Abstract
Stem cell-derived motor neurons (MNs) are increasingly utilized for modeling disease in vitro and for developing cellular replacement strategies for spinal cord injury and diseases such as spinal muscular atrophy (SMA) and amyotrophic lateral sclerosis (ALS). Human embryonic stem cell (hESC) differentiation into MNs, which involves retinoic acid (RA) and activation of the sonic hedgehog (SHH) pathway is inefficient and requires up to 60 days to develop MNs with electrophysiological properties. This prolonged differentiation process has hampered the use of hESCs, in particular for high-throughput screening. We evaluated the MN gene expression profile of RA/SHH-differentiated hESCs to identify rate-limiting factors involved in MN development. Based on this analysis, we developed an adenoviral gene delivery system encoding for MN inducing transcription factors: neurogenin 2 (Ngn2), islet-1 (Isl-1), and LIM/homeobox protein 3 (Lhx3). Strikingly, delivery of these factors induced functional MNs with mature electrophysiological properties, 11-days after gene delivery, with >60–70% efficiency from hESCs and human induced pluripotent stem cells (hiPSCs). This directed programming approach significantly reduces the time required to generate electrophysiologically-active MNs by approximately 30 days in comparison to conventional differentiation techniques. Our results further exemplify the potential to use transcriptional coding for rapid and efficient production of defined cell types from hESCs and hiPSCs.
Introduction
Recent progress in cell-based modeling using human embryonic stem cell (hESC) and induced pluripotent stem cell-(iPSC)-derived motor neurons (MNs) has opened new opportunities to understand the development of the motor system as well as MN disease. MNs are a highly specialized class of neurons that reside in the spinal cord and project axons in organized and discrete patterns to muscles to control their activity. The most prominent MN diseases are spinal muscular atrophy (SMA) and amyotrophic lateral sclerosis (ALS), in which MNs perish in the disease. In the case of SMA, the genetic deficit of reduced SMN levels is known. Indeed gene delivery of SMN in a transgenic model of SMA results in prolonged survival and motor rescue, indicating that methods to increase SMN levels in MNs may be therapeutic.1,2,3,4 Stem cell-derived MNs have been derived from iPSCs from a SMA patient, providing a new population of cells for drug and therapeutic screens.5 In addition, stem cell-based therapies have also demonstrated therapeutic potential in SMA and spinal cord injury. For example, studies have reported that transplanting MN progenitor cells into the spinal cords of SMA mice or into a spinal cord injured rat model partially restores motor decline.6,7 MNs have also been extensively utilized to study ALS and screen potential therapeutic compounds,8,9,10,11,12,13 thereby demonstrating the requirements for generating large numbers of MNs.
To generate large numbers of spinal MNs for use in in vitro-based screens and transplantation studies, neuromuscular researchers have taken advantage of the propensity of mouse ESCs to efficiently differentiate into this cell type. However, progress repeating these studies with hESCs has been slow due to the low efficiency of MN production and tedious, time-consuming culture conditions. The current procedures for generating MNs involve embryoid body formation in serum-free media and subsequent neural rosette formation in the presence of retinoic acid (RA) and sonic hedgehog (SHH), which function to caudalize and ventralize MN progenitors,14,15 respectively. Neurotrophic factors are then added to the medium for further maturation and to aid in cell survival. The entire differentiation process requires up to 2 months to generate MNs, which are electrophysiologically-active, with 10–40% efficiency.16,17,18,19,20,21 To gain insight into the barriers to MN differentiation, we evaluated the gene expression profile of MN codes in RA/SHH-treated cells temporally to determine which factors were rate-limiting in the process. After identifying these rate-limiting factors, we utilized an adenoviral gene delivery strategy to rapidly induce expression of MN codes: neurogenin 2 (NGN2), islet-1 (ISL-1), and LIM/homeobox protein 3 (LHX3) in both hES and hiPS cells. Our differentiation strategy to generate induced MNs from human pluripotent stem cells could be utilized for in vitro MN disease modeling platforms or as a potential source for cell-based therapies.
Results
RA/SHH induction of MNs from hESCs requires a long differentiation and maturation period
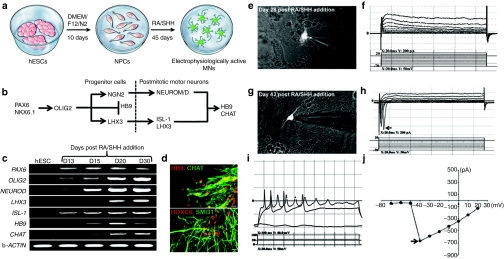
For our initial studies, we utilized the HSF-6 hESC cell line, which was confirmed to have a normal karyotype throughout the course of these studies (Supplementary Figure S1). In addition, these cells expressed high levels of alkaline phosphatase and transcription factors involved in pluripotency such as REX1, OCT3/4, NANOG, and SOX2, as assessed by immunohistochemistry and reverse transcriptase-PCR (RT-PCR) (Supplementary Figure S2a–c). This line also expressed a cell surface marker unique to hESCs, SSEA4, as shown by immunocytochemical analysis (Supplementary Figure S2c), demonstrating that our cells expressed markers for bona fide pluripotent stem cells. To confirm that the HSF-6 cell line could produce MNs, we utilized the prototypical methods of ectopically administering RA and SHH to signal MN development pathways,16,18,22 (Figure 1a–b). The HSF6-hESCs could be induced into a neuroectodermal fate, as previously described,22,23 with >70% of cells positive for neural progenitor cell (NPC) markers PSA-NCAM and OTX2 (Supplementary Figure S3). We then evaluated the MN gene expression profile by semi-quantitative RT-PCR to characterize the timing for MN activation (Figure 1c). Following addition of RA and SHH to embryoid body cultures, we analyzed the transcriptional program that induces MN formation (Figure 1b) temporally over 30 days by semi-quantitative RT-PCR (Figure 1c). In proliferating hESCs, no neuronal specifying factors were present in our cultures, suggesting a homogeneous population of undifferentiated cells. At day 13 of differentiation, we observed expression of early MN-specifying factors, PAX6 and OLIG2 as well as ISL-1 and NEUROD (Figure 1c). Immunocytochemical analysis also confirmed the expression of the former three markers (Supplementary Figure S3). Expression of all MN-specifying genes was evident at day 20 and day 30, including strong expression of LHX3, ISL-1, and HB9 (Figure 1c). Twenty days after RA/SHH, choline acetyltransferase (CHAT) expression appeared and increased at day 30. Immunohistochemistry confirmed the expression of endogenous HB9, LHX3, ISL-1, and CHAT, indicating that these cells were indeed expressing prototypical markers of MNs (Figure 1d, Supplementary Figure S3). Quantification of HB9+ and CHAT+ cells showed ~30% of total cells were indeed mature MNs, an efficiency similar to prior reports of hESC differentiation into MNs.17,18,19 In addition, the hESC-derived MNs expressed SMI31, a nonphosphoryated neurofilament as well as HOXC6, demonstrating cervical MN identity, as expected (Figure 1d).17 Collectively, these results exhibit a slow maturation process of RA/SHH-induced cells into a MN phenotype.
Figure 1.
hESCs require a long differentiation and maturation period to differentiate into functional MNs in vitro. (a) Differentiation paradigm of hESCs into MNs. hESCs were differentiated into neural progenitor cells in DMEM/F12/N2 media followed by RA/SHH treatment to induce MN differentiation. (b) Schematic depicting critical signaling factors in MN development. (c) RT-PCR analysis of markers involved in early and late MN differentiation assayed at multiple time points post RA/SHH treatment. (d) Mature MNs coexpress HB9 (red) and CHAT (green), and also express SMI31 (green) and express a marker of cervical spinal cord identity, HOXC6 (red). (e) Patch-clamped Hb9::RFP MN at 28 days post RA/SHH addition showed no sodium current activity (f). (g) Patch-clamped Hb9::RFP MN 2 weeks later showed sodium current activity (h), action potentials (i), and peak sodium current activity of approximately −700 pA (j). Arrow in panel “h” shows peaks representing sodium currents. Arrow in panel “j” shows the peak sodium current activity. In panel “d” bar = 50 µm. CHAT, choline acetyltransferase; DMEM, Dulbecco's modified Eagle's medium; hESCs, human embryonic stem cells; MN, motor neurons; RA, retinoic acid; RT-PCR, reverse transcriptase-PCR; SHH, sonic hedgehog.
To determine whether the neurons in our cultures, which expressed mature MN markers, were indeed functionally mature, we analyzed their electrophysiological properties to test whether they could fire action potentials. Specifically, we utilized whole-cell perforated patch recording techniques to determine whether they developed the fast inactivating sodium currents and action potentials indicative of mature MNs.24 To easily identify MNs in live cultures, a previously reported lentivirus reporter13 encoding a red fluorescent protein (RFP) under the control of the MN-specific Hb9 promoter was utilized. Transduced MNs were easily visualized by strong RFP expression throughout the whole-cell body and neuritic extensions (Figure 1e,g). We first evaluated the electrophysiological properties of our differentiated MN cultures at day 28, an approximate time point in which we observed high expression levels of HB9 and CHAT. Interestingly, all six recorded MNs at this time point, which had been cultured on astrocytes for an additional 4 days in the presence of neurotrophic factors for further maturation25 were not electrophysiologically mature, despite expressing all of the phenotypic markers for a MN, as no patch-clamped Hb9-RFP+ MN fired action potentials (Figure 1e–f, Figure 2a). This observation suggested additional maturation was required to induce the electrophysiological machinery proteins required for MNs to fire action potentials. Therefore, we cocultured these MNs for an additional 2 weeks on human astrocytes in the presence of RA/SHH and neurotrophic factors. At 42 days post RA/SHH treatment, a total of four Hb9::RFP-positive MNs were patch clamped in culture (Figure 1g) and a current step protocol recorded fast sodium currents within all of these MNs (Figures 1h and 2a). In addition, upon electrical stimulation, patch-clamped Hb9-RFP+ MNs fired action potentials (Figure 1i), with strong peak sodium currents of approximately −700 pA (Figure 1j). All of these results demonstrate that hESCs require up to 52 days under differentiating conditions for complete functional maturation into a MN.16,17,18,19,20
Figure 2.
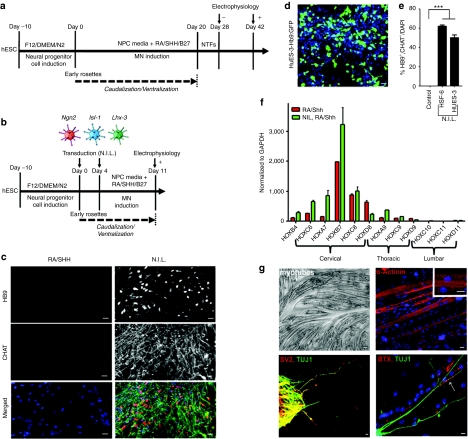
Rapid and efficient MN differentiation of hESCs by the N.I.L. transcription factors. (a) Time line of differentiating human ES or iPS cells towards electrophysiologically-competent MNs utilizing RA/SHH. (b) Time line of accelerating human ES or iPS cell differentiation towards electrophysiologically-competent MNs utilizing N.I.L. (c) N.I.L.-induced MN differentiation showed co-expression of mature MN markers HB9 and CHAT. (d) HuES-3 Hb9::GFP cells differentiated into MNs with the N.I.L. transcription factor code show GFP expression. (e) Quantification of colabeled HB9/CHAT induced MNs. (f) qRT-PCR analysis of HOX genes expressed in the cervical, thoracic, and lumbar regions of the spinal cord assayed from N.I.L.-induced MNs and RA/SHH-induced MNs. (g) Phase contrast picture of C2C12-derived myotubes, which express α-actinin, with a high-power insert showing characteristic striations. (g) N.I.L.-induced MNs express SV2 (red) and colocalize with axons labeled with TUJ1 (green). MN axon terminals form junctions with acetylcholine receptors labeled with bungarotoxin (Red) as shown by an arrow (bottom panel). Arrow in panel “g” shows the colocalization of a MN axon with acetylcholine receptors on a myotube. In panel “c” all bars = 50 µm. In panel “g” all bars = 20 µm. DMEM, Dulbecco's modified Eagle's medium; hESCs, human embryonic stem cells; MN, motor neurons; N.I.L., Ngn2, Isl-1, and Lhx3; RA, retinoic acid; qRT-PCR, quantitative reverse transcriptase-PCR; SHH, sonic hedgehog.
Gene delivery of MN-specifying transcription factors to hESCs results in rapid and efficient generation of MNs
To overcome these barriers related to inefficient and time-intensive MN differentiation from hESCs, we hypothesized that delivery of key MN-specifying transcription factors could rapidly facilitate their differentiation to MNs at a higher efficiency compared to the conventional RA/SHH approach. Based on our temporal gene expression analysis, which identified rate-limiting transcription factors when hESCs were differentiated to MNs upon RA/SHH exposure, we cloned three key MN-specifying genes - Ngn2, Isl-1, and Lhx3. Previous reports have underscored the essential roles of these factors for proper MN development. Specifically, a combinatorial expression of LIM homeodomain transcription factors, Lhx3 and Isl-1, together with the expression of the pro-neural gene, Ngn2, have been shown critical factors to induce MN specification during development.26,27,28,29 Thus, we utilized these factors to efficiently delivery Ngn2, Isl-1, and Lhx3 to our hESC-derived NPCs. We chose adenoviral vectors because of their high infectivity and rapid expression of multiple transgenes within NPCs. Immunohistochemistry revealed that these viruses were capable of infecting hESC-derived NPCs at percentage levels greater than 95% and mediating expression of their respective transcription factor (Supplementary Figure S4). Based on this analysis, our adenoviral delivery approach targeted ~85% of cells in culture. We next tested whether these factors could accelerate the differentiation of our hESC-derived NPCs into MNs with greater efficiency compared solely to utilizing RA/SHH-mediated differentiation (Figure 2a,b). hESCs were induced into EBs, allowed to adhere to poly-ornithine and laminin-coated dishes, and then differentiated for 10 days into an NPC phenotype. Neural rosette structures were subsequently dissociated and then infected with our cocktail of viruses encoding Ngn2, Isl-1, and Lhx3, and hereafter referred to as N.I.L., at a multiplicity of infectivity of 100. We infected cells at two time points (day 0 and day 4 post NPC differentiation) to increase the efficiency and longevity of expression. In addition, media were supplemented during the differentiation procedure with MN factors including RA, SHH, forskolin, and B27 supplement (Figure 2b). Eleven days following infection, we stained cells for phenotypic markers of MNs. At this differentiation stage, cells stained positive for HB9 and CHAT, suggesting that the N.I.L. transcription factors were indeed increasing the rate of MN differentiation within these cells (Figure 2c). Nontransduced control hESC-derived NPCs incubated only with RA/SHH for 11 days showed no HB9 or CHAT immunostaining (Figure 2c), which is also in agreement with our RT-PCR analysis of temporal gene transcription factor induction during MN differentiation with RA/SHH (Figure 1c). To test whether the N.I.L. transcription factors were capable of MN-induced differentiation without any extrinsic neuralizing factors such as RA or SHH, hESC-derived NPCs were infected with N.I.L. and incubated only with B27 supplement to aid in their survival. Without these additional factors, the N.I.L. transcription factors were unable to induce the mature MNs markers, HB9 and CHAT, by this approach, suggesting that the caudalizing effects of RA and ventralizing functions of SHH activate alternate gene expression profiles which are essential for MN differentiation. To verify whether the N.I.L. transcription factors could efficiently direct MN differentiation from an alternative hESC line, we utilized the HuES-3 line, which had been previously engineered with an Hb9::GFP reporter.12 We utilized the same differentiation protocol as described above, and after 11 days of differentiation, we were able to detect GFP expression within 55% of the cells, demonstrating these cells were efficiently being instructed towards a MN fate (Figure 2d). To determine the efficiency of the N.I.L.-directed differentiation approach, we quantified the total number of cells coexpressing HB9 and CHAT. Strikingly, at this early time point, we found that 50 ± 4.5% of our differentiated cells were HB9/CHAT colabeled, indicating that this approach resulted in rapid and efficient generation of MNs (Figure 2e). Quantification of MNs generated from the HSF-6 cell line showed 62 ± 2.5% were HB9/CHAT colabeled (Figure 2e). Collectively, these results show that MNs can be efficiently and rapidly differentiated from two independent hESC lines by the N.I.L. transcription factor code.
N.I.L. induces predominately cervical and thoracic MN subtypes from hESCs
As hundreds of distinct MN subtypes exist throughout the length of the spinal cord, each responsible for controlling specific muscle groups, we next wanted to identify the MN subtypes that were generated from our N.I.L. induced cells. For this analysis, we applied quantitative PCR to analyze expression of members of the HOX family of transcription factors,30,31,32,33 that dictate cervical, thoracic and lumbar segmental identity phenotypes. Consistent with prior results,17,18 the default MN phenotype from RA/SHH differentiation is of a cervical phenotype, and our HOX profiling confirmed high expression of cervical markers, including HOXC6, A7, B7, C8, and D8, with low levels of thoracic HOX gene expression and little to no lumbar HOX genes expressed (Figure 2f). Also as anticipated, the N.I.L. induced cells expressed a very similar profile of HOX gene expression as RA/SHH treated cells, thereby confirming that similar MN subtypes can be generated either from our directed transcription factor based approach or by the conventional RA/SHH treatment protocol (Figure 2f).
N.I.L.-induced MNs form neuromuscular junctions in vitro
To further evaluate the functional maturity of the induced MNs, we tested whether these cells could form neuromuscular junctions when cocultured with differentiated C2C12-derived myotubes, which were confirmed to be expressing the mature myotube marker α-actinin (Figure 2g). Within the cocultures, we detected the synaptic marker, synaptic vesicle protein 2, colocalized with TUJ1-positive stained MN axons (Figure 2g). In addition, motor axons colocalizing with acetylcholine receptors on the myotubes were detected with rhodamine-conjugated α-bungarotoxin (Figure 2g), further indicating that the MNs were capable of forming neuromuscular junctions.
N.I.L.-induced MNs acquire electrophysiological properties in a rapid time frame
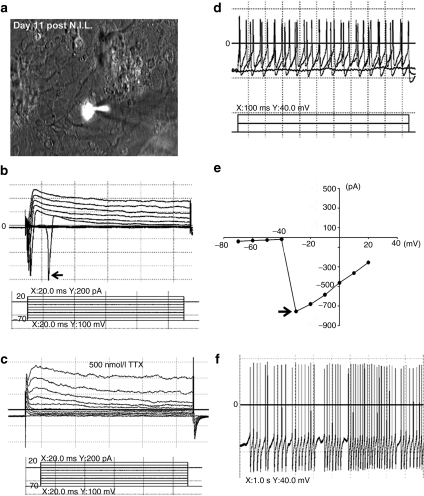
We next evaluated the functional maturity of the MNs by measuring their electrophysiological properties 11 days post-transduction with the N.I.L. cocktail. To visualize MNs, we infected cells with the previously described lentivirus containing an Hb9::RFP reporter and were then transferred to a layer of astrocytes for an additional 4 days before electrophysiological measurements were performed. A total of 10 RFP-positive MNs, differentiated from the HSF-6 cell line, were patch-clamped (Figure 3a), and electrophysiological measurements demonstrated strong sodium currents (Figure 3b), indicative of MN electrophysiological properties in all recorded MNs. Sodium channel currents could also be blocked by administration of 500 nmol/l tetrodotoxin (Figure 3c), further demonstrating the specificity of sodium current channel measurements. In addition, the MNs demonstrated action potentials upon electrical stimulation, indicating their functional maturity (Figure 3d). Sodium current were measured at approximately −800 pA (Figure 3e), with the additional capacity for strong spontaneous activity (Figure 3f). Collectively, these results demonstrate that the N.I.L. instruction rapidly produced MNs with the electrophysiological competence of mature MNs.
Figure 3.
N.I.L.-induced MNs are functionally mature showing characteristic electrophysiological properties. (a) N.I.L.-induced MN labeled with Hb9::RFP and patch clamped showing sodium current activity (b) that could be blocked with tetrodotoxin (TTX) (c). N.I.L.-induced MNs displayed action potentials (d), peak sodium current activity of approximately −800 pA (e), and spontaneous activity (f). Arrow in panel “b” shows peaks representing sodium currents. Arrow in panel “e” shows the peak sodium current activity. MN, motor neurons; N.I.L., Ngn2, Isl-1, and Lhx3.
N.I.L. induces MNs rapidly and efficiently from hiPSCs
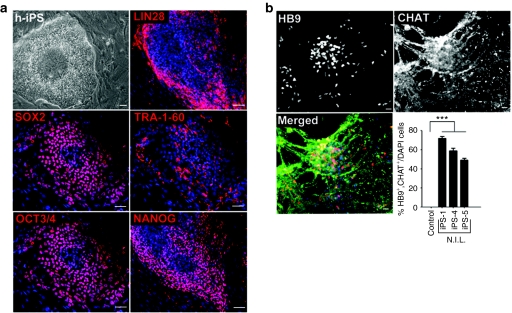
Given the success of using N.I.L. on hESCs for rapidly and efficiently generating electrophysiologically-competent MNs, we next planned to determine whether the same approach could be utilized to generate MNs from human induced pluripotent stem cells (hiPSCs). Numerous reports have demonstrated that hiPSCs can be derived from multiple cell types utilizing various transcription factor cocktail combinations.34,35,36,37,38,39,40,41 We derived a line of hiPSCs from human fibroblasts using retroviral vectors expressing the four Yamanaka factors (OCT3/4, KLF-4, SOX2, and c-MYC)42 (Figure 4a). The hiPSCs were stained with LIN28, OCT3/4, SOX2, TRA-1-60, and NANOG to confirm that they expressed pluripotent characteristics (Figure 4a). These cells were then exposed to the same protocol that we used on hESCs and described in Figure 2b. Similar to the previously described protocol for the N.I.L. induction of MNs, cells were transduced with adenoviruses expressing N.I.L. and evaluated 11 days later for MN markers. The cells exhibited robust MN differentiation, with 72 ± 3.4% of the cells colabeled with HB9 and CHAT (Figure 4b). Previous studies have shown that iPS lines could vary in their ability to differentiate into mature cell types.43 Therefore, we tested two additional iPS lines for their ability to become efficiently differentiated into MNs utilizing our N.I.L. induction strategy.44 Similar to our previous results, we were able to achieve similar MN differentiation efficiencies within iPS lines 4 and 5 showing 59 ± 4.2% and 49 ± 3.3% colabeling with HB9 and CHAT, respectively (Supplementary Figure S5, Figure 4b). Consistent with our previous results on hESC differentiation into MNs with RA/SHH, nontransduced control cells did not stain positive for HB9 or CHAT at this early time point in differentiation. These results further exemplify the utility of the N.I.L directed programming approach on multiple hiPS cell lines showing highly efficient and rapid generation of MNs (Figure 4b).
Figure 4.
N.I.L.-induces rapid and efficient generation of MNs from hiPSCs. (a) Phase contrast picture of a hiPSC clone showing distinct hESC morphology. hiPSCs express markers associated with pluripotency such as LIN28, OCT3/4, SOX2, TRA-1-60, and NANOG. (b) N.I.L.-induced MNs express HB9 and CHAT, and are colocalized in the same cells merged with the nuclear dye, DAPI. Quantification of induced MNs colabeled with HB9/CHAT from hiPS cells. In panel “a” bars = 100 µm. In panel “b” bars = 50 µm. hESC, human embryonic stem cell; hiPSCs, human induced pluripotent stem cells; MN, motor neurons; N.I.L., Ngn2, Isl-1, and Lhx3.
Discussion
Pluripotent cells have proven to be an invaluable stem cell source for deriving MNs. However, generating MNs is time intensive and current approaches yield 10–40% MN differentiation along with a heterogeneous population of neuronal and non-neuronal cells.16,17,18,19,20,21 In part, this heterogeneity is caused by the way in which RA and SHH impart neuronal diversity within the embryonic spinal cord.45 To more accurately and efficiently guide hESCs into MNs, we utilized critical MN-specifying transcription factors that act downstream of RA/SHH signaling.29 Our results demonstrate that MN-specifying transcription factors are capable of producing functional MNs in a rapid and efficient manner. A future direction for this technology would be to program alternate neuronal types that exist throughout the CNS. Indeed, recent work has shown that delivery of a critical transcription factor, Lmx1a, to hES cells can direct their differentiation into mesencephalic dopaminergic neurons.46 In addition, it may be possible to use transcription factor codes to generate alternate MN subtype fates such as those that innervate limb muscles.
Although we have not observed any untoward effects of sustained expression of these factors within MNs, it will be of interest to determine whether the MN genetic program is retained after silencing viral-mediated transcription factor expression postdifferentiation. Furthermore, we have observed that N.I.L.-directed MNs could survive up to 6 weeks in culture demonstrating their high applicability as a drug screening tool.
MNs have been increasingly utilized to model neurodegenerative diseases in vitro such as SMA and ALS.5,8,9,10,11,12,13 For example, Ebert et al. demonstrated that MNs differentiated from iPS cells, which were derived from an SMA patient, showed that SMA MNs exhibited shortened neurite extensions and decreased survival in culture compared to healthy MNs.5 Additional reports modeling ALS in vitro have exemplified the susceptibility of MNs to ALS-derived astrocytes.8,9,10,11,12,13 Although ALS patient-derived MNs have been generated through iPS cell technology,47,48 no report has documented any defect in these MNs. To overcome the technical challenges of generating MNs from large numbers of iPS lines from patients with MN disease, a more rapid and efficient method of MN differentiation would be highly invaluable to stem cell biologists over currently available techniques. Our induced MN differentiation method from either hES or iPS cells bypasses these technical constraints of MN differentiation by generating a large supply of these cells within a rapid and accelerated time frame. We envision this technology would be highly applicable for drug screening platforms for MN disease such as SMA or ALS. Pluripotent stem cell lines could be engineered with an Hb9::GFP or Hb9::luciferase reporter system for monitoring and quantification purposes during the drug screening process. In addition, toxicology studies could also be performed on highly enriched MNs to test for candidate drug targets.
In sum, this transcriptional coding strategy allows for the generation of functional MNs with characteristic electrophysiological properties at high efficiency within ~11 days from hESCs or hiPSCs, a strategy that should prove valuable to those studying basic MN biology in vitro, developing screening assays for MN disease, or advancing applications in regenerative medicine.
Materials and Methods
Cell culture. The HSF-6 hESC line (P65) was obtained from the National Stem Cell Bank (NSCB) and was cultured and passaged on irradiated primary mouse embryonic fibroblasts (Millipore, Billerica, MA) as previously described.49 hiPSCs were cultured similarly to hESCs. Human iPS lines, iPS-4 and iPS-5, were obtained from Children's Hospital Boston Stem Cell Program, George Q. Daley's laboratory. The HuES-3 Hb9::GFP cell line was a kind gift from Kevin Eggan (Harvard University, Cambridge, MA).
Adenoviral and lentiviral constructs. The AdEasy Adenoviral Vector System (Stratagene, La Jolla, CA) was utilized to construct adenoviruses for the mouse Lhx3, Isl1, and Ngn2 genes. All cDNAs corresponding to the coding regions of these genes were subcloned into the BamHI and Xho I sites of the pShuttle-CMV adenoviral vector, and all subsequent steps were performed according to the manufacturer's instructions using human 293 cells for viral propagation. Viral titers were between 1011 and 1012 viral particles/ml, as determined by the TCID50 assay.
A lentiviral reporter construct driving RFP under the control of the Hb9 promoter (kind gift of Sam Pfaff, Carol Marchetto, and Fred Gage) was utilized to identify MNs. Lentivirus, pseudotyped with vesicular stomatis virus glycoprotein, was produced by quadruple transfection in HEK-293 cells, as previously described.50
hiPSC generation. pMXs retroviral vectors (Addgene, Cambridge, MA) encoding OCT3/4, SOX2, KLF-4, and c-MYC were transfected using the CaCl2 method separately with CMV-GP and VSVG plasmids into human embryonic kidney carcinoma 293 cells (HEK-293) to produce retroviruses. Forty-eight hours postinfection, supernatant was collected, mixed, and filtered through a 0.22-micron filter and supplemented with polybrene at 5 µg/ml. 60,000 normal fibroblasts (Coriell, Camden, NJ) were infected with retroviruses on a 10 cm2 tissue culture plate. Media was changed to Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and cells were allowed to grow for 1 week until media was changed to hESC media supplemented with the rho kinase inhibitor, Y-27632 (Calbiochem, Gibbston, NJ).
Karyotyping. Karyotyping of the HSF6 hESC line was performed at the Molecular Cytogenetics shared resource at the Ohio State University Comprehensive Cancer Center.
qRT-PCR. RNA was harvested from proliferating hESCs, RA/SHH-induced, and N.I.L.-induced MNs. Total RNA was reverse transcribed with RT2 First Strand Kit (SABiosciences, Frederick, MD) according to the manufacturer's instructions. Real-time quantitative PCR reactions were performed using RT2 Real-Time SYBR Green/Rox PCR Master Mix (SABiosciences) and run on an RT2 Profiler PCR Array for Human HOX genes (SABiosciences). Primers used in this study are shown in Supplementary Table S1.
MN differentiation. To induce MN differentiation, hESC or iPSC colonies were first removed from a mouse feeder layer with 1 mg/ml collagenase IV. Colonies were then resuspended in Dulbecco's modified Eagle's medium/F12 supplemented with 1% N2 and 10% knockout serum (Invitrogen, Carlsbad, CA), and plated within a Petri dish to induce embryoid body formation for the first 2 days. After 2 days of incubation, serum-free media was utilized as described above without 10% knockout serum for an additional 2 days. After 4 days of differentiation, EBs were then allowed to attach as distinct noncontacting colonies to polyornithine/laminin coated dishes for an additional 6 days to induce the formation of neural rosettes. Neural rosettes were then manually removed by a small pipettor, dissociated with accutase (Invitrogen), plated on poly-ornithine/laminin coated dishes, and then treated either with RA (2 µmol/l) and SHH (500 ng/ml), for conventional MN differentiation.
N.I.L.-induced MN differentiation. NPCs were differentiated from hESCs or iPSCs by the same protocol as described above. After 10 days of differentiation into neural rosettes containing NPCs, neural rosettes were manually removed with a pipettor, dissociated, and plated on poly-ornithine/laminin coated dishes as described above. After 24 hours, NPCs were infected with N.I.L., which is considered day 1 on the differentiation time-line (Figure 2a–b) (multiplicity of infection = 100), and supplemented with RA (2 µmol/l), forskolin (5 µmol/l), SHH (500 ng/ml), and B27 (0.2%). On day 4, NPCs were infected an additional time at a similar multiplicity of infection and supplemented with the same factors as described above.
Myotube coculture experiments. C2C12 cells were differentiated in 10% horse serum to induce myotube formation. MNs were then plated to the myotube monolayer and showed the neuromuscular synapses within 1 week of coculture.
Immunocytochemistry and alkaline phosphatase activity staining. Cells were stained with the antibodies shown in Supplementary Table S1. All images were captured using a Zeiss LSM510-META confocal laser-scanning microscope (Zeiss, Thornwood, NY). Staining for alkaline phosphatase activity was performed using the alkaline phosphatase activity kit (Millipore).
Electrophysiology recording. Whole-cell patch recordings were performed from hESC-derived MNs that express Hb9::RFP. The recording micropipettes were pulled to have tip resistance of 2–4 MΩ when filled with an internal solution (115 mmol/l K-gluconate, 4 mmol/l NaCl, 1.5 mmol/l MgCl2, 20 mmol/l HEPES, and 0.5 mmol/l EGTA, pH 7.3) and placed in a bath solution (115 mmol/l NaCl, 2 mmol/l KCl, 10 mmol/l HEPES, 3 mmol/l CaCl2, 10 mmol/l glucose, and 1.5 mmol/l MgCl2, pH 7.3). Recordings were made using a EPC10 amplifier (HEKA Instruments, Southboro, MA). Signals were filtered at 3 kHz and sampled at 20 kHz. The whole-cell capacitance was fully compensated, whereas the series resistance was uncompensated but monitored during the experiment by the amplitude of the capacitive current in response to a 5-mV pulse. For current-clamp recordings, cells were either held at the level without current injection to observe resting membrane potential and spontaneous firing or held at −60 ~ −70 mV level through current injection prior to the current step protocol. For voltage-clamp recordings, cells were clamped at −70 mV before the voltage step protocol was performed. All recordings were performed at room temperature.
SUPPLEMENTARY MATERIAL Figure S1. HSF-6 hESCs show a normal 46XX karyotype. Figure S2. The HSF-6 hESC line shows markers of pluripotency. hESCs show alkaline phosphatase (AP) activity (a), pluripotency gene expression profiles (b), and pluripotency transcription factor expression such as OCT3/4, NANOG, SOX2, and surface antigen, SSEA-4 (c). All scale bars are equal to 100 μM. Figure S3. HSF-6 hESCs transit through progenitor cell intermediates towards a MN fate. Immunocytochemical analysis of NPCs differentiated from hESCs expressing OTX2 (red) and NCAM (green). Upon addition of RA/SHH, progenitor cells express PAX6 (red) and OLIG2 (red). NGN2 (red) was also detected, which is expressed in early MN progenitor cells and is co-localized with SMI31 (green). Post-mitotic MNs express ISL-1 (red) and LHX3 (red) and both proteins are co-localized with CHAT (green). All scale bars are equal to 50 μM. Figure S4. Adenoviral transcription factors Ngn2, Isl-1, and Lhx3 can infect hESC-derived NPCs. Immunocytochemical analysis of Ngn2, Isl-1, and Lhx3 after adenoviral infection stained red and merged with DAPI. All scale bars are equal to 50 μM. Figure S5. N.I.L.-induced MNs express HB9 and CHAT, and are colocalized in the same cells merged with the nuclear dye, DAPI in iPS lines 4 and 5. Scale bars are equal to 50 μM. Table S1. All antibodies and sequences of primers utilized in this study.
Acknowledgments
We are thankful to Kevin Eggan for providing the HuES-3 Hb9::GFP cell line. This work was funded by NIH R01 NS6449l2-1A1, RC2 NS69476-01, Helping Link Foundation to B.K.K., The Ohio State University Electrophysiology Core supported by NINDS core grant P30 NS045758., and Project A.L.S. to F.H.G. and B.K.K. and F.H.G. is supported in part by California Institute for Regenerative Medicine (CIRM). K.M. is supported by a fellowship from the Swiss National Science Foundation (SNSF), and C.J.M. is supported by a fellowship from the Craig H. Neilsen Foundation.
Supplementary Material
HSF-6 hESCs show a normal 46XX karyotype.
The HSF-6 hESC line shows markers of pluripotency. hESCs show alkaline phosphatase (AP) activity (a), pluripotency gene expression profiles (b), and pluripotency transcription factor expression such as OCT3/4, NANOG, SOX2, and surface antigen, SSEA-4 (c). All scale bars are equal to 100 μM.
HSF-6 hESCs transit through progenitor cell intermediates towards a MN fate. Immunocytochemical analysis of NPCs differentiated from hESCs expressing OTX2 (red) and NCAM (green). Upon addition of RA/SHH, progenitor cells express PAX6 (red) and OLIG2 (red). NGN2 (red) was also detected, which is expressed in early MN progenitor cells and is co-localized with SMI31 (green). Post-mitotic MNs express ISL-1 (red) and LHX3 (red) and both proteins are co-localized with CHAT (green). All scale bars are equal to 50 μM.
Adenoviral transcription factors Ngn2, Isl-1, and Lhx3 can infect hESC-derived NPCs. Immunocytochemical analysis of Ngn2, Isl-1, and Lhx3 after adenoviral infection stained red and merged with DAPI. All scale bars are equal to 50 μM.
N.I.L.-induced MNs express HB9 and CHAT, and are colocalized in the same cells merged with the nuclear dye, DAPI in iPS lines 4 and 5. Scale bars are equal to 50 μM.
All antibodies and sequences of primers utilized in this study.
REFERENCES
- Foust KD, Wang X, McGovern VL, Braun L, Bevan AK, Haidet AM.et al. (2010Rescue of the spinal muscular atrophy phenotype in a mouse model by early postnatal delivery of SMN Nat Biotechnol 28271–274. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Dominguez E, Marais T, Chatauret N, Benkhelifa-Ziyyat S, Duque S, Ravassard P.et al. (2011Intravenous scAAV9 delivery of a codon-optimized SMN1 sequence rescues SMA mice Hum Mol Genet 20681–693. [DOI] [PubMed] [Google Scholar]
- Passini MA, Bu J, Roskelley EM, Richards AM, Sardi SP, O'Riordan CR.et al. (2010CNS-targeted gene therapy improves survival and motor function in a mouse model of spinal muscular atrophy J Clin Invest 1201253–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valori CF, Ning K, Wyles M, Mead RJ, Grierson AJ, Shaw PJ.et al. (2010Systemic delivery of scAAV9 expressing SMN prolongs survival in a model of spinal muscular atrophy Sci Transl Med 235ra42. [DOI] [PubMed] [Google Scholar]
- Ebert AD, Yu J, Rose FF, Jr, Mattis VB, Lorson CL, Thomson JA.et al. (2009Induced pluripotent stem cells from a spinal muscular atrophy patient Nature 457277–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi SL, Nistor G, Wyatt T, Yin HZ, Poole AJ, Weiss JH.et al. (2010Histological and functional benefit following transplantation of motor neuron progenitors to the injured rat spinal cord PLoS ONE 5e11852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti S, Nizzardo M, Nardini M, Donadoni C, Salani S, Ronchi D.et al. (2008Neural stem cell transplantation can ameliorate the phenotype of a mouse model of spinal muscular atrophy J Clin Invest 1183316–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai M, Re DB, Nagata T, Chalazonitis A, Jessell TM, Wichterle H.et al. (2007Astrocytes expressing ALS-linked mutated SOD1 release factors selectively toxic to motor neurons Nat Neurosci 10615–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giorgio FP, Carrasco MA, Siao MC, Maniatis T., and, Eggan K. Non-cell autonomous effect of glia on motor neurons in an embryonic stem cell-based ALS model. Nat Neurosci. 2007;10:608–614. doi: 10.1038/nn1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodge JC, Haidet AM, Yang W, Passini MA, Hester M, Clarke J.et al. (2008Delivery of AAV-IGF-1 to the CNS extends survival in ALS mice through modification of aberrant glial cell activity Mol Ther 161056–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodge JC, Treleaven CM, Fidler JA, Hester M, Haidet A, Handy C.et al. (2010AAV4-mediated expression of IGF-1 and VEGF within cellular components of the ventricular system improves survival outcome in familial ALS mice Mol Ther 182075–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giorgio FP, Boulting GL, Bobrowicz S., and, Eggan KC. Human embryonic stem cell-derived motor neurons are sensitive to the toxic effect of glial cells carrying an ALS-causing mutation. Cell Stem Cell. 2008;3:637–648. doi: 10.1016/j.stem.2008.09.017. [DOI] [PubMed] [Google Scholar]
- Marchetto MC, Muotri AR, Mu Y, Smith AM, Cezar GG., and, Gage FH. Non-cell-autonomous effect of human SOD1 G37R astrocytes on motor neurons derived from human embryonic stem cells. Cell Stem Cell. 2008;3:649–657. doi: 10.1016/j.stem.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Wichterle H, Lieberam I, Porter JA., and, Jessell TM. Directed differentiation of embryonic stem cells into motor neurons. Cell. 2002;110:385–397. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- Briscoe J., and, Ericson J. Specification of neuronal fates in the ventral neural tube. Curr Opin Neurobiol. 2001;11:43–49. doi: 10.1016/s0959-4388(00)00172-0. [DOI] [PubMed] [Google Scholar]
- Singh Roy N, Nakano T, Xuing L, Kang J, Nedergaard M., and, Goldman SA. Enhancer-specified GFP-based FACS purification of human spinal motor neurons from embryonic stem cells. Exp Neurol. 2005;196:224–234. doi: 10.1016/j.expneurol.2005.06.021. [DOI] [PubMed] [Google Scholar]
- Lee H, Shamy GA, Elkabetz Y, Schofield CM, Harrsion NL, Panagiotakos G.et al. (2007Directed differentiation and transplantation of human embryonic stem cell-derived motoneurons Stem Cells 251931–1939. [DOI] [PubMed] [Google Scholar]
- Li XJ, Du ZW, Zarnowska ED, Pankratz M, Hansen LO, Pearce RA.et al. (2005Specification of motoneurons from human embryonic stem cells Nat Biotechnol 23215–221. [DOI] [PubMed] [Google Scholar]
- Shin S, Dalton S., and, Stice SL. Human motor neuron differentiation from human embryonic stem cells. Stem Cells Dev. 2005;14:266–269. doi: 10.1089/scd.2005.14.266. [DOI] [PubMed] [Google Scholar]
- Lim UM, Sidhu KS., and, Tuch BE. Derivation of Motor Neurons from three Clonal Human Embryonic Stem Cell Lines. Curr Neurovasc Res. 2006;3:281–288. doi: 10.2174/156720206778792902. [DOI] [PubMed] [Google Scholar]
- Hu BY., and, Zhang SC. Differentiation of spinal motor neurons from pluripotent human stem cells. Nat Protoc. 2009;4:1295–1304. doi: 10.1038/nprot.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhara SK., and, Stice SL. Neural differentiation of human embryonic stem cells. J Cell Biochem. 2008;105:633–640. doi: 10.1002/jcb.21891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Xu J, Pang ZP, Ge W, Kim KJ, Blanchi B.et al. (2007Integrative genomic and functional analyses reveal neuronal subtype differentiation bias in human embryonic stem cell lines Proc Natl Acad Sci USA 10413821–13826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles GB, Yohn DC, Wichterle H, Jessell TM, Rafuse VF., and, Brownstone RM. Functional properties of motoneurons derived from mouse embryonic stem cells. J Neurosci. 2004;24:7848–7858. doi: 10.1523/JNEUROSCI.1972-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MA, Weick JP, Pearce RA., and, Zhang SC. Functional neural development from human embryonic stem cells: accelerated synaptic activity via astrocyte coculture. J Neurosci. 2007;27:3069–3077. doi: 10.1523/JNEUROSCI.4562-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK., and, Pfaff SL. Transcriptional networks regulating neuronal identity in the developing spinal cord. Nat Neurosci. 2001;4 Suppl:1183–1191. doi: 10.1038/nn750. [DOI] [PubMed] [Google Scholar]
- Pfaff SL, Mendelsohn M, Stewart CL, Edlund T., and, Jessell TM. Requirement for LIM homeobox gene Isl1 in motor neuron generation reveals a motor neuron-dependent step in interneuron differentiation. Cell. 1996;84:309–320. doi: 10.1016/s0092-8674(00)80985-x. [DOI] [PubMed] [Google Scholar]
- Thaler JP, Lee SK, Jurata LW, Gill GN., and, Pfaff SL. LIM factor Lhx3 contributes to the specification of motor neuron and interneuron identity through cell-type-specific protein-protein interactions. Cell. 2002;110:237–249. doi: 10.1016/s0092-8674(02)00823-1. [DOI] [PubMed] [Google Scholar]
- Shirasaki R., and, Pfaff SL. Transcriptional codes and the control of neuronal identity. Annu Rev Neurosci. 2002;25:251–281. doi: 10.1146/annurev.neuro.25.112701.142916. [DOI] [PubMed] [Google Scholar]
- Dasen JS, De Camilli A, Wang B, Tucker PW., and, Jessell TM. Hox repertoires for motor neuron diversity and connectivity gated by a single accessory factor, FoxP1. Cell. 2008;134:304–316. doi: 10.1016/j.cell.2008.06.019. [DOI] [PubMed] [Google Scholar]
- Dasen JS, Tice BC, Brenner-Morton S., and, Jessell TM. A Hox regulatory network establishes motor neuron pool identity and target-muscle connectivity. Cell. 2005;123:477–491. doi: 10.1016/j.cell.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Dasen JS, Liu JP., and, Jessell TM. Motor neuron columnar fate imposed by sequential phases of Hox-c activity. Nature. 2003;425:926–933. doi: 10.1038/nature02051. [DOI] [PubMed] [Google Scholar]
- Rousso DL, Gaber ZB, Wellik D, Morrisey EE., and, Novitch BG. Coordinated actions of the forkhead protein Foxp1 and Hox proteins in the columnar organization of spinal motor neurons. Neuron. 2008;59:226–240. doi: 10.1016/j.neuron.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki T, Ohnishi H, Oda Y, Tadokoro M, Sasao M, Kato H.et al. (2010Generation of induced pluripotent stem cells from human adipose-derived stem cells without c-MYC Tissue Eng Part A 162197–2206. [DOI] [PubMed] [Google Scholar]
- Hester ME, Song S, Miranda CJ, Eagle A, Schwartz PH., and, Kaspar BK. Two factor reprogramming of human neural stem cells into pluripotency. PLoS ONE. 2009;4:e7044. doi: 10.1371/journal.pone.0007044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S.et al. (2007Induced pluripotent stem cell lines derived from human somatic cells Science 3181917–1920. [DOI] [PubMed] [Google Scholar]
- Kim JB, Greber B, Araúzo-Bravo MJ, Meyer J, Park KI, Zaehres H.et al. (2009Direct reprogramming of human neural stem cells by OCT4 Nature 461649–643. [DOI] [PubMed] [Google Scholar]
- Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T.et al. (2008Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts Nat Biotechnol 26101–106. [DOI] [PubMed] [Google Scholar]
- Haase A, Olmer R, Schwanke K, Wunderlich S, Merkert S, Hess C.et al. (2009Generation of induced pluripotent stem cells from human cord blood Cell Stem Cell 5434–441. [DOI] [PubMed] [Google Scholar]
- Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P., and, Woltjen K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JB, Zaehres H, Araúzo-Bravo MJ., and, Schöler HR. Generation of induced pluripotent stem cells from neural stem cells. Nat Protoc. 2009;4:1464–1470. doi: 10.1038/nprot.2009.173. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K.et al. (2007Induction of pluripotent stem cells from adult human fibroblasts by defined factors Cell 131861–872. [DOI] [PubMed] [Google Scholar]
- Bock C, Kiskinis E, Verstappen G, Gu H, Boulting G, Smith ZD.et al. (2011Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines Cell 144439–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A.et al. (2008Disease-specific induced pluripotent stem cells Cell 134877–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessell TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- Friling S, Andersson E, Thompson LH, Jönsson ME, Hebsgaard JB, Nanou E.et al. (2009Efficient production of mesencephalic dopamine neurons by Lmx1a expression in embryonic stem cells Proc Natl Acad Sci USA 1067613–7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimos JT, Rodolfa KT, Niakan KK, Weisenthal LM, Mitsumoto H, Chung W.et al. (2008Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons Science 3211218–1221. [DOI] [PubMed] [Google Scholar]
- Boulting GL, Kiskinis E, Croft GF, Amoroso MW, Oakley DH, Wainger BJ.et al. (2011A functionally characterized test set of human induced pluripotent stem cells Nat Biotechnol 29279–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS.et al. (1998Embryonic stem cell lines derived from human blastocysts Science 2821145–1147. [DOI] [PubMed] [Google Scholar]
- Kafri T, Blömer U, Peterson DA, Gage FH., and, Verma IM. Sustained expression of genes delivered directly into liver and muscle by lentiviral vectors. Nat Genet. 1997;17:314–317. doi: 10.1038/ng1197-314. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HSF-6 hESCs show a normal 46XX karyotype.
The HSF-6 hESC line shows markers of pluripotency. hESCs show alkaline phosphatase (AP) activity (a), pluripotency gene expression profiles (b), and pluripotency transcription factor expression such as OCT3/4, NANOG, SOX2, and surface antigen, SSEA-4 (c). All scale bars are equal to 100 μM.
HSF-6 hESCs transit through progenitor cell intermediates towards a MN fate. Immunocytochemical analysis of NPCs differentiated from hESCs expressing OTX2 (red) and NCAM (green). Upon addition of RA/SHH, progenitor cells express PAX6 (red) and OLIG2 (red). NGN2 (red) was also detected, which is expressed in early MN progenitor cells and is co-localized with SMI31 (green). Post-mitotic MNs express ISL-1 (red) and LHX3 (red) and both proteins are co-localized with CHAT (green). All scale bars are equal to 50 μM.
Adenoviral transcription factors Ngn2, Isl-1, and Lhx3 can infect hESC-derived NPCs. Immunocytochemical analysis of Ngn2, Isl-1, and Lhx3 after adenoviral infection stained red and merged with DAPI. All scale bars are equal to 50 μM.
N.I.L.-induced MNs express HB9 and CHAT, and are colocalized in the same cells merged with the nuclear dye, DAPI in iPS lines 4 and 5. Scale bars are equal to 50 μM.
All antibodies and sequences of primers utilized in this study.