Abstract
The weighted stochastic simulation algorithm (wSSA) was developed by Kuwahara and Mura [J. Chem. Phys. 129, 165101 (2008)] to efficiently estimate the probabilities of rare events in discrete stochastic systems. The wSSA uses importance sampling to enhance the statistical accuracy in the estimation of the probability of the rare event. The original algorithm biases the reaction selection step with a fixed importance sampling parameter. In this paper, we introduce a novel method where the biasing parameter is state-dependent. The new method features improved accuracy, efficiency, and robustness.
INTRODUCTION
The stochastic simulation algorithm (SSA) is widely used for the discrete stochastic simulation of chemically reacting systems. Although ensemble simulation by SSA and its variants has been successful in the computation of probability density functions in many chemically reacting systems, the ensemble size needed to compute the probabilities of rare events can be prohibitive.
The weighted SSA (wSSA) was developed by Kuwahara and Mura1 to efficiently estimate the probabilities of rare events in stochastic chemical systems. The wSSA was developed to estimate p(x0,ε;t), which is the probability that the system, starting at x0, will reach any state in the set of states ε before time t. The estimation procedure is a carefully biased version of the SSA, which in theory can be used to estimate any expectation of the system. However, it is important to note that, in contrast to SSA trajectories, wSSA trajectories should not be regarded as valid representations of the actual system behavior.
The key element in the wSSA is importance sampling (IS), which is used to bias the reaction selection procedure. The wSSA introduced in Ref. 1 uses a fixed constant as the IS parameter to multiply the original propensities. In this paper, we introduce a state-dependent IS method that has several advantages over the original fixed parameter IS method. The new method features improved accuracy, efficiency, and robustness.
In Sec. 2 we describe the current status of the wSSA. In Sec. 3 we present the new state-dependent biasing method. We apply the new biasing method to several examples in Sec. 4 and compare its performance with that of the original wSSA. In Sec. 5 we summarize the results and discuss areas for future work.
CURRENT STATUS OF THE WSSA
In this section, we briefly describe the weighted stochastic simulation algorithm. A more detailed explanation of the algorithm can be found in Refs. 1, 2.
To begin, consider a well-stirred system of molecules of N species (S1,…,SN) which interact through M reaction channels (R1,…,RM). We specify the state of the system at current time t by the vector x=(x1,…,xN), where xi is the number of molecules of species Si. The propensity function aj of reaction Rj is defined so that aj(x)dt is the probability that one Rj reaction will occur in the next infinitesimal time interval [t,t+dt), given that its current state is X(t)=x. The propensity sum a0(x) is defined as . The SSA is based on the fact that the probability that the next reaction will carry the system to x+vj, where vj is the state change vector for reaction j, between times t+τ and t+τ+∂τ is
| (1) |
In the direct method implementation of the SSA, we choose τ, the time to the next reaction, by sampling an exponential random variable with mean 1∕a0(x). The next reaction index j is chosen with probability aj(x)∕a0(x).
In the wSSA the time increment τ is chosen as we would in the SSA, but we bias the selection of reaction index j: for that we use, instead of the true propensities aj(x), an alternate set of propensities bj(x). We then correct the resulting bias with appropriate weights wj(x). In the original wSSA, bj(x)=γjaj(x), where γj>0 is called the importance sampling parameter for Rj. Choosing γj>1 will make Rj more likely to be selected while choosing γj<1 will have the opposite effect. When γj=1 for all j, i.e., we do not bias the reaction selection process, the wSSA simply turns into the SSA. Thus, in the wSSA, the right side of Eq. 1 becomes
| (2) |
where . This biased probability can be restored by multiplying Eq. 2 by the weighting factor,
| (3) |
Together, we have
| (4) |
We can extend this statistical weighting of a single-reaction jump to an entire trajectory by using the memoryless Markovian property—each jump depends on its starting state but not on the history of the state; therefore, the probability of a single trajectory is the product of all the individual jumps that make up the trajectory. Since each jump in the wSSA requires a correction factor of wj(x) in Eq. 3, the entire trajectory needs to be weighted by w=∏jwj(x).
The aim of the wSSA is to estimate
| (5) |
It is important to note that p(x0,ε;t) is not the probability that the system will reach some state in ε at time t, but will have reached that set at least once before time t.
Estimating p(x0,ε;t) with the SSA is straightforward. After running n simulations of SSA, we record mn, the number of trajectories that reached any state in ε before time t. Since each trajectory in the ensemble is equally statistically significant, we can estimate p(x0,ε;t) as mn∕n, which approaches the true probability as n→∞. While estimating p(x0,ε;t) with the SSA is a simple procedure, an extremely large n is required to obtain an estimate with low uncertainty when p(x0,ε;t)⪡1.
Knowing the uncertainty of an estimate is crucial because it provides quantitative information about the accuracy of the estimate. The one standard deviation uncertainty is given by , where σ is the square root of the sample variance. For sufficiently large n, the true value is 68% likely to fall within one-standard-deviation of the estimate (within the range of ). Increasing the uncertainty interval by a factor of 2 raises the confidence level to 95%; increasing it by a factor of 3 gives us a confidence level of 99.7%. Therefore, it is desirable to obtain an estimate with a small uncertainty (i.e., small sample variance) because it signifies that the estimate is close to the true probability.
For unweighted SSA trajectories, the relative uncertainty is (see Ref. 2)
| (6) |
When p(x0,ε;t)⪡1 (and mn∕n⪡1), Eq. 6 reduces to
| (7) |
This shows that to we need to observe 10 000 successful trajectories to achieve 1% relative accuracy. Since the average rate of observing a state in ε using SSA is 1∕p(x0,ε;t), we need about 1∕p(x0,ε;t)×10 000 SSA simulations, which quickly becomes computationally infeasible as p(x0,ε;t) decreases, especially for large systems.
The wSSA resolves the inefficiency of the SSA by assigning a different weight to every trajectory. In estimating the same probability with a given n, we can observe many more successful trajectories using the wSSA than the SSA. Each successful trajectory is likely to have a very small weight, which results from using an alternate set of propensities b(x), instead of the original propensities a(x). Since each trajectory in the wSSA has a different weight, we redefine , where wk=0 if the kth trajectory did not reach ε before time t. We also keep track of the second moment of the trajectory weights, in order to calculate the sample variance, given by . Note that different algorithms, such as described in Ref. 3, can be used to compute a running variance to avoid cancellation error.
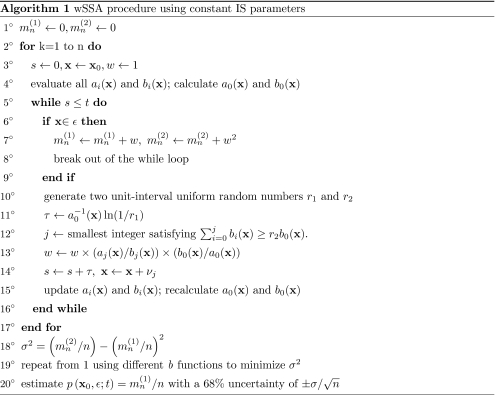
The current algorithm can be seen in Fig. 1.
Figure 1.
The weighted stochastic simulation algorithm using constant importance sampling parameters.
THE STATE-DEPENDENT BIASING METHOD
The key element of the wSSA is importance sampling, which is a general technique often used with a Monte Carlo method to reduce the variance of an estimate of interest. The rationale for using this technique is that not all regions of the sample space have the same importance in simulation. When we have some knowledge about which sampling values are more important than others, we can use important sampling to improve efficiency as well as accuracy. The technique involves choosing an alternative distribution from which to sample the random numbers. This alternative distribution is chosen such that the important samples are chosen more frequently than they are in the original distribution. After using the alternative distribution to sample, a correction is applied to ensure that the new estimate is unbiased. The wSSA employs this technique in the reaction selection procedure. The next reaction is chosen using b(x) (12° in Algorithm 1), and the bias is corrected with an appropriate weight, (aj(x)∕bj(x))×(b0(x)∕a0(x)) (13°). Mathematical details of importance sampling and Monte Carlo averaging can be found in Appendix of Ref. 2.
The current method for selecting b functions is to simply multiply the original propensities by a positive scalar γj, i.e., bj(x)=γjaj(x). The fixed multiplier facilitates the wSSA implementation, but there are several associated drawbacks. The most obvious drawback is increased variance due to over- and underperturbations. The changes in species population during the simulation cause the relative propensity of reaction j, aj(x)∕a0(x), to fluctuate as well, changing the probability of selecting the jth reaction at each time step. If γj is constrained to be a constant for all possible values of aj(x)∕a0(x), then the optimal value for γj would be the one that perturbed aj “just right” for most values of aj(x). However, if aj(x)∕a0(x) varied widely, then γj would over- or underperturb aj(x) near the extreme values of aj(x)∕a0(x). A related drawback concerns a narrow parameter space of γj that produces an accurate estimate for p(x0,ε;t). If aj(x)∕a0(x) reaches values near 0 as well as 1 during a single simulation, then it is impossible to avoid both over- and underperturbations because the jth importance sampling parameter can have only a single value which can avoid only one of the two cases. Thus, there will be very few values for γj that produce an accurate estimate. Unless the initial value for γj is set near these few values, the resulting estimate is guaranteed to have high variance. This unreliable estimate is not useful in deciding how to perturb γj to obtain an estimate with lower variance (step 19° of the Algorithm 1). Consequently, one may waste a lot of computational power searching for a value of γj that produces an accurate estimate.
Since the above problems are due to fluctuations in the propensity, which is caused by fluctuations in population size, an intuitive solution would be to vary γj according to the current state x. Although an arbitrary function may be used to achieve such goal, we proceed as follows.
First, we partition reactions into three groups:GE, GD, and GN. We define GE as the set of reactions that are to be encouraged. This set may include reactions that directly or indirectly increase the likelihood of rare event observation. The IS parameters for this group have values greater than 1 to increase the reaction firing frequency. Similarly we define GD as the set of reactions that are to be discouraged. The IS parameters for GD will be between 0 and 1, to decrease the likelihood of firing a reaction in GD. All reactions that do not influence the rare event observation are grouped into GN. Since the reactions in GN do not need to be perturbed, the IS parameters for these reactions are set to 1. We note that in practice, optimal partitioning of the reactions requires knowledge of the system, which may not always be available.
Second, we define the relative propensity of reaction j as ρj(x)≡aj(x)∕a0(x). ρj(x) is a fractional propensity between 0 and 1 that roughly indicates the likelihood of choosing Rj as the next reaction from the current state x. Since the value of ρj at each time step gives a qualitative indication of the amount of perturbation needed by Rj, the new biasing method will define γj to be a function of ρj.
When ρj→0, the probability of choosing Rj as the next reaction decreases. Thus, for Rj∊GE, more encouragement is needed as its relative propensity decreases. However, when ρj≈1, Rj is likely to be selected as the next reaction without any additional encouragement. In this case, taking bj(x)>aj(x) will overperturb the system and thus increase the variance in the estimate. Therefore, for Rj∊GE, there must be a value of ρj at which no further encouragement is applied. Similarly for Rj∊GD, no further discouragement is necessary when ρj≈0. The value of ρj for which we stop the perturbation is defined as ρj0, i.e., γj=1 for ρj≥ρj0(Rj∊GE) and for ρj≤ρj0(Rj∊GD). Lastly for Rj∊GD, more discouragement is necessary as ρj increases. This corresponds to γj→0 as ρj→1.
Based on above considerations, we take the jth IS parameter of Rj∊GE as
| (8) |
where gj(ρj) is a parabolic function of ρj that has the following properties:
| (9) |
For Rj∊GD, we select γj as the following:
| (10) |
where hj(ρj) is a parabolic function of ρj that has the following properties:
| (11) |
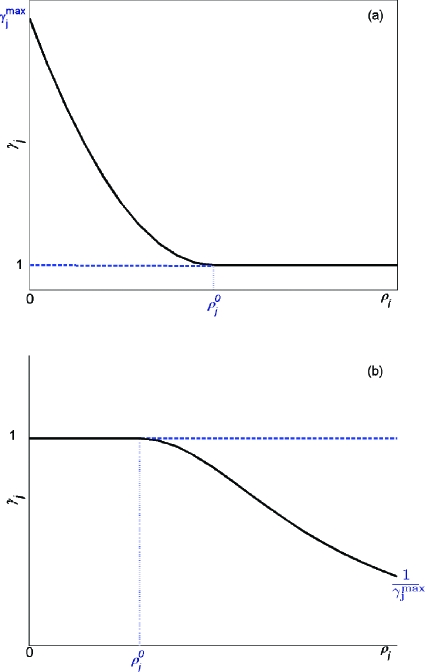
Graphical representations of the function γj(ρj(x)) are illustrated in Fig. 2, for Rj∊GE and Rj∊GD.
Figure 2.
(a) A graphical representation of γj(ρj(x)) for Rj∊GE. (b) A graphical representation of γj(ρj(x)) for Rj∊GD.
There are two parameters, and ρj0, for each γj(ρj(x)). is defined as the parameter associated with the maximum perturbation allowed on the jth reaction. Given the values for and ρj0, gj(ρj), and hj(ρj) as defined in Eqs. 9, 11 are unique parabolas whose formula is
| (12) |
An important point to keep in mind is that perturbing aj(x) by an unnecessarily large amount not only potentially increases the number of rare event observations but also increases the variance of an estimate as well. We want to observe enough rare events to calculate necessary statistics, yet at the same time to minimize the variance. The advantage of using γj(ρj) is that a user has the freedom to choose and ρj0, whose optimal values are problem dependent. Currently, there is no fully automated method to find the optimal value for the two parameters. For the examples in Sec. 4, we first choose a value for ρj0 by assigning a value near 0.15±0.05 (Rj∊GD) or 0.55±0.05 (Rj∊GE). We then vary to find an estimate with the lowest parameter.
After incorporating the new strategy for choosing γj, we obtain the algorithm in Fig. 3.
Figure 3.
The weighted stochastic simulation algorithm using state-dependent importance sampling parameters. Changes from Fig. 1 are highlighted in red.
NUMERICAL EXAMPLES
In this section we illustrate our new biasing algorithm with three examples, comparing the results with those obtained using the original algorithm. As we will see, the new biasing method increases the computational speed not only relative to the SSA but also relative to the scheme used in the original wSSA papers.1, 2 The measure used to calculate gain in computational efficiency is the same as in Ref. 2, given by
| (13) |
where nSSA and nwSSA are the numbers of runs in each of the two methods required to achieve comparable accuracy.
Two-state conformational transition
Consider the following system:
| (14) |
The initial state is set to x0=[100 0], i.e., all 100 molecules are initially in A form. This model concerns two conformational isomers—isomers that can be interconverted by rotation about single bonds. For this system we are interested in p(0,30;10) for B; that is, the probability that given no B molecules at time 0, its population reaches 30 before time 10. The steady-state population of B for the rate constants in Eq. 14 is approximately 11. The rare event description of x2=30 is about three times its steady-state value, so we expect its probability to be very small. Because this is a simple closed system, it is possible to calculate the exact probability of p(0,30;10) using a generator matrix (or a probability transition matrix).4 Using MATLAB’s matrix exponential function, we have calculated p(0,30;10)=1.191×10−5.
Since the steady-state population of B is much less than the rare event description of 30, it is necessary to bias the system so that the population of B increases. This can be achieved by either encouraging R1 or discouraging R2. Note that changing the propensities of both reactions is not necessary, as only the relative ratio of the two propensities matters in reaction selection.
For simulation of system 14 with the original wSSA, R1 was encouraged with different values of γ1>1. Thus, the b functions are given by
| (15) |
To find the γ1 that produces the minimum variance, γ1 was varied from 1.05 to 1.65 in increments of 0.05. Of these values, γ1=1.4 produced the lowest variance. Taking n=107 and γ1=1.4, the following estimate±twice the uncertainty (95% confidence interval) was obtained:
| (16) |
In the new biasing method, we replicated the reaction partition as done in the original system, i.e., GE={R1}, GD={∅}, GN={R2}. After assigning ρ10=0.5, was varied from 8 to 34 in increments of 2. Using n=105 and the optimal , the following estimate was obtained with a 95% confidence interval:
| (17) |
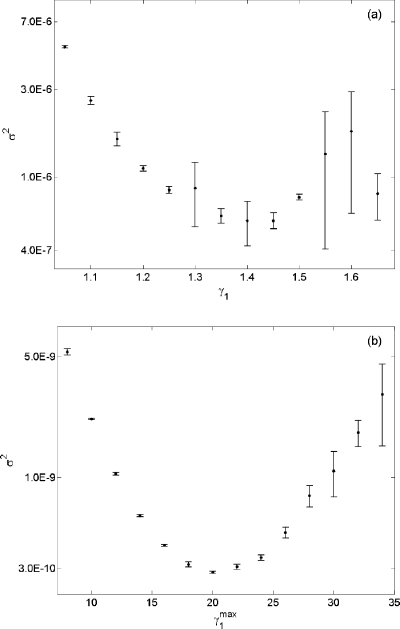
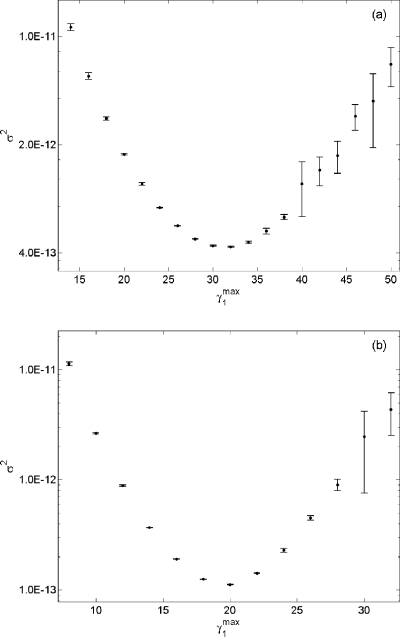
Although both estimates contain the true probability in their 95% confidence interval, the new biasing method required only 1% of the total number of simulations used in the original algorithm. Furthermore, the new biasing method produced a more accurate estimate with a much tighter confidence interval. Figure 4 shows a side by side comparison of σ2 for p(x0,30;10) using both biasing methods. We note that the new algorithm not only decreased the variance by a factor of 1000 but also produced an accurate estimate with a larger range of . Given n=107, the previous algorithm was able to produce an accurate estimate with a narrow range of γ1∊[1.10 1.60]. In contrast, any estimate from using the new biasing method with produced more accurate estimate than Eq. 16.
Figure 4.
(a). A plot of σ2 vs γ1 obtained in wSSA runs of reaction Eq. 14 using the algorithm in Fig. 1. Here we estimate p(0,30;10) for B. Each vertical bar shows the estimated mean and one standard deviation of σ2 at that γ1 value as found in four n=107 runs. (b) A plot of σ2 vs obtained from using the algorithm in Fig. 3. Each vertical bar was obtained from four n=105 runs.
Using the SSA to obtain an estimate with similar variance as in Eq. 17 would require a much greater computational expense. For our particular simulation, the calculated efficiency gain of the new algorithm over the SSA was 4.1×104, i.e., it would have taken a computer 4.1×104 times longer to obtain a similar result using the unweighted SSA.
Single species production and degradation
Our next example is taken from Refs. 1, 2 and consists of the following two reactions:
| (18) |
The initial state of the system is x0=[1 40], and we are interested in p(40,80;100) for S2—the probability that x2 reaches 80 before time 100 given x2=40 at t=0. This particular reaction set is well-studied, and it is known that the population of S2 in its steady-state follows the Poisson distribution with mean (and variance) of k1x1∕k2. Since the initial state of S2 is also its steady-state mean, x2 is expected to fluctuate around 40 with a standard deviation of 6.3. To advance the system toward the rare event, R1 was chosen to be encouraged in both of the following wSSA simulations.
First, 4×107 wSSA simulations were performed for each γ1 ranging from 1.45 to 2.25 in increment of 0.05. The following estimate was obtained using the optimal γ1=1.85:
| (19) |
We repeated the simulation with the new state-dependent biasing method which encouraged R1 as was done with the original algorithm. After running only 105 simulations with ρ10=0.6 and different values of , we obtained the following result using the optimal :
| (20) |
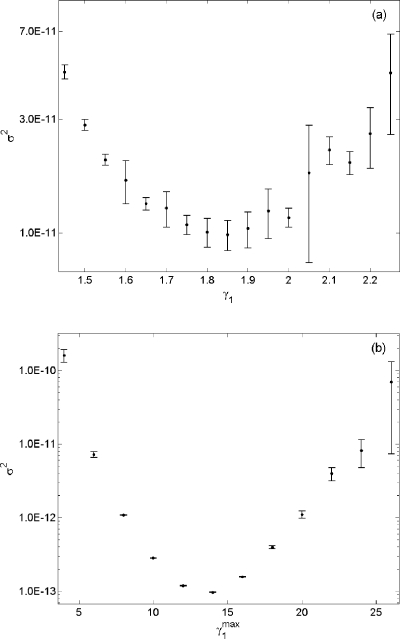
Figure 5 shows a side by side comparison of the variance using both biasing methods. As is shown, the biasing method with a state-dependent importance sampling parameter yielded estimates with variance two orders of magnitude less than that produced using the constant parameter importance sampling parameter. We also note that the latter simulation required 100 times fewer simulations to obtain a result with equivalent accuracy and uncertainty 20, which is a significant improvement over Eq. 19. Furthermore, the variance of an estimate generated by using any is lowerthan the variance of estimate 19. Lastly, the computational efficiency gain of Eq. 20 over SSA was 3.1×106, which is more than 100 times the gain from using the original biasing method.
Figure 5.
(a) A plot of σ2 vs γ1 obtained in wSSA runs of reaction 18 using the algorithm in Fig. 1. Here we estimate p(40,80;100) for S2. Each vertical bar shows the estimated mean and one standard deviation of σ2 at that γ1 value as found in four n=4×107 runs. (b) A plot of σ2 vs obtained from using the algorithm in Fig. 3. Each vertical bar was obtained from four n=105 runs.
Modified yeast polarization
Our last example concerns pheromone induced G-protein cycle in Saccharomyces cerevisia5 with a constant population of ligand, L=2. The model description was modified from Ref. 5, such that the system does not reach equilibrium. There are six species in this model, x=[RGRLGaGbgGd] and eight reactions as follows:
| (21) |
The kinetic parameters are
The state representing the initial condition is x0=[50 50 0 0 0 0]—there are 50 molecules of R and G, but none of the other species are initially present. For this system, we define the rare event to be p(x0,εGbg;20), where εGbg is the set of all states x for which the population of Gbg is equal to 50. We first partition the reactions as GE={∅}, GD={R6}, and GN={R1,..,R5,R7,R8}. The only reaction chosen to be perturbed is R6, which indirectly discourages the consumption of Gbg by delaying the production of Gd. Here we note that a more intuitive choice of reactions to include in GD would be R7, since it directly consumes a Gbg molecule. Upon numerical testing, however, we found that the estimate from perturbing R7 showed much higher variance than the one obtained from discouraging R6. This difference in performance is due to the four orders of magnitude separating the reaction constants kGd and kG. Because the reaction constant kG is so large, an extremely small IS parameter is required to effectively discourage R7. The results from our testing indicate that such a small IS parameter confers high variance. In contrast, the IS parameter needed by R6 was more modest and led to much better performance.
Following the partitioning with GD={R6}, the b functions for the constant parameter biasing method are
| (22) |
First, we ran 108 simulations of wSSA for each of the constant IS parameter γ6, where γ6 ranged from 1.2 to 2.0 in increments of 0.1. Then 108 simulations with the state-dependent biasing method was conducted for each with ρ60=0.15, where the b functions are given by
| (23) |
Because R6∊GD, the value of γ6 at each time step is chosen according to hj(ρj) in Eq. 12. The best IS parameter, γ=1.5, from the first set of simulations yields the following estimate:
| (24) |
The best estimate from using the state-dependent IS method with γmax=3 yields
| (25) |
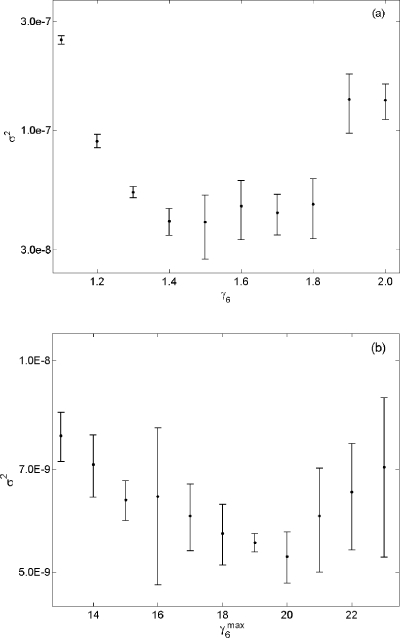
System 21 exhibits high intrinsic stochasticity, which causes difficulty in obtaining an estimate with low variance unless a large n is used. Despite this difficulty, we see that the uncertainty of the estimate obtained from using the state-dependent biasing algorithm is about three times less than the uncertainty in Eq. 24. Figure 6 shows a side by side comparison of σ2 for p(x0,εGbg;20), and we see that the variance in Fig. 6b is less than the variance in Fig. 6a by a factor of 10. In addition to increased accuracy, the new method also provides increased robustness, in that it provides a broader range of acceptable values for its parameter than for the old method’s parameter γ6(1.3–1.8).
Figure 6.
(a) A plot of σ2 vs γ6 obtained in wSSA runs of reaction 21 using the algorithm in Fig. 1. Here we estimate p(0,εGbg;20), where εGbg is the set of all states x in which the population of Gbg is 50. Each vertical bar shows the estimated mean and one standard deviation of σ2 at that γ6 value as found in four n=108 runs. (b) A plot of σ2 vs obtained from using the algorithm in Fig. 3. Each vertical bar was obtained from four n=108 runs.
The computational gain of wSSA from using a constant importance sampling parameter is 21, while that of the state-dependent importance sampling method is 250. Both gains imply a significant speed up against the SSA considering that the simulation time for 108 wSSA trajectories of system 21 is several days.
Lastly, we comment that although the value of ρj0 is chosen arbitrarily from a specified range (0.15±0.05 for Rj∊GD and 0.55±0.05 for Rj∊GE), the performance of the state-dependent biasing algorithm remains almost the same for different values of ρj0. For Rj∊GE, the lower and upper boundary values for ρj0 are 0.5 and 0.6, respectively. Figure 5b of system 18 was obtained using ρ10=0.6, which is the upper boundary value of ρj0 range. To compare the performance of the new algorithm for system 18 using different values of ρ10, we have created a plot of variance versus with two different values of ρ10∊{0.5,0.55}, which are lower boundary and median value [Figs. 7a, 7b]. We see that the minimum variance from both subplots of Fig. 7 is of similar magnitude compared to the minimum variance in Fig. 5b. Similar observation can be made for the other two examples.
Figure 7.
(a) A plot of σ2 vs obtained in wSSA runs of reaction 18 using Here we estimate p(40,80;100) for S2. Each vertical bar shows the estimated mean and one standard deviation of σ2 at that value as found in four n=105 runs. (b) A plot of σ2 vs obtained using the same setting as in Fig. 7a, except ρ10=0.55.
CONCLUSIONS
In this paper we have introduced a state-dependent biasing method for the weighted stochastic simulation algorithm. As numerical results from Sec. 4 support, the new state-dependent biasing method improves the accuracy of a rare event probability estimate and speeds up the simulation time. While the state-dependent biasing method excels in many aspects, it involves twice the number of parameters than the constant parameter biasing method. Currently, there is no automated method to assign an appropriate value to these parameters, and thus the computational effort associated with this task can be challenging as system size increases. It may be thought that using a line instead of a parabola in Eq. 12 would simplify the algorithm. However, owing probably to the nonlinearity of propensities that involve more than one species, we have found that the parabola usually works better. We have also observed in our numerical experiments that using a line has a negative impact on robustness, as compared to a parabola.
As noted in Sec. 3, the new biasing algorithm toggles between the original propensities and the biased propensities to select the next reaction, depending on the value of ρj(x). Therefore, the wSSA using the state-dependent biasing method can be regarded as an efficient adaptive algorithm. However, the value for two parameters ρj0 and must be determined prior to the simulation, and correctly partitioning reactions into three groups (GE, GD, and GN) can be challenging for large systems. These issues will be explored in future work.
ACKNOWLEDGMENTS
The authors acknowledge with thanks financial support as follows: D.T.G. was supported by the California Institute of Technology through Consulting Agreement No. 102-1080890 pursuant to Grant No. R01GM078992 from the National Institute of General Medical Sciences and through Contract No. 82-1083250 pursuant to Grant No. R01EB007511 from the National Institute of Biomedical Imaging and Bioengineering, and also from the University of California at Santa Barbara under Consulting Agreement No. 054281A20 pursuant to funding from the National Institutes of Health. M.R. and L.R.P. were supported by Grant No. R01EB007511 from the National Institute of Biomedical Imaging and Bioengineering, DOE Grant No. DEFG02-04ER25621, NSF IGERT Grant No. DG02-21715, and the Institute for Collaborative Biotechnologies through Grant No. DFR3A-8-447850-23002 from the U.S. Army Research Office.
References
- Kuwahara H. and Mura I., J. Chem. Phys. 129, 165101 (2008). 10.1063/1.2987701 [DOI] [PubMed] [Google Scholar]
- Gillespie D. T., Roh M., and Petzold L. R., J. Chem. Phys. 130, 174103 (2009). 10.1063/1.3116791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan T. F., Golub G. H., and Leveque R. J., Am. Stat. 37, 242 (1983). 10.2307/2683386 [DOI] [Google Scholar]
- Nelson R., Probability, Stochastic Processes, and Queuing Theory: The Mathematics of Computer Performance Modeling (Springer, New York, 1995). [Google Scholar]
- Drawert B., Lawson M. J., Petzold L. R., and Khammash M., J. Chem. Phys. 132, 074101 (2010). 10.1063/1.3310809 [DOI] [PMC free article] [PubMed] [Google Scholar]