Abstract
Nuclear factor-κB (NF-κB) is a central regulator of immune response and a potential target for developing anti-inflammatory agents. Mechanistic studies suggest that compounds that directly inhibit NF-κB DNA binding may block inflammation and the associated tissue damage. Thus, we attempted to discover peptides that could interfere with NF-κB signaling based on a highly conserved DNA-binding domain found in all NF-κB members. One such small peptide, designated as anti-inflammatory peptide-6 (AIP6), was characterized in the current study. AIP6 directly interacted with p65 and displayed an intrinsic cell-penetrating property. This peptide demonstrated significant anti-inflammatory effects in vitro and in vivo. In vitro, AIP6 inhibited the DNA-binding and transcriptional activities of the p65 NF-κB subunit as well as the production of inflammatory mediators in macrophages upon stimulation. Local administration of AIP6 significantly inhibited inflammation induced by zymosan in mice. Collectively, our results suggest that AIP6 is a promising lead peptide for the development of specific NF-κB inhibitors as potential anti-inflammatory agents.
Introduction
Nuclear factor-κB (NF-κB) represents a group of five structurally related and evolutionarily conserved mammalian proteins, namely RelA (p65), RelB, c-Rel, p50 (and its precursor p105), and p52 (and its precursor p100).1 Most members of this family can homodimerize as well as heterodimerize with one another. The most prevalent form of NF-κB is a heterodimer consisting of the p50 and p65 subunits. As a ubiquitous transcription factor, NF-κB plays a key role in regulating the transcription of many genes related to inflammation, such as cytokines, chemokines, and inflammatory enzymes.2,3,4 A number of proteins regulated by NF-κB also induce the activation of NF-κB, thereby forming a positive regulatory loop that amplifies inflammatory responses. Abnormal activation of NF-κB has been implicated in the pathogenesis of many inflammatory diseases, such as rheumatoid arthritis, asthma, sepsis, viral infections, and inflammatory bowel diseases, and its role has been confirmed in experimental models involving both acute and chronic inflammation.5,6,7,8,9 Suppression of NF-κB is likely to be effective for the treatment of inflammatory diseases.
NF-κB is one of the points involved in cross-talk between multiple signal transduction pathways. Activated NF-κB leads to transcriptional upregulation of multiple genes. In unstimulated cells, NF-κB is located in the cytoplasm in an inactive form and is bound to inhibitor proteins, namely IκBs, including IκBα, IκBβ, and IκBε. A variety of molecules, including proinflammatory cytokines such as tumor necrosis factor-α (TNF-α) and interleukin-1β (IL-1β) and bacterial products such as lipopolysaccharides, and viral infection induce phosphorylation of IκB at specific NH2-terminal serine residues. Phosphorylated IκB is then ubiquitinated and degraded by the proteasome; thus, the bound NF-κB is released. NF-κB then translocates to the nucleus, binds to κB elements with a consensus sequence of 5′-GGGRNYYYCC-3′, and induces the expression of target genes.2,10 The recognition and binding of NF-κB to κB elements is a key step in the process of NF-κB activation. The p50 subunit of the p50-p65 heterodimer mainly interacts with the κB elements.11,12 The RXXRXRXXC motif in p50, conserved in all Rel/NF-κB proteins, is essential in DNA recognition and binding.13
Most NF-κB inhibitors developed to date inhibit either NF-κB protein activation or IκB phosphorylation and degradation, thus preventing the release of free NF-κB and its entry into the nucleus. Another approach that directly inhibits NF-κB DNA binding, by interfering with the DNA binding of NF-κB to the promoter of targeted genes, also appears amenable to designing specific inhibitors. However, a limited number of agents have been developed based on this approach.14 We reasoned that short peptides resembling the DNA-binding motif RXXRXRXXC in p50 might selectively bind with κB elements and prevent the binding of active NF-κB complexes. With this in mind, we designed and synthesized a series of peptides. Intriguingly, one of these peptides, designated anti-inflammatory peptide-6 (AIP6), inhibited NF-κB transcriptional activity but did not bind with the κB motif.
In the current study, we show that the peptide AIP6 interacts directly with p65 to inhibit the DNA-binding and transcriptional activities of NF-κB and the production of inflammatory mediators in vitro. Interestingly, AIP6 possesses an intrinsic cell-penetrating property. Local administration of AIP6 inhibited inflammatory responses induced by zymosan in the joints and soft tissues in mice. Taken together, our findings suggest that AIP6 can be used as a lead peptide to develop anti-inflammatory agents for the treatment of diseases caused by abnormal activation of NF-κB.
Results
AIP6 binds to and inhibits the DNA-binding activity of the NF-κB p65 subunit
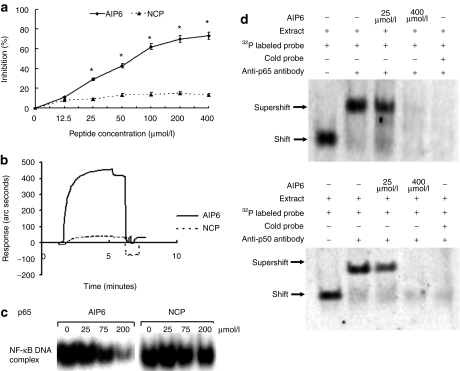
We discovered that AIP6 is a potential inhibitor of NF-κB signaling by using a bioinformatic approach based on the conserved RXXRXRXXC motif that is required for DNA recognition and binding of NF-κB family members.13 We first determined whether AIP6 could block the binding of NF-κB and κB elements in vitro by using an assay based on the enzyme-linked immunosorbent assay (ELISA). AIP6 or negative control peptide (NCP) at various concentrations was preincubated with Jurkat nuclear extract [phorbol, 12-myristate, 13 acetate (TPA) + calcium ionophore-stimulated] for 30 minutes before being added to microtiter plates coated with κB site oligonucleotides. AIP6 at ≥25 µmol/l was found to inhibit the DNA-binding activity of NF-κB in a dose-dependent manner but NCP did not (Figure 1a). Interestingly, when AIP6, even at 400 µmol/l, was preincubated in microtiter plates coated with κB oligonucleotides before the addition of Jurkat nuclear extract (TPA + calcium ionophore-stimulated), no inhibition of the DNA-binding activity of NF-κB was found (Supplementary Figure S1).
Figure 1.
Effects of AIP6 on the DNA-binding activity of NF-κB p65. (a) The effect of AIP6 on the DNA-binding activity of NF-κB was measured by ELISA. AIP6s were preincubated with 2.5 µg Jurkat nuclear extracts for 30 minutes. The mixture was then added to each well to detect the DNA-binding activity of NF-κB. The inhibition ratio (%) of various concentrations of peptides was calculated and plotted by ELISA analysis. (b) Interaction analysis of AIP6 with the p65 NF-κB subunit by using surface plasmon resonance measurements. Recombinant NF-κB p65 was used in EMSA and surface plasmon resonance measurement. (c) The effect of AIP6 on the DNA-binding activity of p65 measured by EMSA. (d) Effect of AIP6 on the DNA-binding activity of the NF-κB p50/p65 heterodimer was analyzed by supershift assay by using Jurkat nuclear extracts with p65 or p50 antibody. Results are expressed as mean ± SEM (n = 3). *P < 0.05 versus inhibition ratio of NCP. AIP6, anti-inflammatory peptide-6; ELISA, enzyme-linked immunosorbent assay; EMSA, electrophoretic mobility shift assays; NCP, negative control peptide; NF-κB, nuclear factor-κB.
This suggested that AIP6 binds to one or more NF-κB subunits rather than to the κB site. We first used surface plasmon resonance spectroscopy to measure the binding of AIP6 with recombinant p65 or p50. AIP6 bound to p65 (Figure 1b) but not to p50 (Supplementary Figure S2). Gel shift assay showed that AIP6 interfered with the binding activity of NF-κB subunit p65 to the κB sites in a dose-dependent manner (Figure 1c) but did not affect that of the p50 subunit (Supplementary Figure S2). Next, we performed supershift assays to analyze the effects of AIP6 on DNA binding of p50/p65 heterodimers, which make up the predominant NF-κB complex. AIP6 inhibited the interactions between the p50/p65 heterodimers and DNA (Figure 1d). As expected, an excess of cold probe completely blocked this interaction (Figure 1d). These results suggested that AIP6 does not bind to the κB element but disrupts the binding between NF-κB and the κB element through direct interaction with p65, not with p50.
AIP6 effectively transduces cells in vitro and in vivo
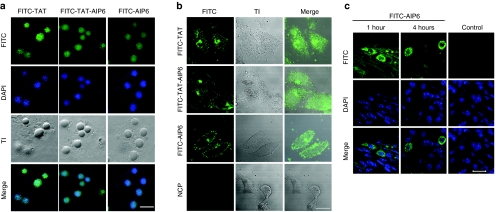
To study the effects of AIP6 on cells, we first used a cell-permeable peptide transactivator of transcription (TAT) derived from the HIV to synthesize a TAT-AIP6 fusion peptide. AIP6 and TAT-AIP6 were labeled with fluorescein isothiocyanate (FITC) and analyzed for uptake in RAW 264.7 macrophages. Confocal microscopy detected strong fluorescence in the cytoplasm and nucleus of almost all RAW 264.7 cells 60 minutes after incubation with FITC-labeled peptides (Figure 2a). The cellular uptake was similar between the AIP6 and TAT-AIP6 groups. To exclude the possibility that transduction was due to phagocytosis in macrophages, similar experiments were conducted in LoVo cells derived from human colorectal cancer line. Confocal microscopy showed that TAT-AIP6, AIP6, and TAT, but not NCP, effectively entered the LoVo cells (Figure 2b). These results indicated that AIP6 has an intrinsic cell-penetrating property similar to that of TAT. We therefore continued to use AIP6 in our study.
Figure 2.
Cellular uptake of AIP6 with or without TAT sequence. (a) Cellular uptake of TAT-AIP6 and AIP6 in RAW 264.7 macrophages. Cells were treated with FITC-labeled TAT, FITC-labeled AIP6, or FITC-labeled TAT-AIP6 (green) for 60 minutes with nuclei counterstained with DAPI (blue). Representative confocal images with FITC, DAPI, phase contrast, and merged FITC/DAPI channels. Bar = 20 µm. (b) Transduction of AIP6 into colon cancer cells. The cells were treated and analyzed similarly as a. Bar = 20 µm. (c) Cellular uptake of AIP6 in mice. After 1-hour and 4-hour injection intervals of AIP6 or PBS (control), soft tissues were removed from the paws of zymosan-treated mice. Frozen sections were analyzed by confocal microscopy with nuclei counterstained with DAPI. Bar = 20 µm. AIP6, anti-inflammatory peptide-6; CPP, cell-permeable peptide; DAPI, 4′,6-diamidino-2-phenylindole; FITC, fluorescein isothiocyanate; PBS, phosphate-buffered saline; TAT, transactivator of transcription; TI, transmission image.
To examine the uptake of AIP6 in vivo, we used a mouse model with zymosan-induced local inflammation. FITC-labeled AIP6 (6 µg/paw, in 50 µl) or phosphate-buffered saline (PBS) was injected into the hind paws of mice that had been challenged with zymosan before 24 hours. Frozen tissue sections were evaluated by confocal microscopy. FITC-labeled AIP6 efficiently entered the cells 1 hour or 4 hours after injection. No fluorescence signal was detected in samples obtained from PBS-treated mice. Therefore, AIP6 efficiently transduces cells in vitro and in vivo (Figure 2c).
AIP6 inhibits NF-κB activation and production of proinflammmatory mediators
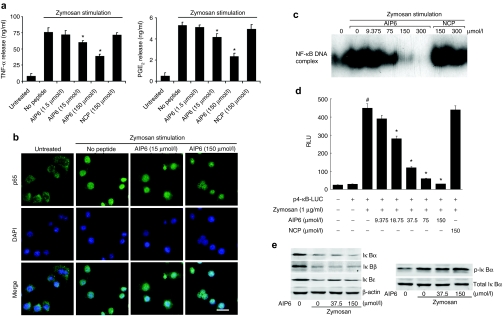
Knowing that AIP6 transduces cells and interferes with the DNA-binding activity of NF-κB, we determined the potential anti-inflammatory activity of AIP6 in zymosan-activated macrophages. The levels of two representative proinflammatory mediators, TNF-α and prostaglandin E2 (PGE2), in the medium of RAW 264.7 cells were measured by ELISA. Zymosan treatment significantly increased the levels of TNF-α and PGE2. Pretreatment with AIP6, but not NCP, decreased the production of TNF-α and PGE2 in a dose-dependent manner (Figure 3a).
Figure 3.
Effect of AIP6 on transcriptional activity of NF-κB in zymosan-activated macrophages. (a) Effects of AIP6s on zymosan-induced production of inflammatory mediators. RAW 264.7 cells were treated with AIP6 at indicated concentration or NCP (150 µmol/l) and stimulated with zymosan (0.1 mg/ml) for 24 hours. The production of TNF-α and PGE2 in culture supernatants was measured by ELISA. Results are expressed as mean ± SEM (n = 3), *P < 0.05 zymosan versus untreated; #P < 0.05 zymosan + AIP6 versus zymosan. (b) Effects of AIP6 on nuclear translocation of p65. Representative confocal images of p65 (green) localization with nuclei counterstained with DAPI (blue) in control (untreated) RAW 264.7 cells and zymosan-treated RAW 264.7 cells for 30 minutes with or without AIP6 at indicated concentrations. Bar = 20 µm. (c) Effects of AIP6 on the DNA-binding activity of p65 was measured by EMSA in RAW 264.7 cells. Cells were incubated at various concentrations of AIP6s or NCPs for 2 hours, followed by zymosan treatment for 1 hour. Nuclear extracts were prepared to analyze NF-κB activation by EMSA. (d) The effect of AIP6 on the expression of an NF-κB–driven luciferase reporter. RAW 264.7 cells transfected with p4-κB-luciferase reporter were pretreated with different doses of AIP6 or NCP (150 µmol/l) for 2 hours and stimulated with zymosan for 16 hours. The luciferase activity and NF-κB transcriptional activity were plotted as relative luminescence units (RLU). *P < 0.05 zymosan versus untreated; #P < 0.05 zymosan + AIP6 versus zymosan. (e) Effects of AIP6 on the expression and phosphorylation of IκB. RAW 264.7 cells were treated with AIP6 at indicated concentrations for 1 hour and stimulated with zymosan for 45 minutes (left) or 15 minutes (right). The expressions of various IκBs or p-IκBα were analyzed by western blot analysis. AIP6, anti-inflammatory peptide-6; DAPI, 4′,6-diamidino-2-phenylindole; ELISA, enzyme-linked immunosorbent assay; EMSA, electrophoretic mobility shift assay; I-κB, inhibitory κB; NCP, negative control peptide; NF-κB, nuclear factor-κB; TNF, tumor necrosis factor; PGE2, prostaglandin E2; TNF-α, tumor necrosis factor-α.
Zymosan-induced production of cytokines is the result of a signaling cascade that culminates in the activation, nuclear translocation, and transcriptional activation of NF-κB.15 Therefore, we next investigated whether AIP6 interferes with the NF-κB activation cascade. In untreated cells, p65 was mainly found in the cytoplasm. Zymosan (0.1 mg/ml) treatment for 1 hour induced nuclear translocation of NF-κB. Pretreatment of cells with AIP6s did not significantly affect zymosan-induced nuclear import of NF-κB p65 in the cells (Figure 3b). Nonetheless, AIP6 inhibited the DNA-binding activity of NF-κB in stimulated RAW 264.7 nuclear extract in a dose-dependent manner. NCP had no effect on the DNA-binding ability of NF-κB at all concentrations tested (Figure 3c).
Experiments with transfected NF-κB luciferase reporter also indicated that zymosan-stimulated NF-κB transcriptional activity diminished upon treatment with AIP6 (Figure 3d). NCP at 150 µmol/l did not exert any inhibitory effect on NF-κB transcriptional activation. Furthermore, AIP6 did not affect activator protein (AP)-1- or signal transducer and activator of transcription-1/3-mediated transcription (Supplementary Figures S3a,b). AIP6 also effectively inhibited NF-κB transcriptional activation induced by TPA, a strong activator of NF-κB, through the protein kinase C pathway16 (Supplementary Figure S3c).
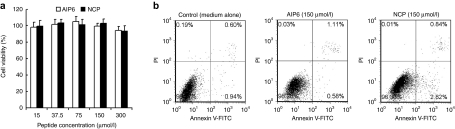
Pretreatment with AIP6 did not affect zymosan-induced degradation of IκB proteins or phosphorylation of IκBα in RAW 264.7 cells (Figure 3e). AIP6 and NCP at 150 µmol/l did not significantly affect cell proliferation or apoptosis, as measured by the water-soluble tetrazolium assay (Figure 4a) or by flow cytometry (Figure 4b). Together, these findings indicated that AIP6 selectively inhibits NF-κB–mediated transcription and has low nonspecific cytotoxicity at effective concentrations.
Figure 4.
Effects of AIP6 and negative control peptide on cell proliferation and viability. RAW 264.7 cells were incubated with zymosan (0.1 mg/ml) with AIP6 (white) or NCP (black) at indicated concentrations for 24 hours. (a) Cell proliferation was analyzed by WST-1 assay. Results are expressed as mean ± SEM (n = 3) and normalized to control (%) (cells not treated with peptide as 100%). (b) Apoptosis was analyzed by flow cytometry after staining with annexin V and PI. Cells were treated with no peptide, AIP6 (150 µmol/l), or NCP (150 µmol/l). AIP6, anti-inflammatory peptide-6; FITC, fluorescein isothiocyanate; NCP, negative control peptide; PI: propidium iodide; WST-1, water-soluble tetrazolium.
AIP6 suppresses zymosan-induced inflammation in mice
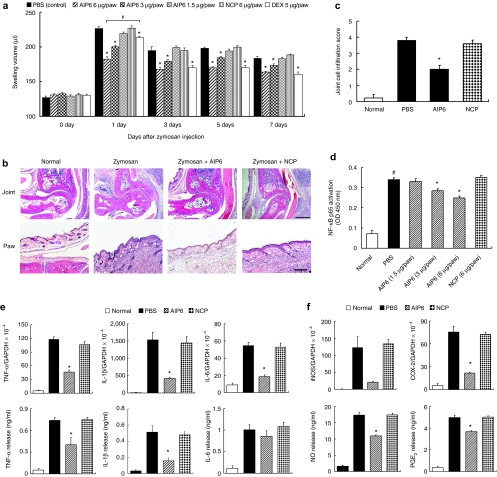
Based on the above anti-inflammatory activities of AIP6 in vitro, we decided to analyze its efficacy in an experimental inflammation model in mice.12 Zymosan injection in the feet of mice led to a time-dependent swelling of the joints and paws, which peaked at 24 hours. Swelling started to decrease and remained above control levels until 7 days (Figure 5a). From day 1 to day 7, inhibition of swelling by AIP6 (6 µg/paw) was better than or comparable to that induced by dexamethasone (5 µg/paw). AIP6 injection significantly inhibited the formation of swelling at different time points, including the swelling peak at 24 hours, in a dose-dependent manner. The optimal inhibitory effect of dexamethasone was observed 3 days after zymosan injection. NCP did not show any effect at any time point. We also histologically assessed inflammation in the joints and paws. Zymosan induced significant mononuclear cell infiltration, thickening of the synovial membrane, and severe inflammation of soft tissues, all of which was suppressed by AIP6 (6 µg/paw) (Figure 5b). Lower levels of inflammatory cell infiltration were found in the joints of mice treated with AIP6 compared with other groups (Figure 5c).
Figure 5.
Effects of AIP6 on zymosan-induced inflammation in vivo. (a) Effects of AIP6 on joint and paw edema. PBS, AIP6, or NCP in 50 µl was administered to the hindpaws (both left and right) of mice at different time points (8 hours, 1 day, 3 days, and 5 days) after zymosan injection. Results are expressed as mean ± SEM (n = 9). *P < 0.05 AIP6/dexamethasone versus PBS at the same point; #P < 0.05, AIP6 versus dexamethasone. (b) Representative HE-stained sections of ankle joints (upper) and hindpaws (lower) from mice with indicated treatments at day 7: normal group, zymosan injected group, zymosan + AIP6 (6 µg/paw)-treated group, and zymosan + NCP (6 µg/paw)-treated group. Bar = 50 µm for joint pictures (upper) and 10 µm for hindpaw pictures (lower). (c) Effects of AIP6 on the inflammation in joints. Inflammatory cell infiltration was scored at a scale from 0 (no cells) to 4 (severe cell influx in joint cavity and synovium). Results are expressed as mean ± SEM (n = 5).*P < 0.05 AIP6 versus PBS or NCP. (d) Effects of AIP6 on NF-κB activation in vivo. Nuclear extracts were prepared from soft tissues from each experiment group at day 7, and the DNA-binding activity was measured by ELISA by using the p65 nuclear transfactor kit, as in Figure 1a. Results are expressed as mean ± SEM (n = 4). *P < 0.05 AIP6 versus PBS or NCP; #P < 0.05 zymosan + PBS versus untreated. (e,f) Effects of AIP6 on the expression of inflammatory mediators. Total RNA or protein extracts isolated from each experiment group were collected 7 days after zymosan injection. AIP6 or NCP (6 µg/paw) was injected at 8 hours, 1 day, 3 days, and 5 days after zymosan treatment. The mRNA levels of indicated genes were measured by real-time PCR. Protein levels were measured by ELISA. Results are expressed as mean ± SEM (n = 4). *P < 0.05 AIP6 versus PBS or NCP. AIP6, anti-inflammatory peptide-6; COX-2, cyclooxygenase-2; Dex, dexamethasone; ELISA, enzyme-linked immunosorbent assay; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HE, hematoxylin and eosin; IL, interleukin; iNOS, inducible nitric oxide synthase; NCP, negative control peptide; OD, Optical density; PBS, phosphate-buffered saline; PGE2, prostaglandin E2; TNF, tumor necrosis factor.
The reduced inflammatory response in mice treated with AIP6 was associated with suppressed NF-κB p65 activation. Using nuclear extracts prepared from mice in various treatment groups at day 7, we measured the DNA-binding activity of NF-κB p65 with an ELISA-based assay. Zymosan caused nearly fourfold increase in DNA binding, which was suppressed by AIP6, but not NCP (Figure 5d). As expected, AIP6 injection strongly inhibited zymosan-induced expression of NF-κB target genes and inflammatory mediators, including TNF-α, IL-1β, IL-6, inducible nitric oxide synthase, cyclooxygenase-2, PGE2, and nitric oxide (Figure 5e,f).
Discussion
NF-κB regulates the transcription of many genes involved in immune response and is considered a potential therapeutic target in inflammatory diseases. In this study, we have discovered and characterized a pentapeptide AIP6 (RLRWR) that prevents the binding of NF-κB with κB elements through specific interaction with p65. AIP6 suppressed zymosan- or TPA-induced NF-κB activation and the production of inflammatory mediators in macrophages. The inhibitory effects of AIP6 on NF-κB appeared specific and did not involve IκB degradation, IκBα phosphorylation, or p65 nuclear import. Using a zymosan-induced inflammation model, we showed that local AIP6 injection impaired acute inflammation responses through inhibition of NF-κB activation and production of proinflammatory mediators. Importantly, AIP6 displays an intrinsic and strong cell-penetrating property in vitro and in vivo.
A variety of agents exhibit various degrees of effectiveness in suppressing NF-κB signaling. However, few of these compounds are specific, and some side effects have been reported. For example, glucocorticoids, which are considered as the most powerful nonspecific inhibitors of NF-κB, resulted in reduced bone formation and suppression of hypothalamic–pituitary–adrenal axis function.18,19 Therefore, there is a clear need for anti-inflammatory agents that lack steroid action and have reduced side effects. NF-κB activation or function can be inhibited by more specific means. For example, the NF-κB essential modifier (NEMO)-binding domain peptide inhibits p65 phosphorylation, and the SN50 peptide inhibits NF-κB nuclear transport.20,21,22 Among various strategies aimed at inhibiting NF-κB activation, blocking the binding of the p65 or p50 subunit with the κB site may be more specific23,24 because a higher degree of specificity may be achieved by the inhibition of NF-κB–mediated transcription. In this study, we designed a peptide based on a sequence required for DNA binding, which is conserved in NF-κB family members. Unexpectedly, this peptide selectively interacted with the p65 NF-κB subunit but not the κB motif, and it functioned as a selective inhibitor of NF-κB–mediated transcription. AIP6 did not appear to affect the phosphorylation or degradation of IκB proteins or the nuclear import of p65. The molecular mechanism of AIP6-mediated inhibition of the DNA-binding activity of p65 is not fully understood; AIP6 may induce conformational changes in the protein25 or prevent the formation of the transcriptionally competent p65/p50 complex. The results of our ELISA-based transfactor assays make us speculate AIP6 is a competitive inhibitor against NF-κB transcription activity that will require a relatively higher quantity to be effective. Thus, to keep anti-inflammatory action of AIP6, AIP6 has to be delivered frequently to maintain its in vivo concentration. The exact mechanism may be determined through additional molecular and structural studies. Nevertheless, this property may help avoid undesirable effects by reducing interference with the basal activity of NF-κB, which may be essential for the survival of several cell types.20
Inhibitors of transcription factors can only exert biological inhibitory effects after their efficient uptake into cells. Transduction mediated by cell-penetrating peptides (CPPs) can deliver bioactive reagents directly into living cells. Commonly used CPPs include the HIV-1 TAT protein, penetratin, and polyarginine.26 Our data showed that AIP6 itself possesses an intrinsic cell-penetrating property independent of phagocytosis. This unique property may be attributed to the presence of several positively charged arginine residues and the low molecular weight and moderate hydrophobicity of AIP6.27,28 Other preferred mechanisms for CPP-mediated transduction have been proposed, including energy-independent membrane translocation, endocytosis, and specific receptors.29 In addition, the toxicity and unsuspected side effects of several CPPs have been reported in recent years.30,31,32,33 A similar study reported that 16-AA, a CPP, was found to suppress inflammatory response in vitro and in vivo by inhibiting the activation of NF-κB.34 The fact that AIP6 is cell penetrating and contains only 5-AA residues with no obvious toxicity makes it an attractive lead structure for future drug development. It might also be a practical short CPP. Of course, we cannot deny the potential toxicity of AIP6 that we did not test in this study, and we therefore need more work to assess the safety of AIP6 before it can be developed to a therapeutic agent.
AIP6 suppressed zymosan-induced inflammation that is associated with reduced levels of inflammatory mediators and well-known NF-κB targets, including TNF-α, IL-1 β, IL-6, inducible nitric oxide synthase, and cyclooxygenase-2.4,35,36 Local treatment with AIP6 inhibited NF-κB activation in this model, which suggested that AIP6 blocks excessive NF-κB activation and highlighted the importance of the inhibition of NF-κB–mediated transcription in overall anti-inflammatory effects. Because AIP6 did not affect AP-1– or signal transducer and activator of transcription-mediated transcription of reporters, it is ruled out as a general inhibitor of transcription. However, our study could not rule out the possibility that AIP6 interferes with other pathways. As discussed before, few inhibitors of NF-κB are specific. For example, most IκB kinase-β inhibitors can exert their function via ATP competition, and SN50 also prevents nuclear translocation of AP-1, nuclear factor of activated T-cells and signal transducer and activator of transcription-1.23,37 Gene expression profiling coupled with chromatin immunoprecipitation assay could lead to better understanding of the anti-inflammatory mechanisms of AIP6.
In summary, our study shows that AIP6, a cell-penetrating pentapeptide, inhibits the DNA-binding activity of NF-κB and efficiently attenuates inflammatory responses in vitro and in vivo. These results suggest that AIP6 is a promising lead structure for the development of specific NF-κB inhibitors as potential anti-inflammatory agents.
Materials and Methods
Chemicals and materials. All peptides, including FITC-labeled TAT protein of HIV, FITC-labeled AIP6, and FITC-labeled NCP, used in this study were synthesized by HD Biosciences Company, Shanghai, China. AIP6 and NCP were purified to more than 95% purity by using high pressure liquid chromatography. The sequences of the peptides were as follows: AIP6, Arg-Leu-Arg-Trp-Arg (RLRWR); NCP, Leu-Arg-Trp-Arg-Lys (LRWRK); and TAT, Arg-Leu-Arg-Trp-Arg (YGRKKRRQRR-RLRWR). Other chemicals and materials are listed in Supplementary Materials and Methods.
Cell lines. RAW 264.7 macrophages and LoVo colon cancer cells were initially purchased from the American Type Culture Collection (Manassas, VA). RAW 264.7 macrophages and LoVo cells were cultured in Dulbecco's modified Eagle's media supplemented with 10% heat-inactivated fetal bovine serum, benzylpenicillin potassium (143 U/ml), and streptomycin sulfate (100 µg/ml) under 37 °C and 5% CO2 atmosphere.
ELISA-based NF-κB p65 transfactor assay. To determine whether AIP6 has the ability to bind with κB elements, we designed an experiment using an ELISA-based Transfactor p65 kit (Active Motif, Carlsbad, CA). About 30 µl of binding buffer (Active Motif) containing AIP6 or NCP at various concentrations (25, 50, 100, 200, and 400 µmol/l) was added to each well of the corresponding microtiter plate coated with an oligonucleotide containing the κB site of the immunoglobulin light chain gene promoter (GGGACTTTCC) and incubated for 30 minutes at room temperature. After washing three times with binding buffer, 30 µl binding buffer and 20 µl complete lysis buffer (Active Motif) containing 2.5 µg Jurkat nuclear extract (TPA + calcium ionophore) were added to the corresponding microtiter plate. The following steps were performed according to the manufacturer's instructions. Briefly, the plate was incubated for 1 hour at room temperature on a rocking platform. After washing, NF-κB p65 antibody (1:1,000) was added and incubated for 1 hour at room temperature. The wells were then washed, and horseradish peroxidase-conjugated antibody (1:1,000) was added and incubated for 1 hour at room temperature. After washing, a substrate solution was added to the wells and incubated at room temperature for 10 minutes, protected from light. Stop solution (2NH2SO4) was added, and the optical density was measured at 450 nm. Inhibition ratio (%) = [(optical density value of positive control group − optical density value of peptide-treated group)/optical density value of positive control group] × 100. The wells to which only Jurkat nuclear extract was added were set as the positive control group.
To detect the inhibitory effect of AIP6 on the binding activity of NF-κB, another experiment was performed. To detect the binding activity of AIP6 to p65, 30 µl binding buffer containing AIP6 or NCP at various concentrations (25, 50, 100, 200, and 400 µmol/l) was mixed with 20 µl complete lysis buffer containing 2.5 µg Jurkat nuclear extract and incubated for 30 minutes at room temperature. The mixtures were then added to the corresponding microtiter plate, and the plate was incubated for 1 hour at room temperature on a rocking platform. The steps described above were then performed.
The effect of AIP6 on the nuclear levels of NF-κB p65 in local inflammatory tissue was also determined using ELISA Transfactor p65 kits. Nuclear extracts were collected using the reagents supplied in the Nuclear Extract Kit (Active Motif). Proteins were quantified using the bicinchoninic acid assay and subjected to ELISA-based NF-κB p65 transfactor assay. All the procedures were performed according to the manufacturer's instructions.
Electrophoretic mobility shift assay. To identify the inhibitory effect of AIP6 on the binding activity of the p65 or p50 subunit, NF-κB probe (5′-AGTTGAGGGGACTTTCCCAGGC-3′) was phosphorylated with T4 polynucleotide kinase in the presence of [γ-32P] ATP and was purified. Recombinant p65 protein (20 ng) and recombinant p50 protein were used for electrophoretic mobility shift assay. Reaction mixtures containing binding buffer (15 mmol/l Tris-HCl (pH 7.5), 75 mmol/l NaCl, 1.5 mmol/l EDTA, 1.5 mmol/l dithiothreitol, 7.52% glycerol, and 0.3% Nonidet P-40), 1 µg of poly (dI-dC)-(dI-dC), and either recombinant p65 protein or recombinant p50 protein were kept on ice for 10 minutes. AIP6 (25, 50, 100, and 200 µmol/l) or NCP (25, 50, 100, and 200 µmol/l) was added to the mixtures and incubated at room temperature. After 30 minutes, 1 µmol/l 32P-labeled NF-κB probe was added to the mixtures and further incubated at room temperature for 30 minutes. For the supershift assays, 2.5 µl anti-p65 or anti-p50 antibody was added to the reaction mixture simultaneously with the Jurkat nuclear extract, and AIP6 (25 and 400 µmol/l) was added to the mixtures. The mixture was incubated as described above. Subsequently, 20 µl of the mixture was loaded onto a 5% native polyacrylamide gel prepared in 0.5× tris-borate-EDTA and electrophoresed for 2.5 hours. The gel was dried and then subjected to autoradiography. In another experiment, RAW 264.7 cells were pretreated with AIP6s (9.375, 75, 150, and 300 µmol/l) or NCPs (150 and 300 µmol/l) for 2 hours and stimulated with zymosan (0.1 mg/ml) for 1 hour. The nuclear extract was prepared from these cells and then subjected to electrophoretic mobility shift assay.
Surface plasmon resonance spectroscopy. To identify the specific interaction between AIP6 and the p65 or p50 NF-κB subunit, surface plasmon resonance measurements were performed using a BIAcore 3000 instrument (Biacore, Piscataway, NJ). The running buffer was protein binding buffer [50 mmol/l Tris-HCl (pH 7.2), 0.1 mol/l NaCl, 10 mmol/l MgCl2, 10 µmol/l ZnCL2, 1 mmol/l dithiothreitol, 0.1% (v/v) NP40]. Data acquisition was then performed, and baseline data was gathered for several minutes. The carboxymethylated dextran surface of a CM5 sensor chip (BIAcore AB) was activated by injecting a coupling solution of N-hydroxysuccinimide and 1-ethyl-3-(dimethylaminopropyl) carbodiimide hydrochloride (BIAcore AB). Recombinant p65 or p50 proteins were diluted to 25 µg/ml in 10 mmol/l acetate buffer (pH 5.0) and injected over the surface of the sensor chip for 8 minutes at a flow rate of 5 µl/minute. The unreacted sites of the sensor chip surface were then quenched by injection of 40 µl of 1 M Tris-HCl (pH 8.0). Unimmobilized p65 and p50 were washed 3 times with 40 µl of 10 mmol/l HCl. AIP6 and NCP were diluted to 1 µg/ml in protein binding buffer and injected over the surface of the sensor chip for 5 min at a flow rate of 10 µl/min. The unbound AIP6 and NCP were washed with protein binding buffer. The resonant angle response was recorded. The chip surface was then regenerated by adding 10 mmol/l HCl until the response signal returned to baseline to proceed with another binding cycle.
Confocal fluorescence microscopy. All the samples were analyzed under a laser scanning confocal microscope (Zeiss LSM510, Oberkochen, Germany). Cells were treated under a standard condition of 5% CO2 in a humidified incubator at 37 °C. To investigate the transduction of AIP6 in vitro, RAW 264.7 cells and LoVo cells were cultured in the presence of FITC-labeled TAT, FITC-labeled TAT-AIP6, FITC-labeled AIP6, and FITC-labeled NCP at a concentration of 150 µmol/l for 60 minutes. After washing three times, nuclei were stained with 4′,6-diamidino-2-phenylindole. These cells were then washed two times, and the transduction of AIP6 was assayed by confocal fluorescence analysis.
Confocal fluorescence analysis was also performed to investigate the transduction of AIP6 in vivo. Subdermal injections of 50 µl of 150 µmol/l FITC-labeled AIP6 (6 µg/paw) were performed in the inflamed hindpaws of mice (at 24 hours after zymosan injection). Control animals received subdermal injections of 50 µl of PBS into inflamed hindpaws. Mice were anesthetized with chloral hydrate and killed 1 hour or 4 hours after the injection. Soft samples from hindpaws were removed immediately and fixed in 4% paraformaldehyde. Samples were cut on a cryostat at 20 µm, and DNA dye (4′,6-diamidino-2-phenylindole) was added. The sections were then analyzed under confocal microscopy.
Confocal laser scanning microscopy was also used to observe whether AIP6 affected the nuclear import of NF-κB p65. RAW 264.7 cells were pretreated with 15 or 150 µmol/l AIP6 for 2 hours and stimulated with zymosan (0.1 mg/ml) for 1 hour. These cells were then fixed in 4% paraformaldehyde, permeabilized in 0.5% Triton X-100, and blocked in 1% bovine serum albumin in PBS. For immunostaining, the cells were incubated with anti-NF-κB p65 antibody (1:300) for 2 hours and then stained with FITC-labeled secondary antibody (1:800) for 1 hour. For staining the nuclei, the cells were incubated with 4′,6-diamidino-2-phenylindole solution.
Luciferase reporter assays. RAW 264.7 cells were plated at a density of 0.5 × 105 cells/well in 24-well plates and allowed to reach up to 70–80% confluence. The p4-κB-luciferase plasmid (1 µg) was transfected using Lipofectamine Plus reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Thereafter, the cells were treated for 24 hours with AIP6 (0, 9.375, 18.75, 37.5, 75, and 150 µmol/l) or NCP (150 µmol/l) and then stimulated with 1 µg/ml zymosan. After 16 hours of incubation, cells were lysed and luciferase activity was measured using a luminometer (Promega, Fitchburg, WI) according to the manufacturer's instructions. AP-1-Luc and m67-Luc vectors were used to detect AP-1 or STAT-dependent transcription activities. The effect of AIP6 on NF-κB transcription activities in TPA-stimulated macrophages are described in Supplemental Materials and Methods.
Western blot analysis. The cells were pretreated with 37.5 or 150 µmol/l AIP6 for 2 hours, stimulated with zymosan (0.1 mg/ml) for 15 minutes or 45 minutes, and then lysed in a lysis buffer. Whole cell lysates were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis and then transferred to a polyvinylidene difluoride membrane. The blots were usually incubated at 4 °C overnight with primary antibodies in 5% bovine serum albumin in 10 mmol/l tris-buffered saline Tween-20 [10 mmol/l Tris (pH 8.0), 50 mmol/l NaCl, 0.05% (v/v) Tween-20]. The primary antibodies were anti-IκBα (dilution, 1:500), anti-IκBβ (1:500), anti-IκBε (1:500), anti-p-IκBα (1:200), and anti-glyceraldehyde-3-phosphate dehydrogenase (1:2,000). The blots were then incubated with a secondary antibody, namely, horseradish peroxidase-labeled anti-rabbit IgG antibody (1:1,000), at room temperature for 2–5 hours. Immune complexes on the blots were finally visualized by radiography after reaction with an enhanced chemiluminescence reagent (GE Healthcare, Little Chalfont, UK). Each blot presented is representative of findings in at least three similar independent experiments.
Cell proliferation and apoptosis assays. The cytotoxicity of AIP6 and NCP was assessed by using water-soluble tetrazolium assay. RAW 264.7 macrophages (5 × 104/well) were incubated with various concentrations of AIP6 or NCP for 24 hours. They were exposed to water-soluble tetrazolium of 2-(4-iodophenyl)-3- (4-nitrophenyl)-5-(2, 4-′disulfophenyl)-2H-tetrazolium for 3 hours, and then absorbance values were measured at 450 nm.
Apoptosis was assessed by flow cytometry. The RAW 264.7 cells after being incubated with 150 µmol/l of AIP6 or NCP for 24 hours were harvested by trypsin-EDTA followed by two washes with ice-cold PBS and were subsequently stained with antibody to annexin V-FITC and DNA/RNA dye propidium iodide. Samples were analyzed by flow cytometry (FACScan; Becton Dickinson; Mountain View, CA) according to the manufacturer's protocol. The red (610 nm, propidium iodide) and green (525 nm, annexin V) fluorescence emissions were separated optically by separate photomultipliers.
Zymosan-induced inflammation in mice. Female C57BL/6J mice (21–23 g) were provided by the Animal Breeding Center of Third Military Medical University (Chongqing, China). All the animals were maintained in plastic cages at 20 ± 2 °C with free access to food and water and were kept on a 12-hour light/dark cycle. Experiments with animals were conducted in accordance with the guidelines of the National Institutes of Health and Third Military Medical University for laboratory animal care.
For the inflammation model, a 15 mg/ml suspension of zymosan was made in PBS, and the suspension was autoclaved before injection. Animals received subdermal injections of 75 µg zymosan (50 µl) in the left and right hindpaws. The volume of edema (including hindpaw and ankle joint) was measured before zymosan injection (time 0) and at different time points after stimulation using volume meter/plethysmometry (plethysmometer 7140; Ugo Basile; Comerio, Italy). Mice were treated with subdermal administration of the test agents (50 µl) into the hindpaws at 8 hours, 1 day, 3 days, and 5 days. The test agents used in this experiment were 150 µmol/l AIP6 (6 µg/paw), 75 µmol/l AIP6 (3 µg/paw), 37.5 µmol/l AIP6 (1.5 µg/paw), 150 µmol/l NCP (6 µg/paw), and 150 µmol/l dexamethasone (5 µg/paw). For the negative control group, mice were given an equal volume of PBS. The mice were killed by cervical dislocation immediately after the volume was measured (at 7 days after zymosan injection), and the paws and ankle joints were removed. Some soft tissues from paws were recovered by scalpel and immediately processed to obtain tissue extracts or to isolate total RNA.
Histological analysis. Paws and ankle joints were dissected and fixed for 2 weeks in 10% buffered formalin. Fixed tissues were decalcified for 2 weeks in 15% EDTA, dehydrated, and embedded in paraffin. Tissue sections (10 µm) were cut and stained with hematoxylin/eosin for the detection of inflammation. Joint inflammation was scored by the influx of inflammatory cells in the joint cavity and synovium. Histological assessment of joints was scored by two observers blinded to the treatment, according to methods described earlier.38 A score of 0 indicated no cell influx, and 1–4 was scored according to the degree of cell influx.
Quantitative real-time reverse transcription PCR. Total RNA was isolated from paw soft tissues by using TRIzol, according to the manufacturer's instructions. The extracted RNA was kept at −80 °C before use. Real-time PCR was performed at a final volume of 20 µl in capillary tubes in a LightCycler instrument (Roche Diagnostics, Mannheim, Germany). The primers used for PCR are given in Supplementary Table S1. Reaction mixtures contained 2 µl of LightCycler Fast-Start DNA mastermix for SYBR Green I (Roche Diagnostics), 0.5 µmol/l of each primer, 4 mmol/l MgCl2, and 2 µl of template DNA. All capillaries were sealed, centrifuged at 500g for 5 seconds, and then amplified in a LightCycler instrument with activation of polymerase (95 °C for 10 minutes), followed by 45 cycles of 10 seconds at 95 °C, 10 seconds at 60 °C, and 10 seconds at 72 °C. The temperature transition rate was 20 °C/second for all steps. Double-stranded PCR products were measured during the extension step at 72 °C by detection of fluorescence associated with the binding of SYBR Green I to the product. Fluorescence curves were analyzed with LightCycler software v. 3.0. For quantitative analysis of TNF-α, IL-1β, IL-6, inducible nitric oxide synthase, and cyclooxygenase-2 mRNA, LightCycler (Roche Diagnostics) was used. The expression levels of TNF-α, IL-1β, IL-6, inducible nitric oxide synthase, and cyclooxygenase-2 were calculated and corrected for the values of the endogenously expressed housekeeping gene control (glyceraldehyde-3-phosphate dehydrogenase). Melting curve analysis was performed immediately after the amplification protocol under the following conditions: 0 second (hold time) at 95 °C, 15 seconds at 60 °C, and 0 second (hold time) at 95 °C. Temperature change rates were 20 °C/second, except in the final step, in which it was 0.1 °C/second. The melting peak generated represented the specific amplified product. The crossing point was defined as the maximum of the second derivative from the fluorescence curve. Negative controls were also included and contained all the elements of the reaction mixture, except template DNA. All samples were processed in duplicate.
ELISA. The macrophages were pretreated with AIP6s or NCPs for 2 hours and then stimulated with zymosan (0.1 mg/ml) for 24 hours. The levels of TNF-α and PGE2 were determined in the harvested supernatants using corresponding ELISA kits, according to the manufacturer protocols. Levels of TNF-α, IL-1β, IL-6, PGE2, and nitric oxide in the tissue extracts were evaluated by ELISA using corresponding anti-mouse antibodies and biotinylated secondary antibodies, according to the manufacturer's instructions.
Statistical analysis. Results are expressed as means ± SE. The inhibitory effect of AIP6 and NCP at various concentrations was compared by using a two-tailed Student's t-test. Differences between multiple groups were evaluated by using one-way analysis of variance, followed by Dunnett's post hoc or Tukey comparisons. Results were considered statistically significant at P < 0.05.
SUPPLEMENTARY MATERIAL Figure S1. Effects of AIP6 on the DNA-binding activity of NF-κB was measured by ELISA. AIP6 was preincubated with microtiter plates coated with the κB motif oligonucleotide for 30 min. After washing 3 times by binding buffer, 2.5 μg stimulated Jurkat nuclear extract was added to each well to detect DNA binding activity of NF-κB. Results are mean ± SEM (n = 3). Figure S2. Effects of AIP6 on DNA binding activity of NF-κB p50. (a) Interaction analysis of AIP6 with NF-κB p50 subunit using Surface plasmon resonance measurements. (b) Effects of AIP6 on the DNA binding activity of p50 was measured by EMSA. Recombinant NF-κB p50 was used in the Surface plasmon resonance measurements and EMSA. Results are mean ± SEM (n = 3). Figure S3. Effects of AIP6 on TPA (phorbol, 12-myristate, 13 acetate)-induced AP-1 (a), TPA-induced NF-κB (b), and interferon (IFN)-γ–induced signal transducer and activator of transcription (STAT) activation (c) in RAW 264.7 macrophages. Transcriptional activity is represented as relative luminescence units (RLU). #p <0.05 vs. pAP-1-Luc or p4-κB-Luc added and TPA untreated group, or pSTAT1/3-Luc added and IFN-γ untreated group; *p <0.05 vs. luciferase reporter vector added and no peptide treated group. Results are mean ± SEM (n = 3). Table S1. Primers and parameters used in real-time PCR. Materials and Methods.
Acknowledgments
We thank Java Vatsyayan, Department of Cell Biology and Physiology, University of Pittsburgh School of Medicine, Rangos Research Center at Children's Hospital of Pittsburgh of UPMC, for helpful discussion and critical review. This study was supported by an establishment grant from the National High Technology Research and Development Program (“863” Program, No. 2008AA02Z440) of China, the National Nature Science Foundation of China (NSFC, No. 30672414; No. 81001320), National Institutes of Health grants R01CA129829 and U01DK085570, ACS grant RGS-10-124-01-CCE, and FAMRI grant 032029_YCSA. The authors declared no conflict of interest.
Supplementary Material
Effects of AIP6 on the DNA-binding activity of NF-κB was measured by ELISA. AIP6 was preincubated with microtiter plates coated with the κB motif oligonucleotide for 30 min. After washing 3 times by binding buffer, 2.5 μg stimulated Jurkat nuclear extract was added to each well to detect DNA binding activity of NF-κB. Results are mean ± SEM (n = 3).
Effects of AIP6 on DNA binding activity of NF-κB p50. (a) Interaction analysis of AIP6 with NF-κB p50 subunit using Surface plasmon resonance measurements. (b) Effects of AIP6 on the DNA binding activity of p50 was measured by EMSA. Recombinant NF-κB p50 was used in the Surface plasmon resonance measurements and EMSA. Results are mean ± SEM (n = 3).
Effects of AIP6 on TPA (phorbol, 12-myristate, 13 acetate)-induced AP-1 (a), TPA-induced NF-κB (b), and interferon (IFN)-γ–induced signal transducer and activator of transcription (STAT) activation (c) in RAW 264.7 macrophages. Transcriptional activity is represented as relative luminescence units (RLU). #p <0.05 vs. pAP-1-Luc or p4-κB-Luc added and TPA untreated group, or pSTAT1/3-Luc added and IFN-γ untreated group; *p <0.05 vs. luciferase reporter vector added and no peptide treated group. Results are mean ± SEM (n = 3).
Primers and parameters used in real-time PCR.
REFERENCES
- Ghosh S., and, Karin M. Missing pieces in the NF-kappaB puzzle. Cell. 2002;109 Suppl:S81–S96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- Baldwin AS., Jr Series introduction: the transcription factor NF-kappaB and human disease. J Clin Invest. 2001;107:3–6. doi: 10.1172/JCI11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar FH, Li Y, Wang Z., and, Kong D. NF-kappaB signaling pathway and its therapeutic implications in human diseases. Int Rev Immunol. 2008;27:293–319. doi: 10.1080/08830180802276179. [DOI] [PubMed] [Google Scholar]
- Tian B., and, Brasier AR. Identification of a nuclear factor kappa B-dependent gene network. Recent Prog Horm Res. 2003;58:95–130. doi: 10.1210/rp.58.1.95. [DOI] [PubMed] [Google Scholar]
- Hart LA, Krishnan VL, Adcock IM, Barnes PJ., and, Chung KF. Activation and localization of transcription factor, nuclear factor-kappaB, in asthma. Am J Respir Crit Care Med. 1998;158 5 Pt 1:1585–1592. doi: 10.1164/ajrccm.158.5.9706116. [DOI] [PubMed] [Google Scholar]
- Han Z, Boyle DL, Manning AM., and, Firestein GS. AP-1 and NF-kappaB regulation in rheumatoid arthritis and murine collagen-induced arthritis. Autoimmunity. 1998;28:197–208. doi: 10.3109/08916939808995367. [DOI] [PubMed] [Google Scholar]
- Courtois G., and, Gilmore TD. Mutations in the NF-kappaB signaling pathway: implications for human disease. Oncogene. 2006;25:6831–6843. doi: 10.1038/sj.onc.1209939. [DOI] [PubMed] [Google Scholar]
- Hajra L, Evans AI, Chen M, Hyduk SJ, Collins T., and, Cybulsky MI. The NF-kappa B signal transduction pathway in aortic endothelial cells is primed for activation in regions predisposed to atherosclerotic lesion formation. Proc Natl Acad Sci USA. 2000;97:9052–9057. doi: 10.1073/pnas.97.16.9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MA., and, Jones WK. NF-kappaB action in sepsis: the innate immune system and the heart. Front Biosci. 2004;9:1201–1217. doi: 10.2741/1304. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y., and, Gaynor RB. Therapeutic potential of inhibition of the NF-kappaB pathway in the treatment of inflammation and cancer. J Clin Invest. 2001;107:135–142. doi: 10.1172/JCI11914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscott J, Kwon H., and, Génin P. Hostile takeovers: viral appropriation of the NF-kappaB pathway. J Clin Invest. 2001;107:143–151. doi: 10.1172/JCI11918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson LA, Trauger JW, Baird EE, Dervan PB, Graves BJ., and, Gottesfeld JM. Inhibition of Ets-1 DNA binding and ternary complex formation between Ets-1, NF-kappaB, and DNA by a designed DNA-binding ligand. J Biol Chem. 1999;274:12765–12773. doi: 10.1074/jbc.274.18.12765. [DOI] [PubMed] [Google Scholar]
- Matthews JR, Watson E, Buckley S., and, Hay RT. Interaction of the C-terminal region of p105 with the nuclear localisation signal of p50 is required for inhibition of NF-kappa B DNA binding activity. Nucleic Acids Res. 1993;21:4516–4523. doi: 10.1093/nar/21.19.4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pande V., and, Ramos MJ. Nuclear factor kappa B: a potential target for anti-HIV chemotherapy. Curr Med Chem. 2003;10:1603–1615. doi: 10.2174/0929867033457250. [DOI] [PubMed] [Google Scholar]
- Kim BH, Hong SS, Kwon SW, Lee HY, Sung H, Lee IJ.et al. (2008Diarctigenin, a lignan constituent from Arctium lappa, down-regulated zymosan-induced transcription of inflammatory genes through suppression of DNA binding ability of nuclear factor-kappaB in macrophages J Pharmacol Exp Ther 327393–401. [DOI] [PubMed] [Google Scholar]
- Umezawa K. Inhibition of tumor growth by NF-kappaB inhibitors. Cancer Sci. 2006;97:990–995. doi: 10.1111/j.1349-7006.2006.00285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SB, Lee CW, Yoon YD, Lee JH, Kang JS, Lee KH.et al. (2005Prevention of arthritic inflammation using an oriental herbal combination BDX-1 isolated from Achyranthes bidentata and Atractylodes japonica Arch Pharm Res 28902–908. [DOI] [PubMed] [Google Scholar]
- Mader R, Lavi I., and, Luboshitzky R. Evaluation of the pituitary-adrenal axis function following single intraarticular injection of methylprednisolone. Arthritis Rheum. 2005;52:924–928. doi: 10.1002/art.20884. [DOI] [PubMed] [Google Scholar]
- Weitoft T, Larsson A, Saxne T., and, Rönnblom L. Changes of cartilage and bone markers after intra-articular glucocorticoid treatment with and without postinjection rest in patients with rheumatoid arthritis. Ann Rheum Dis. 2005;64:1750–1753. doi: 10.1136/ard.2004.035022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimi E, Aoki K, Saito H, D'Acquisto F, May MJ, Nakamura I.et al. (2004Selective inhibition of NF-kappa B blocks osteoclastogenesis and prevents inflammatory bone destruction in vivo Nat Med 10617–624. [DOI] [PubMed] [Google Scholar]
- Takada Y, Singh S., and, Aggarwal BB. Identification of a p65 peptide that selectively inhibits NF-kappa B activation induced by various inflammatory stimuli and its role in down-regulation of NF-kappaB-mediated gene expression and up-regulation of apoptosis. J Biol Chem. 2004;279:15096–15104. doi: 10.1074/jbc.M311192200. [DOI] [PubMed] [Google Scholar]
- Kang MI, Henrich CJ, Bokesch HR, Gustafson KR, McMahon JB, Baker AR.et al. (2009A selective small-molecule nuclear factor-kappaB inhibitor from a high-throughput cell-based assay for “activator protein-1 hits” Mol Cancer Ther 8571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uwe S. Anti-inflammatory interventions of NF-kappaB signaling: potential applications and risks. Biochem Pharmacol. 2008;75:1567–1579. doi: 10.1016/j.bcp.2007.10.027. [DOI] [PubMed] [Google Scholar]
- Piccagli L, Fabbri E, Borgatti M, Bianchi N, Bezzerri V, Mancini I.et al. (2009Virtual screening against p50 NF-kappaB transcription factor for the identification of inhibitors of the NF-kappaB-DNA interaction and expression of NF-kappaB upregulated genes ChemMedChem 42024–2033. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Yoshimori A, Kino K, Komori R, Miyazawa H., and, Tanuma SI. A new small molecule that directly inhibits the DNA binding of NF-kappaB. Bioorg Med Chem. 2009;17:5293–5297. doi: 10.1016/j.bmc.2009.05.030. [DOI] [PubMed] [Google Scholar]
- Orange JS., and, May MJ. Cell penetrating peptide inhibitors of nuclear factor-kappa B. Cell Mol Life Sci. 2008;65:3564–3591. doi: 10.1007/s00018-008-8222-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Kilk K., and, Langel U. Predicting cell-penetrating peptides. Adv Drug Deliv Rev. 2008;60:572–579. doi: 10.1016/j.addr.2007.09.003. [DOI] [PubMed] [Google Scholar]
- Futaki S. Membrane-permeable arginine-rich peptides and the translocation mechanisms. Adv Drug Deliv Rev. 2005;57:547–558. doi: 10.1016/j.addr.2004.10.009. [DOI] [PubMed] [Google Scholar]
- Heitz F, Morris MC., and, Divita G. Twenty years of cell-penetrating peptides: from molecular mechanisms to therapeutics. Br J Pharmacol. 2009;157:195–206. doi: 10.1111/j.1476-5381.2009.00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton SJ, Jones DN, Darker JG, Eggleston DS, Hunter AJ., and, Walsh FS. Cellular uptake and spread of the cell-permeable peptide penetratin in adult rat brain. Eur J Neurosci. 2000;12:2847–2855. doi: 10.1046/j.1460-9568.2000.00171.x. [DOI] [PubMed] [Google Scholar]
- Saar K, Lindgren M, Hansen M, Eiríksdóttir E, Jiang Y, Rosenthal-Aizman K.et al. (2005Cell-penetrating peptides: a comparative membrane toxicity study Anal Biochem 34555–65. [DOI] [PubMed] [Google Scholar]
- Kilk K, Mahlapuu R, Soomets U., and, Langel U. Analysis of in vitro toxicity of five cell-penetrating peptides by metabolic profiling. Toxicology. 2009;265:87–95. doi: 10.1016/j.tox.2009.09.016. [DOI] [PubMed] [Google Scholar]
- Ward B, Seal BL, Brophy CM., and, Panitch A. Design of a bioactive cell-penetrating peptide: when a transduction domain does more than transduce. J Pept Sci. 2009;15:668–674. doi: 10.1002/psc.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letoha T, Kusz E, Pápai G, Szabolcs A, Kaszaki J, Varga I.et al. (2006In vitro and in vivo nuclear factor-kappaB inhibitory effects of the cell-penetrating penetratin peptide Mol Pharmacol 692027–2036. [DOI] [PubMed] [Google Scholar]
- Surh YJ, Chun KS, Cha HH, Han SS, Keum YS, Park KK.et al. (2001Molecular mechanisms underlying chemopreventive activities of anti-inflammatory phytochemicals: down-regulation of COX-2 and iNOS through suppression of NF-kappa B activation Mutat Res 480481243–268. [DOI] [PubMed] [Google Scholar]
- Feldmann M, Brennan FM., and, Maini R. Cytokines in autoimmune disorders. Int Rev Immunol. 1998;17:217–228. doi: 10.3109/08830189809084493. [DOI] [PubMed] [Google Scholar]
- Torgerson TR, Colosia AD, Donahue JP, Lin YZ., and, Hawiger J. Regulation of NF-kappa B, AP-1, NFAT, and STAT1 nuclear import in T lymphocytes by noninvasive delivery of peptide carrying the nuclear localization sequence of NF-kappa B p50. J Immunol. 1998;161:6084–6092. [PubMed] [Google Scholar]
- Hartog A, Leenders I, van der Kraan PM., and, Garssen J. Anti-inflammatory effects of orally ingested lactoferrin and glycine in different zymosan-induced inflammation models: evidence for synergistic activity. Int Immunopharmacol. 2007;7:1784–1792. doi: 10.1016/j.intimp.2007.09.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Effects of AIP6 on the DNA-binding activity of NF-κB was measured by ELISA. AIP6 was preincubated with microtiter plates coated with the κB motif oligonucleotide for 30 min. After washing 3 times by binding buffer, 2.5 μg stimulated Jurkat nuclear extract was added to each well to detect DNA binding activity of NF-κB. Results are mean ± SEM (n = 3).
Effects of AIP6 on DNA binding activity of NF-κB p50. (a) Interaction analysis of AIP6 with NF-κB p50 subunit using Surface plasmon resonance measurements. (b) Effects of AIP6 on the DNA binding activity of p50 was measured by EMSA. Recombinant NF-κB p50 was used in the Surface plasmon resonance measurements and EMSA. Results are mean ± SEM (n = 3).
Effects of AIP6 on TPA (phorbol, 12-myristate, 13 acetate)-induced AP-1 (a), TPA-induced NF-κB (b), and interferon (IFN)-γ–induced signal transducer and activator of transcription (STAT) activation (c) in RAW 264.7 macrophages. Transcriptional activity is represented as relative luminescence units (RLU). #p <0.05 vs. pAP-1-Luc or p4-κB-Luc added and TPA untreated group, or pSTAT1/3-Luc added and IFN-γ untreated group; *p <0.05 vs. luciferase reporter vector added and no peptide treated group. Results are mean ± SEM (n = 3).
Primers and parameters used in real-time PCR.