sclerosis, diabetes, and autoimmune arthritis (21–23). We report the identification of retroviral sequences in cerebrospinal fluids (CSFs) obtained from individuals with recent-onset schizophrenia, and the differential transcriptional up-regulation of members of the HERV-W family of endogenous retroviruses in the postmortem frontal cortex of individuals with schizophrenia.
Materials and Methods
CSF Samples.
Patients with new-onset schizophrenia.
We obtained CSF samples from 35 previously healthy individuals (presenting to the Department of Psychiatry at the University of Heidelberg) with symptoms consistent with new-onset schizophrenia or schizoaffective disorder as defined by the Diagnostic and Statistical Manual of Mental Disorders, 4th Ed. (25). These individuals had not been admitted previously to the hospital for schizophrenia and were without manifestations of acute infectious, inflammatory, or neurological diseases before their admission. The median age of these individuals was 25 years (range 18–48). CSF samples were collected by standard lumbar puncture procedures after admission to the hospital, clinical stabilization, and the obtaining of informed consent. The samples were obtained a median of 14 days (range 1–43) after admission, immediately aliquoted, and stored at −80°C until analyzed.
Patients with chronic schizophrenia.
Samples were obtained also from 20 individuals with chronic schizophrenia or schizoaffective disorder who were inpatients at St. Patrick's Hospital, County Roscommon, Ireland (24). All of these individuals were clinically stable and on medication at the time consent was obtained. The age range of the patients was 23–65 years, and they had been ill for a mean of 14.6 years (range 1–37) before the samples were obtained. None of the individuals had significant medical problems other than their psychiatric disorder. Samples were stored at −80°C.
Patients with noninflammatory neurological disorders.
CSF samples were obtained also from 22 individuals (who presented to the Department of Neurology of the Johns Hopkins School of Medicine) for the evaluation of noninflammatory neurological disorders. A total of 14 of these individuals had a diagnosis of pseudotumor cerebri and 8 had a diagnosis of normal-pressure hydrocephalus. Samples were obtained by using standard lumbar puncture techniques, were aliquoted, and were stored at −80°C until testing.
Controls.
To address the question of potential regional differences, two control groups were recruited. (i) CSF samples were obtained from 12 individuals undergoing spinal anesthesia at the Department of Surgery, University of Heidelberg. None of these patients had evidence of mental illness after thorough clinical and standardized psychiatric evaluation, including the German version of the structured clinical interview for the Diagnostic and Statistical Manual of Mental Disorders, 4th Ed. (25). These samples were processed and stored in a manner identical to that of the samples obtained from the individuals with recent-onset schizophrenia. (ii) At St. Patrick's Hospital, County Roscommon, Ireland, 18 healthy staff members volunteered to undergo lumbar puncture. The age range of the volunteers was 21–46 years, and the group was composed of an equal number of males and females. None of these individuals had any evidence of psychiatric, neurological, or acute medical illness. These samples were processed and stored in a manner identical to that of the samples from the individuals with chronic schizophrenia.
Informed consent was obtained from all study participants. All studies were approved by the local ethical committees.
CSF Preparation.
Cell-free supernatant was prepared from a 400-μl aliquot of CSF by centrifugation at 1,000 × g for 30 min at 4°C. Particle-associated RNA was pelleted from the cell-free supernatant by centrifugation at 100,000 × g for 60 min at 4°C, extracted from the pellet by reaction with guanidine isothiocyanate (Qiagen, Chatsworth, CA), homogenized by spinning through a shredding device (QIAshredder, Qiagen), and purified by using silica gel-based spin columns (RNeasy kit, Qiagen). The RNA was eluted in 50 μl of RNase-free water and stored at −70°C until further processing.
Brain Tissue Samples.
Frontal cortex tissue, Brodmann's area 10, was obtained postmortem from five individuals with schizophrenia. This brain region was chosen because the involvement of the frontal cortices in schizophrenia is accepted generally (8, 26).
These individuals had a median age of death of 35 years (range 30–56) and mean duration of illness of 18.6 years (range 8–32). Corresponding frontal cortex tissue also was obtained from six individuals with no history of psychiatric disease. The median age of death of these individuals was 46 years (range 34–59). These samples were part of the Stanley Brain Collection (Bethesda, MD). The methods used for obtaining and processing brain samples from this collection have been described (27).
Tissue Preparation.
Total RNA was isolated from 0.5 g of tissue by reaction with guanidinium isothiocyanate and phenol (GIBCO/BRL) and homogenization, according to the manufacturer's instructions. Contaminating DNA was removed by treatment with 30 units of RNase-free DNase I (Roche Molecular Biochemicals) in DNase buffer for 15 min at 37°C followed by the addition of 20 mM EDTA and extraction with phenol/chloroform/isoamyl alcohol and subsequent precipitation with ethanol. Pelleted RNA was reconstituted in RNase-free water. The brain samples were documented to be free of DNA by the demonstration of the absence of intron-containing c-myc DNA by nested PCR (28).
cDNA Synthesis.
First-strand cDNA was generated from a 10-μl aliquot (CSF) or 1 μg of RNA (tissue) by using Moloney murine leukemia virus reverse transcriptase (Super Script II, GIBCO/BRL) and an oligo(dT) primer in combination with the SMART II oligonucleotide (SMART PCR cDNA synthesis kit, CLONTECH). The second strand was then generated in a 15- (tissue) or 22-cycle (CSF) PCR to yield 100 μl of double-stranded cDNA, according to the manufacturer's instructions.
Pan-Retroviral PCR.
The oligonucleotide primers and conditions used were derived from those described by Tuke et al. (29). The first PCR mixture was performed by amplifying 1 μl of the double-stranded cDNA reaction with the following reagents: 1 μM primer PAN-UO (5′-CTTGGATCCTGGAAAGTGC/TTA/GCCC/AC-3′)/1 μM primer PAN-Dl (5′-CTCAAGCTTCAGC/GAT/GGTCATCCAT/CGTA-3′)/0.5 unit of thermostable DNA polymerase (AmpliTaq Gold, Perkin–Elmer). The above was brought to a final volume of 25 μl with a PCR mix containing 0.2 mM dNTPs/2.5 mM MgCl2/1× PER buffer II (Perkin–Elmer). The PCR was performed in a Thermal Cycler 480 (Perkin–Elmer) by using the following cycling conditions: heat activation of the polymerase for 10 min at 95°C; followed by 35 cycles of 95°C for 1 min, 34°C for 1 min, and 72°C for 1 min; with a final extension at 72°C for 10 min. One microliter of this reaction was reamplified in a seminested reaction by using the PAN-UI (5′-CTTGGATCCAGTGT/CTA/GCCC/ACAA/GGG-3′) primer in combination with PAN-DI and the following cycling conditions: 10 min at 95°C; followed by 30 cycles of 95°C for 1 min, 45°C for 30 sec, and 72°C for 1 min; with a final extension at 72°C for 10 min. Both PCR mixes also were treated with 3 units of RNase-free DNase I (Roche Molecular Biochemicals), followed by heat inactivation before the addition of template, primers, and DNA polymerase to eliminate spurious reactions caused by contaminating genomic DNA. A 10-μl aliquot of the resulting PCR product was analyzed after electrophoresis on a 2.5% low-melting-temperature agarose gel (NuSieve, FMC) in 40 mM Tris–acetic acid/1 mM EDTA, pH 8.3 (TAE), staining with 1× SYBR Green (Molecular Probes) in TAE and visualization with a fluorescent image analyzer (FluorImager SI, Molecular Dynamics).
Cloning and Sequencing.
PCR products were ligated directly into a plasmid vector (pCR II-TOPO, Invitrogen). Recombinant plasmids found to contain a retroviral insert by PCR with the PAN-UI and PAN-DI primers were sequenced by using the fluorescent dideoxynucleotide terminator method of cycle sequencing on a Perkin–Elmer 373A or 377 automated DNA sequencer. The resulting sequences were compared with each other by using the pileup program (Wisconsin Package Version 9.0, Genetics Computer Group, Madison, WI) and to previously reported sequences by using blastn and blastx algorithms. At least seven independent clones were sequenced from each CSF sample. A CSF sample was considered to contain a species of retroviral RNA if all of the sequences from each individual were identical. CSF samples that contained mixed sequences were not included in the analysis, because at this point, a clear distinction between possible viral RNA copackaging (33) and contamination by genomic DNA cannot be made. Excessive DNase treatment of the eluted RNA was avoided because of the risk of hydrolyzing small amounts of RNA present in the samples. In a few samples, regardless of clinical diagnosis, the no-reverse-transcriptase control PCR indicated the presence of minute quantities of contaminating genomic DNA, which necessitated these criteria. DNA-free brain tissues were analyzed regardless of the makeup of the retroviral sequences.
Results
We detected nucleotide sequences homologous to those of known retroviruses in the CSFs of 10 of 35 (28.6%) individuals tested in the acute phase of newly diagnosed schizophrenia (Fig. 1, Table 1). The 10 individuals who had retroviral sequences detected in the acute phase of schizophrenia did not differ significantly from the 25 acutely ill individuals without detectable retroviral sequences in terms of age, gender, length of illness, or clinical symptoms at disease presentation. In addition, the groups did not differ in terms of the levels of total IgG, IgM, IgA, albumin, or cells detected in the CSF. We also found retroviral sequences in the CSF of 1 of 20 individuals with chronic schizophrenia. On the other hand, we did not find retroviral RNA in any of the CSFs obtained from 32 individuals without psychiatric disease nor from the 22 individuals with noninflammatory neurological diseases (χ2 = 19.25, df = 3, P < 0.001).
Figure 1.
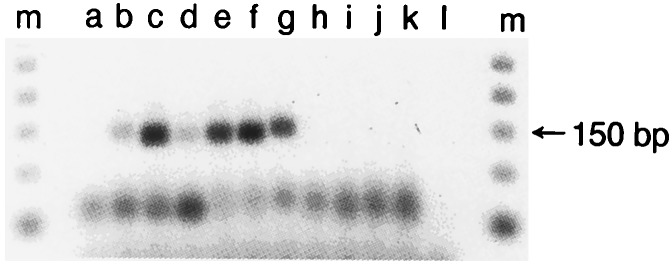
Agarose gel electrophoresis of PCR products generated by the reactions described in the text. The source of the samples is as follows: lanes a and l, negative controls; lanes b–f, CSFs of five individuals with recent-onset schizophrenia (b, A1; c, A2; d, A3; e, A4; f , A5); lane g, genomic human DNA; lanes h–k, CSFs from four unaffected control individuals; and lane m, 50-bp markers. The nucleotide sequences corresponding to the visualized bands from the CSFs are presented in Fig. 2.
Table 1.
Results from analysis of CSF from individuals with recent onset schizophrenia, chronic schizophrenia, individuals with noninflammatory neurological disorder and individuals with no history of psychiatric disease
| Patient group | n | No. with retrovirus sequence
|
|||
|---|---|---|---|---|---|
| HERV-W | ERV9 | ERV-FRD | Total | ||
| Recent onset | 35 | 7 | 2 | 1 | 10 (28.6%)* |
| Chronic | 20 | 1 | ND | ND | 1 (5%) |
| Neurological | 22 | ND | ND | ND | 0 |
| Unaffected† | 30 | ND | ND | ND | 0 |
Retroviral pol RNA was detected as described in Materials and Methods. Homologies, ≥90%, to previously described sequences were identified by using the blast algorithm. ND, not detected.
P = 0.0012 vs. unaffected, Fisher's exact test (two-tailed).
The two groups of Irish (n = 18) and German (n = 12) controls were merged because they did not differ with respect to the outcome of this analysis.
The retroviral sequences identified in the CSFs of the 11 individuals with schizophrenia were compared with the corresponding sequences in the pol genes of known retroviruses. A total of 8 of the 11 sequences showed ≥90% nucleotide identity to the HERV-W species of endogenous retrovirus (Fig. 2). Multiple sclerosis-associated retrovirus (MSRV)-associated RNA sequences (30) have permitted the identification and characterization of a family of homologous HERV elements, named HERV-W, on the basis of the tryptophan tRNA motif identified in MSRV/HERV-W sequences (31). Nucleotide sequences amplified from an additional two CSF samples were approximately 92% identical to members of the endogenous retroviral-9 (ERV9) family of retroviruses. ERV9, which was identified originally in undifferentiated embryonal cells (32), is 70–75% identical to HERV-W in the pol gene. The remaining CSF-derived sequence was identical to the FRD strain of endogenous retrovirus, which was identified originally in a human breast cancer cell line (33). The pol genes of the viruses HERV-W, ERV9, and FRD are all related to other mammalian C-type retroviruses, such as murine leukemia virus, gibbon ape leukemia virus, and feline leukemia virus, on the basis of nucleotide and amino acid sequence homology.
Figure 2.
Alignment of nucleotide sequences (A) and predicted amino acids (B) amplified from the CSFs of 10 individuals with acute schizophrenia (A1–10) and one individual with chronic schizophrenia (C1) compared with corresponding sequences from MSRV, ERV9, and FRD retroviruses (GenBank accession nos. AF009668, X57147, and U27240, respectively). The depicted sequences amplified from the CSF samples represent identical sequences obtained from at least seven independent clones. Dashes indicate nucleotides or amino acids that are identical to the reference sequence depicted on the top line. The open circles indicate gaps in sequence. The asterisks indicate the presence of stop codons.
The retroviral sequences that we identified in the CSFs of individuals with schizophrenia are similar to endogenous retroviral sequences found in human genomic DNA. We thus examined the possibility that our results could be explained by the differential presence of DNA in the CSFs of some individuals with acute onset schizophrenia. As depicted in Table 2, we found that the degenerate primers used in our study amplify a diverse set of sequences from DNA derived from these individuals, as well as from other individuals with schizophrenia, and unaffected controls. This pattern contrasts with that found after the amplification of reverse-transcribed particle-associated RNA extracted from the CSFs depicted in Fig. 2, in which all of the clones from a single individual were identical to each other and displayed homology to a single retroviral species. Furthermore, the CSF samples that contained detectable retroviral sequences did not differ from those that did not in terms of the concentrations of erythrocytes, leukocytes, or albumin. It is also unlikely that the finding of retroviral sequences in the CSF of acutely affected individuals is an artifact of drug therapy or incidental environmental exposures. All but one of the acutely psychotic individuals with retroviral RNA detected in the CSF had received antipsychotic medications for less than 2 weeks at the time of CSF sampling; one of the individuals had not received any antipsychotic medications before testing. All of these individuals had been free of nonpsychiatric diseases and living in their normal home environments before the onset of their first psychotic episode.
Table 2.
Nucleotide homologies of DNA and RNA amplified from individuals with schizophrenia (SCZ) and unaffected controls
| Patient group | New-onset SCZ (A4) | New-onset SCZ (A5) | Chronic SCZ (n = 4) | Unaffected (n = 6) | |||
|---|---|---|---|---|---|---|---|
| Template | DNA | RNA | DNA | RNA | DNA | DNA | |
| No. clones analyzed | 16 | 7 | 8 | 9 | 30 | 53 | |
| Retrivirus homology | |||||||
| HERV-W | 4 (25%) | 7 (100%)* | 1 (12.5%) | 9 (100%)† | 10 (33.3%) | 14 (26.4%) | |
| FRD | 3 (19.0%) | 0 | 1 (12.5%) | 0 | 10 (33.3%) | 14 (26.4%) | |
| ERV9 | 4 (25.0%) | 0 | 2 (25.0%) | 0 | 5 (16.7%) | 13 (24.5%) | |
| HC2 | 3 (19.0%) | 0 | 1 (12.5%) | 0 | 5 (16.7%) | 4 (7.5%) | |
| MLN | 1 (6.0%) | 0 | 0 | 0 | 0 | 5 (9.5%) | |
| HERV-K | 0 | 0 | 1 (12.5%) | 0 | 0 | 2 (3.8%) | |
| Other | 1 (6.0%) | 0 | 2 (25.0%)‡ | 0 | 0 | 1 (1.9%) | |
DNA was isolated from peripheral blood mononuclear cells from individuals A4 and A5 with newly diagnosed schizophrenia as well as from four individuals with chronic schizophrenia and six unaffected individuals. DNA was extracted and amplified by PCR without prior reverse transcription. RNA was extracted from the cell-free CSFs of patients A4 and A5, reverse transcribed, and amplified by PCR, as described above. Homologies shown represent those with ≥90% nucleotide identity. Data presented relating to new-onset schizophrenia denote the homologies of clones derived from the indicated individuals. Data for the chronic schizophrenia and unaffected groups represent the homologies of clones derived from four and six individuals, respectively.
P = 0.0013 compared to corresponding A4 DNA, Fisher's exact test (two-tailed).
P = 0.0004 compared to corresponding A5 DNA, Fisher's exact test (two-tailed).
Represents homology to two different uncharacterized retroviral sequences.
Analyzing the frontal cortex tissue cDNA for retroviral pol transcripts, we found that all samples contained retroviral RNA. However, after sequencing of a large number of clones from each brain and pooling the data within the two groups, two very different frequency-distribution patterns appeared (Table 3). Of a total of 48 clones from the five schizophrenic brains, 65% showed greatest homology to members of the HERV-W family of retroviruses. In two of the five brains analyzed in this group, HERV-W was the only pol transcript detected. In the normal brains, only 6.7% of the pol transcripts showed homology to this family of endogenous retroviruses (P < 0.0001, Fisher's exact test, two-tailed).
Table 3.
Distribution of retroviral pol transcripts (species homologies ≥90%) in frontal cortex tissue obtained postmortem from unaffected individuals (n = 6) and individuals with a diagnosis of schizophrenia (n = 5)
| Species | No.
homologous clones
|
|
|---|---|---|
| Unaffected | Schizophrenia | |
| HERV-W | 6 (6.7%) | 31 (64.6%)* |
| ERV-MLN | 10 (11.1%) | ND |
| ERV-FRD | 10 (11.1%) | 4 (8.3%) |
| ERV-FTD | ND | 1 (2.1%) |
| HERV-K | 8 (8.9%) | 7 (14.6%) |
| ERV9 | 52 (57.8%) | 1 (2.1%)* |
| HC2 | 2 (2.2%) | 1 (2.1%) |
| Other | 1 (1.1%) | ND |
| No match | 1 (1.1%) | 3 (6.2%) |
| Total clones | 90 | 48 |
Retroviral pol RNA was detected as described in Materials and Methods. Homologies to previously reported sequences were analyzed by using the blastn algorithm. ND, not detected.
P < 0.0001, Fisher's exact test (two-tailed).
Discussion
We found that CSF obtained from several individuals with recent-onset schizophrenia contains retroviral sequences not found in control samples. There are several mechanisms by which retroviral sequences might be transcribed within the central nervous systems of individuals with recent-onset schizophrenia. For example, the long terminal repeat regions of many retroviral RNAs contain binding sites for a number of different transcription factors and enhancers, including ones for hormonal and inflammatory mediators. These promoters can activate not only retroviral sequences but also human genes located downstream from the site of retroviral integration (34–36). The interaction between these transcriptional activators and endogenous retroviruses is of interest in light of the neuroendocrine and immunological abnormalities that have been reported in some individuals with schizophrenia (37, 38). Retroviral RNA can also be transcribed differentially under the control of the promoters of human genes located adjacent to integrated retroviral elements (39). The identification of the promoter mechanisms involved in the activation of the endogenous retroviruses in individuals with schizophrenia might result in the characterization of aspects of disease pathogenesis.
It is also possible that some of the retroviral RNAs identified in the CSFs of individuals with schizophrenia are derived from infection with exogenous retroviruses that have sequence homology with HERVs. The occurrence of exogenous and endogenous viruses with similar sequences has been documented previously in murine, ovine, and feline species. Furthermore, exogenous and endogenous retroviruses can recombine to generate viruses of altered pathogenicity (40, 41). It is of note in this regard that 3 of the 11 sequences found in the CSFs of individuals with schizophrenia would not be expected, by themselves, to be replication competent, because of the presence of stop codons in the polymerase region (Fig. 2).
We found retroviral sequences in the CSFs of 28.6% of individuals with acute onset schizophrenia and in 5% of individuals in later stages of the disease process; however, we found no retroviral sequences in individuals with noninflammatory neurological diseases or without neurological or psychiatric diseases. The differences in the prevalence of retroviral RNA in the CSFs among the different patient groups should be interpreted with some caution in light of the fact that the samples had been stored for different lengths of time and were obtained from geographically distinct study populations.
Similarly, our finding of the absence of retroviral RNA in CSFs from individuals without psychiatric diseases should also be interpreted with some caution because of the fact that some of the control samples also were obtained and stored under conditions different from those of the individuals with psychiatric diseases. However, the 12 CSFs from control individuals, which were matched to the group with recent-onset schizophrenia in terms of geographical origin and handling, had been stored for the shortest length of time before analysis. This fact argues against our findings being caused solely by artifacts because of handling, storage, or other regional differences.
Many of the retroviral sequences we have identified in the CSFs of individuals with schizophrenia are homologous to the HERV-W family of endogenous retroviruses, a member of which (MSRV) was identified originally in the CSFs of individuals with multiple sclerosis. We also found evidence of increased transcription of HERV-W in brain tissues obtained postmortem from individuals with schizophrenia. A possible role for HERV-W in the pathogenesis of schizophrenia is supported by the identification of an HERV-W sequence by representational difference analysis in the DNA from monozygotic twins discordant for schizophrenia, indicating that individuals with schizophrenia may differ from unaffected individuals in terms of de novo integrations of viral genomes homologous to the HERV-W family into their genomic DNA (42). It has been found recently that the envelope protein of HERV-W is capable of causing cell fusion and the generation of syncytia (43). It is not known whether this function of HERV-W is related to the association with schizophrenia.
The reason for finding similar retroviral sequences in the CSFs of individuals with both schizophrenia and multiple sclerosis is not known with certainty. It is possible that, although similar in the polymerase region, the retroviruses activated in the two diseases differ in the genes encoding other viral proteins or in the viral long terminal repeats. It is also possible that individuals with schizophrenia and multiple sclerosis undergo the activation of similar retroviral sequences but differ in terms of genetically determined responses to the retroviral activation. Schizophrenia and multiple sclerosis are distinct clinical entities and have different pathological manifestations, gender ratios, and clinical courses. However, the diseases share a number of epidemiological features (44, 45). These include similarities in ages of onset, seasons of birth, and geographic distributions. In addition, some patients can display clinical manifestations of both diseases (46–49). The further characterization of retroviral sequences transcribed within the central nervous system of individuals with schizophrenia and multiple sclerosis might better define the relationships between retroviral activation and human brain diseases and lead to improved methods for the diagnosis and treatment of these disorders.
Acknowledgments
We thank Mrs. B. Zwissler and Drs. S. Demisch, J. Pantel, K. Kronmüller, and D. Weimer for their assistance in the management of the patients. We thank Drs. J. Boeke, C. Ross, H. Perron, R. Löwer, R. Weiss, and D. Griffiths for their comments and suggestions relating to the manuscript. We thank Ms. Flora Leister, Shuojia Li, and Ann Cusic for their assistance with the project. We thank the Stanley Foundation, the medical faculty of the University of Heidelberg, and the State of Baden-Württemberg for their support of this project.
Abbreviations
- CSF
cerebrospinal fluid
- ERV
endogenous retrovirus
- HERV
human ERV
Footnotes
References
- 1.Rice D P. J Clin Psychiatry. 1999;60:4–6. [PubMed] [Google Scholar]
- 2.Wyatt R J, Henter I, Leary M C, Taylor E. Soc Psychiatry Psychiatr Epidemiol. 1995;30:196–205. doi: 10.1007/BF00789054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mortensen P B, Pederson C B, Westergaard T, Wohlfahrt J, Ewald H, Mors O, Andersen P K, Melbye M. N Engl J Med. 1999;340:603–608. doi: 10.1056/NEJM199902253400803. [DOI] [PubMed] [Google Scholar]
- 4.Torrey E F, Miller J, Rawlings R, Yolken R H. Schizophr Res. 1997;28:1–38. doi: 10.1016/s0920-9964(97)00092-3. [DOI] [PubMed] [Google Scholar]
- 5.Barondes S H, Alberts B M, Andreasen N C, Bargmann C, Benes F, Goldman-Rakic P, Gottesman I, Heinemann S F, Jones E G, Kirschner M, et al. Proc Natl Acad Sci USA. 1997;94:1612–1614. doi: 10.1073/pnas.94.5.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yolken R H, Torrey E F. Clin Microbiol Rev. 1995;8:131–145. doi: 10.1128/cmr.8.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yolken R H, Karlsson H, Yee F, Johnston-Wilson N L, Torrey E F. Brain Res Brain Res Rev. 2000;31:193–199. doi: 10.1016/s0165-0173(99)00037-5. [DOI] [PubMed] [Google Scholar]
- 8.Andreasen N C. N Engl J Med. 1999;340:645–647. doi: 10.1056/NEJM199902253400811. [DOI] [PubMed] [Google Scholar]
- 9.Crow T J. Br J Psychiatry. 1984;145:243–253. doi: 10.1192/bjp.145.3.243. [DOI] [PubMed] [Google Scholar]
- 10.O'Reilly R L, Singh S M. Am J Med Genet. 1996;67:19–24. doi: 10.1002/(SICI)1096-8628(19960216)67:1<19::AID-AJMG3>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 11.Torrey E F, Yolken R H. Brain Res Brain Res Rev. 2000;31:113–117. doi: 10.1016/s0165-0173(99)00028-4. [DOI] [PubMed] [Google Scholar]
- 12.Atwood W J, Berger J R, Kaderman R, Tornatore C S, Major E O. Clin Microbiol Rev. 1993;6:339–366. doi: 10.1128/cmr.6.4.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winkler C, Modi W, Smith M W, Nelson G W, Wu X, Carrington M. Science. 1998;279:389–393. doi: 10.1126/science.279.5349.389. [DOI] [PubMed] [Google Scholar]
- 14.Martin M P, Dean M, Smith M W, Winkler C, Gerrard B, Michael N L, Lee B, Doms R W, Margolick J, Buchbinder S, et al. Science. 1998;282:1907–1911. doi: 10.1126/science.282.5395.1907. [DOI] [PubMed] [Google Scholar]
- 15.Patience C, Wilkinson D A, Weiss R A. Trends Genet. 1997;13:116–120. doi: 10.1016/s0168-9525(97)01057-3. [DOI] [PubMed] [Google Scholar]
- 16.Lower R, Boller K, Hasenmaier B, Korbmacher C, Muller-Lantzsch N, Lower J, Kurth R. Proc Natl Acad Sci USA. 1993;90:4480–4484. doi: 10.1073/pnas.90.10.4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mueller-Lantzsch N, Sauter M, Weiskircher A, Kramer K, Best B, Buck M, Grasser F. AIDS Res Hum Retroviruses. 1993;9:343–350. doi: 10.1089/aid.1993.9.343. [DOI] [PubMed] [Google Scholar]
- 18.Venables P J, Brookes S M, Griffiths D, Weiss R A, Boyd M T. Virology. 1995;211:589–592. doi: 10.1006/viro.1995.1442. [DOI] [PubMed] [Google Scholar]
- 19.Mayer J, Meese E, Mueller-Lantzsch N. Cytogenet Cell Genet. 1979;79:157–161. doi: 10.1159/000134709. [DOI] [PubMed] [Google Scholar]
- 20.Löwer R, Löwer J, Kurth R. Proc Natl Acad Sci USA. 1996;93:5177–5184. doi: 10.1073/pnas.93.11.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perron H, Garson J A, Bedin F, Beseme F, Paranhos-Baccala G, Komurian-Pradel F, Mallet F, Tuke P W, Voisset C, Blond J L, Lalande B, et al. Proc Natl Acad Sci USA. 1997;94:7583–7588. doi: 10.1073/pnas.94.14.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conrad B, Weissmahr R N, Boni J, Arcari R, Schupbach J, Mach B. Cell. 1997;90:303–312. doi: 10.1016/s0092-8674(00)80338-4. [DOI] [PubMed] [Google Scholar]
- 23.Nakagawa K, Brusic V, McColl G, Harrison L C. Arthritis Rheum. 1997;40:627–638. doi: 10.1002/art.1780400407. [DOI] [PubMed] [Google Scholar]
- 24.Torrey E F, McGuire M, O'Hare A, Walsh D, Spellman M P. Am J Psychiatry. 1984;141:966–970. doi: 10.1176/ajp.141.8.966. [DOI] [PubMed] [Google Scholar]
- 25.Wittchen H U, Wunderlich U, Gruschwitz S, Zaudig M, editors. Strukturiertes Klinisches Interview für DSM-IV. Verlag, Göttingen, Germany: Hogrefe; 1997. [Google Scholar]
- 26.Schröder J, Buchsbaum M S, Siegel B V, Geider F J, Lohr J, Tang C H, Wu J, Potkin S G. Schizophr Res. 1996;19:41–53. doi: 10.1016/0920-9964(95)00043-7. [DOI] [PubMed] [Google Scholar]
- 27.Johnston N L, Cervenak J, Shore A D, Torrey E F, Yolken R H. J Neurosci Methods. 1997;77:83–92. doi: 10.1016/s0165-0270(97)00115-5. [DOI] [PubMed] [Google Scholar]
- 28.Dhellin O, Maestre J, Heidmann T. EMBO J. 1997;16:6590–6602. doi: 10.1093/emboj/16.21.6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tuke P W, Perron H, Bedin F, Beseme F, Garson J A. Acta Neurol Scand. 1997;169:16–21. doi: 10.1111/j.1600-0404.1997.tb08145.x. [DOI] [PubMed] [Google Scholar]
- 30.Komurian-Pradel F, Paranhos-Baccala G, Bedin F, Ounanian-Paraz A, Sodoyer M, Ott C, Rajoharison A, Garcia E, Mallet F, Mandrand B, Perron H. Virology. 1999;260:1–9. doi: 10.1006/viro.1999.9792. [DOI] [PubMed] [Google Scholar]
- 31.Blond J L, Beseme F, Duret L, Bouton O, Bedin F, Perron H, Mandrand B, Mallet F. J Virol. 1999;73:1175–1185. doi: 10.1128/jvi.73.2.1175-1185.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.LaMantia G, Maglione D, Pengue G, DiCristofano A, Simeone A, Lanfrancone L, Lania L. Nucleic Acids Res. 1991;19:1513–1520. doi: 10.1093/nar/19.7.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seifarth W, Skladny H, Krieg-Schneider F, Reichart A, Hehlmann R, Leib-Mosch C. J Virol. 1995;69:6408–6416. doi: 10.1128/jvi.69.10.6408-6416.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ora M, Kawakami M, Ushikubo H. J Virol. 1987;61:2059–2062. doi: 10.1128/jvi.61.6.2059-2062.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akopov S B, Nikolaev L G, Khil P P, Lebedev Y B, Sverdlov E D. FEBS Lett. 1998;421:229–233. doi: 10.1016/s0014-5793(97)01569-x. [DOI] [PubMed] [Google Scholar]
- 36.Urnovitz H B, Murphy W H. Clin Microbiol Rev. 1996;9:72–99. doi: 10.1128/cmr.9.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marx C E, Lieberman J A. Psychiatr Clin North Am. 1998;21:413–434. doi: 10.1016/s0193-953x(05)70013-7. [DOI] [PubMed] [Google Scholar]
- 38.Lammers C H, Garcia-Borreguero D, Schmider J, Gotthardt U, Dettling M, Holsboer F, Heuser I J. Biol Psychiatry. 1995;38:803–807. doi: 10.1016/0006-3223(95)00065-8. [DOI] [PubMed] [Google Scholar]
- 39.Kowalski P E, Mager D L. J Virol. 1998;72:6164–6168. doi: 10.1128/jvi.72.7.6164-6168.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu W S, Temin H M. Science. 1990;250:1227–1233. doi: 10.1126/science.1700865. [DOI] [PubMed] [Google Scholar]
- 41.Zhang J, Temin H M. Science. 1993;259:234–238. doi: 10.1126/science.8421784. [DOI] [PubMed] [Google Scholar]
- 42.Deb-Rinker, Klempan T A, O'Reilly R L, Torrey E F, Singh S M. Genomics. 1999;61:133–144. doi: 10.1006/geno.1999.5946. [DOI] [PubMed] [Google Scholar]
- 43.Mi S, Lee X, Li X-P, Veldman G, Finnerty H, Racie L, LaVallie E, Tang X-Y, Edouard P, Howes S, et al. Nature (London) 2000;403:785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 44.Stevens J R. Schizophr Bull. 1988;14:231–241. doi: 10.1093/schbul/14.2.231. [DOI] [PubMed] [Google Scholar]
- 45.Templer D I, Cappelletty G G, Kauffman I. Br J Psychiatry. 1988;153:389–390. doi: 10.1192/bjp.153.3.389. [DOI] [PubMed] [Google Scholar]
- 46.Cazzullo C L, Saresella M, Roda K, Calvo M G, Bertrando P, Doria S, Clerici M, Salvaggio A, Ferrante P. Schizophr Res. 1998;31:49–55. doi: 10.1016/s0920-9964(97)00153-9. [DOI] [PubMed] [Google Scholar]
- 47.Ganguli R, Brar J, Rabin B S. Harv Rev Psychiatry. 1994;2:70–83. doi: 10.3109/10673229409017120. [DOI] [PubMed] [Google Scholar]
- 48.Pine D S, Douglas C J, Charles E, Davies M, Kahn D. J Clin Psychiatry. 1995;56:297–306. [PubMed] [Google Scholar]
- 49.Feinstein A, du Boulay G, Ron M A. Br J Psychiatry. 1992;161:680–685. doi: 10.1192/bjp.161.5.680. [DOI] [PubMed] [Google Scholar]



