Abstract
Background
The alarmone (p)ppGpp mediates a global reprogramming of gene expression upon nutrient limitation and other stresses to cope with these unfavorable conditions. Synthesis of (p)ppGpp is, in most bacteria, controlled by RelA/SpoT (Rsh) proteins. The role of (p)ppGpp has been characterized primarily in Escherichia coli and several Gram-positive bacteria. Here, we report the first in-depth analysis of the (p)ppGpp-regulon in an α-proteobacterium using a high-resolution tiling array to better understand the pleiotropic stress phenotype of a relA/rsh mutant.
Results
We compared gene expression of the Rhizobium etli wild type and rsh (previously rel) mutant during exponential and stationary phase, identifying numerous (p)ppGpp targets, including small non-coding RNAs. The majority of the 834 (p)ppGpp-dependent genes were detected during stationary phase. Unexpectedly, 223 genes were expressed (p)ppGpp-dependently during early exponential phase, indicating the hitherto unrecognized importance of (p)ppGpp during active growth. Furthermore, we identified two (p)ppGpp-dependent key regulators for survival during heat and oxidative stress and one regulator putatively involved in metabolic adaptation, namely extracytoplasmic function sigma factor EcfG2/PF00052, transcription factor CH00371, and serine protein kinase PrkA.
Conclusions
The regulatory role of (p)ppGpp in R. etli stress adaptation is far-reaching in redirecting gene expression during all growth phases. Genome-wide transcriptome analysis of a strain deficient in a global regulator, and exhibiting a pleiotropic phenotype, enables the identification of more specific regulators that control genes associated with a subset of stress phenotypes. This work is an important step toward a full understanding of the regulatory network underlying stress responses in α-proteobacteria.
Background
Rhizobium etli is a soil-dwelling α-proteobacterium that infects the roots of its leguminous host plant Phaseolus vulgaris, the common bean plant, in order to establish a nitrogen-fixing symbiosis [1-4]. Like most microorganisms in nature, R. etli primarily resides in a non-growing state in the soil, where it is confronted with diverse and stressful conditions, such as non-optimal temperatures and pH levels, near-starvation conditions and competition with other microbial populations [5]. Although growth is restricted, long periods of inactivity are sporadically interrupted by proliferation. This cycle of growth and starvation has been likened to a feast and famine lifestyle [6].
Sophisticated regulatory networks allow bacteria to sense and respond to a variety of environmental stresses to rapidly adjust their cellular physiology for survival. These networks comprise transcriptional regulators, sigma factors, proteases and small non-coding RNAs (ncRNAs) that interact in a complex manner in order to control the metabolic changes needed for adaptation [5]. The stringent response is a widespread global regulatory system, activated in response to various unfavorable growth conditions, and mediated by guanosine tetraphosphate (ppGpp) and guanosine pentaphosphate (pppGpp), collectively referred to as (p)ppGpp [7]. This alarmone coordinates entrance into the non-growing state by inducing a general reprogramming of gene regulation, thereby downregulating cellular processes needed for growth and upregulating processes needed for survival. As a result, the available resources are diverted from growth to allow adaptation of the cell to the non-growing state [8,9]. The central role of this alarmone in the general stress response during the stationary phase is also illustrated by the increased sensitivity of (p)ppGpp-deficient mutants in various species to diverse stress factors [10]. Therefore, studying the (p)ppGpp regulon may be useful to identify novel regulators involved in the stress adaptation.
In Escherichia coli, the stress-induced alarmone production depends on two enzymes: RelA and SpoT [7]. When amino acids are limiting, uncharged tRNAs that bind ribosomes stimulate the ribosome-associated RelA to synthesize (p)ppGpp. Subsequent recovery when conditions are favorable again requires degradation of the alarmone, which is catalyzed by SpoT. SpoT is a bifunctional enzyme that can also synthesize (p)ppGpp in response to carbon, iron, phosphorus and fatty acid scarcity. Having two (p)ppGpp synthetases/hydrolases appears to be an exclusive feature of the γ-subdivision of the proteobacteria, as Gram-positive bacteria and most other Gram-negative bacteria, including R. etli, possess only a single RelA/SpoT homolog - usually referred to as Rel or Rsh - that displays both activities [10]. Most Gram-positive species additionally encode small proteins that consist solely of a synthetase domain [11].
(p)ppGpp primarily regulates gene transcription [12,13]. Several models have been proposed to accommodate the effects of (p)ppGpp on transcription. One of these models, the affinity model, argues for an increase in the availability of free RNA polymerase (RNAP) with increasing (p)ppGpp levels. As this alarmone binds near the active site of RNAP, the stability of the ribosomal RNA (rrn) open complexes decreases. Consequently, (p)ppGpp will induce promoters with low RNAP affinity, such as cell maintenance and stress response genes [14,15]. In another model, the σ factor competition model, the binding affinity of alternative sigma factors increases with increasing (p)ppGpp-levels compared to the housekeeping sigma factor σ70. This results in a decrease of σ70-bound RNAP and a downregulation of growth-related promoters that are dependent on high concentrations of σ70-bound RNAP for maximal expression [10,12,16]. In addition to regulating sigma factor activity, (p)ppGpp is also required for sigma factor expression, as is the case for the stationary phase sigma factor σS, the heat shock sigma factor σH and the sigma factor controlling nitrogen metabolism, σ54, in E. coli [17,18]. Hence, these models for gene regulation of (p)ppGpp should be considered as working in concert. Finally, the recently identified cofactor DksA was demonstrated to stabilize binding of RNAP to (p)ppGpp, resulting in enhanced repression or stimulation of transcription in E. coli. However, the interaction between (p)ppGpp and DksA appears to be more complex as both factors also have independent and opposing effects on gene expression in E. coli [13,19,20].
In agreement with (p)ppGpp's central role in stress adaptation, the alarmone was shown to be crucial in many complex physiological processes such as biofilm formation by Listeria monocytogenes, E. coli and Streptococcus mutans, development of multicellular fruiting bodies in Myxococcus xanthus and development of competence in Bacillus subtilis [10]. In addition, a fast growing number of reports demonstrate (p)ppGpp to be important during host interactions in diverse pathogens such as Vibrio cholerae, Pseudomonas aeruginosa, Legionella pneumophila, Francisella novicida, Enterococcus faecalis and Streptococcus pneumoniae [21-24]. Furthermore, various transcriptome studies showed that the alarmone (p)ppGpp is situated high up in the hierarchy of interconnected regulators in E. coli, controlling the expression and/or function of many other regulators such as Lrp, the cAMP receptor protein CRP, the integration host factor IHF, the flagellar master regulator FlhDC, the redox status sensing regulator ArcA and the morphogene BolA [6,8,18,25,26].
(p)ppGpp also affects key aspects of the symbiosis between rhizobia and their leguminous host plants. In Sinorhizobium meliloti, a rsh mutant is defective in nodulation of Medicago sativa and overproduces the exopolysaccharide succinoglycan, which is crucial for root infection [27]. In R. etli, (p)ppGpp controls the physiological adaptation of the bacterium to the endosymbiotic state [28,29]. Although the rsh mutant induces nodulation, the bacteroids are morphologically different compared to the wild type, and nitrogen fixation activity is drastically reduced. Several nitrogen fixation and quorum-sensing genes, essential for symbiosis, were shown to be part of the alarmone regulon, including the symbiotic σN that is required for expression of nitrogen fixation genes [29]. Recently, a detailed phenotypic analysis of the rsh (previously referred to as relA or relRet) mutant showed a prominent role for the alarmone in the general stress response of R. etli during free-living growth and symbiosis [30].
In order to obtain new insights into the molecular basis of adaptation of R. etli to unfavorable growth conditions, we performed a genome-wide transcriptome analysis to compare global gene expression between the wild type and a rsh mutant during different free-living growth phases.
This study is the first in-depth analysis of (p)ppGpp-dependent gene regulation in an α-proteobacterium, revealing notable differences from the well-studied role of (p)ppGpp in E. coli. Of the many detected (p)ppGpp targets that may contribute to the observed stress phenotypes of the rsh mutant, we performed a phenotypic analysis of three specific previously uncharacterized regulators, that is, sigma factor EcfG2/PF00052, DNA-binding transcription factor CH00371 and serine kinase PrkA/CH02817. Our results show that the stress phenotypes of mutants lacking EcfG2 or CH00371 correspond to a subset of the rsh mutant phenotypes, while PrkA may be involved in metabolic adaptation. In addition, we identified several upstream and downstream elements in the stress response pathways of these three novel (p)ppGpp-dependent regulators, providing added detail to the complex picture of the role of (p)ppGpp in R. etli.
Results and Discussion
Experimental design of the transcriptome analysis
Previously, we reported on the crucial role of (p)ppGpp during symbiosis and free-living growth in R. etli CNPAF512 using a rsh mutant [29,30]. Based on these findings, we decided to carry out a transcriptome analysis to characterize to what extent (p)ppGpp deficiency affects gene expression in R. etli. The intracellular (p)ppGpp content of the R. etli wild type, rsh mutant and complemented rsh mutant was determined previously [29], showing the rsh mutant to be (p)ppGpp-deficient. However, due to the sensitivity of the assay, the presence of trace amounts of (p)ppGpp in the rsh mutant, possibly resulting from the presence of an as yet unidentified synthetase gene, cannot be ruled out.
At the time of the experimental setup, only the genomic DNA sequence of R. etli CFN42 was available [31]. Therefore, a custom whole-genome microarray for R. etli CFN42 as well as a CFN42-derived rsh mutant was constructed. Phenotypic analysis of this mutant showed that a lack of (p)ppGpp results in an extended lag phase in different media, an altered morphology and a 75% reduction of nitrogen fixation activity in plants inoculated with the CFN42 rsh mutant compared to the wild type (data not shown). All phenotypes could be fully complemented by providing rsh of CNPAF512 in trans and are in agreement with our previously published rsh mutant analyses [29,30].
To determine the role exerted by the alarmone (p)ppGpp in the regulation of transcription during free-living growth and growth arrest of R. etli, total RNA samples were taken at three different time points corresponding to early and late exponential and stationary phase, respectively (Additional file 1).
Global overview of gene expression
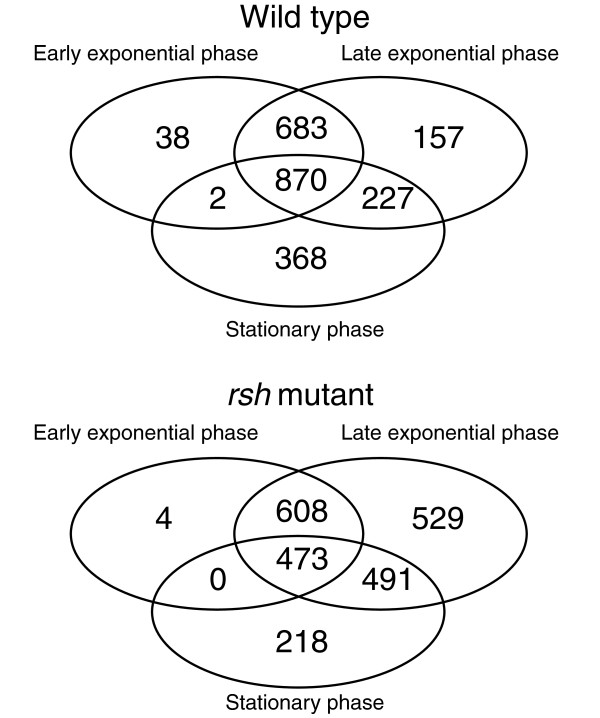
The R. etli CFN42 genome contains 6,030 annotated protein-encoding genes, 67 pseudo genes, 3 rRNA operons and 50 tRNA genes. Recently, we described an additional 89 ncRNA genes [32]. In both the wild type and rsh mutant, over 97% (or (683 + 870)/1,593) of protein-encoding genes that are transcribed above the detection limit (see Materials and methods) during early exponential growth are also expressed during late exponential growth. In addition, numerous genes are induced in the course of growth, as 20% (or (157 + 227)/1,937) of the genes expressed in late exponential phase are not transcribed during early growth (Figure 1).
Figure 1.
Detectable gene expression overview. The number of genes expressed above the detection threshold in each condition and the overlap between the different conditions are shown in Venn diagrams. Upper and lower diagrams represent expression in the wild type and rsh mutant, respectively.
We identified a large number of differentially expressed genes during exponential and stationary phase, both (p)ppGpp-dependent and independent, the former being consistent with the role of rsh as a global regulator described in other species [33-35]. The extent of differential expression is illustrated by the ratio/intensity MA plots in Additional file 2. Alarmone dependency was determined by comparing gene expression of the wild type and rsh mutant during each of the three sampled growth phases (Figure 2a). A total of 834 (p)ppGpp-dependent genes with an expression ratio of at least two-fold were found. Approximately half of these genes (520) were expressed exclusively during stationary phase and only a minority (36) were found to be (p)ppGpp-dependent during all growth conditions.
Figure 2.
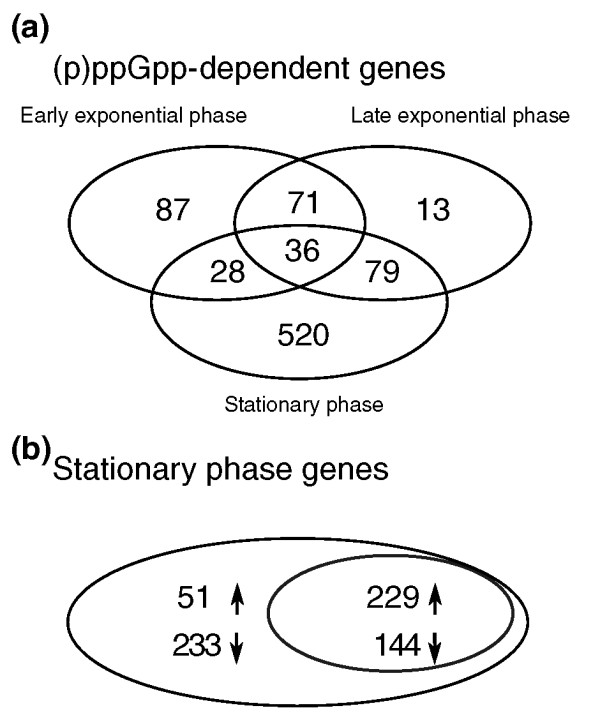
(p)ppGpp-dependent gene expression. (a) Venn diagram of all differentially expressed (p)ppGpp-dependent genes during early exponential phase, late exponential phase and stationary phase. (b) Venn diagram of all genes expressed during stationary phase (large ellipse). The overlap with (p)ppGpp-dependent genes (see (a)) shows all (p)ppGpp-dependent stationary phase genes (small ellipse). Upwards and downwards oriented arrows indicate gene induction and repression, respectively.
By comparing expression in the wild type during stationary and early exponential phase, we identified 657 stationary phase genes (Figure 2b), representing 11% (or 657/6,030) of the annotated protein-coding genes. The overlap of (p)ppGpp-dependent genes and stationary phase genes shows that just over half (57% or (229 + 144)/657) of stationary phase genes are (p)ppGpp-dependent. Because 61% (or 229/373) of these were upregulated (Figure 2b), the alarmone (p)ppGpp seems to have a primarily inducing role in R. etli. A comparable number of (p)ppGpp-dependent genes were found in other bacteria: 490 (11%) of all genes in E. coli and 194 (6%) in Corynebacterium glutamicum after (p)ppGpp-induction by serine hydroxymate [18,34], 589 genes (7%) upon induction of (p)ppGpp synthesis in Streptomyces coelicolor [35] and 373 (18%) of all genes after treatment with mupirocin in Streptococcus pneumoniae [23].
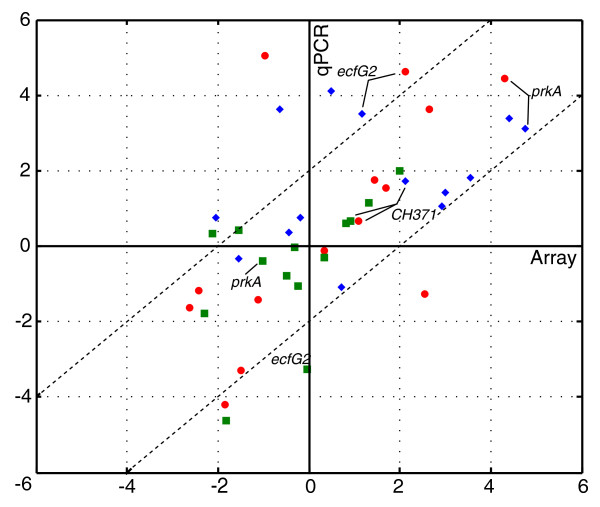
The microarray data were confirmed by analyzing the expression levels of 14 representative genes using reverse transcription-quantitative PCR (RT-qPCR; see Materials and methods). For each gene, expression during early exponential phase and stationary phase in the wild type and rsh mutant was measured so that three different ratios could be plotted versus the respective ratios obtained by microarray analysis (Figure 3), showing the array data to be in good agreement with the RT-qPCR data.
Figure 3.
RT-qPCR validation of the microarray data. Expression of 14 genes was determined using RT-qPCR for the wild type and rsh mutant in early exponential phase and stationary phase. The log2-transformed mean values of two biological replicates were used to report three different fold changes for each gene (Y-axis) compared to the respective microarray fold changes (X-axis). See Additional file 6 for a complete list of the plotted fold change values. Red dots, wild type stationary phase versus early exponential phase. Blue diamonds, wild type versus rsh mutant in stationary phase. Green squares, wild type versus rsh mutant in exponential phase. The fold changes for ecfG2/PF00052, CH00371 and prkA/CH02817 are indicated.
The effect of (p)ppGpp on global gene expression during stationary phase
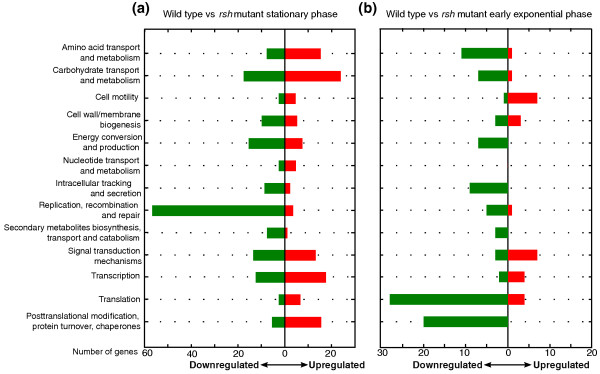
During stationary phase in E. coli, the alarmone (p)ppGpp induces a downregulation of processes involved in cell growth, such as DNA replication and translation, and an upregulation of specific metabolic pathways to cope with certain nutrient deficiencies as well as general stress responses to protect the cell against immediate and future harmful conditions. In order to better understand the role of (p)ppGpp in the global reprogramming of R. etli's transcriptome, we compared the expression of wild type and rsh mutant during stationary phase. As samples were taken approximately 6 hours after growth arrest, the observed differences in expression include both direct and indirect effects caused by a lack of alarmone. Of the 663 differentially expressed genes, 292 and 371 were upregulated and downregulated, respectively, in the wild type compared to the rsh mutant (Figure 2a). These genes were further grouped based on predicted functional role and category (Additional file 3). An overview of the functional categories (Figure 4a) shows the mutant to be less well adapted to the non-growing lifestyle as more growth-associated genes, involved in cell wall biosynthesis, energy production and intracellular trafficking and secretion, are induced in the mutant. Notably, the replication and recombination category is strongly represented in the mutant due to the high number of insertion sequence (IS)-related genes that show expression. An equal number of genes with unknown function were up- and downregulated. In the following paragraphs, selected functional categories, primarily focused on regulation and possible links with the pleiotropic stress phenotype, will be discussed in more detail.
Figure 4.
Differentially expressed genes grouped by functional categories. Up- and downregulated (wild type versus rsh mutant) genes are indicated by red and green bars, respectively, representing the number of genes per functional category. Functional categories of the RhizoBase database were used [92]. (a) Stationary phase data of wild type versus rsh mutant. (b) Early exponential data of wild type versus rsh mutant.
Transcriptional regulators and signal transduction
The link between changes in extracellular conditions and concomitant adaptation of genome expression involves a combination of sensors, transporters, phosphorylation cascades and the modulation of transcription factors [36]. Most of these belong to the 'transcription' and 'signal transduction' categories, of which 29 and 26 genes are differentially expressed, respectively, in the wild type compared to the (p)ppGpp-deficient mutant at onset of growth arrest (Additional file 3).
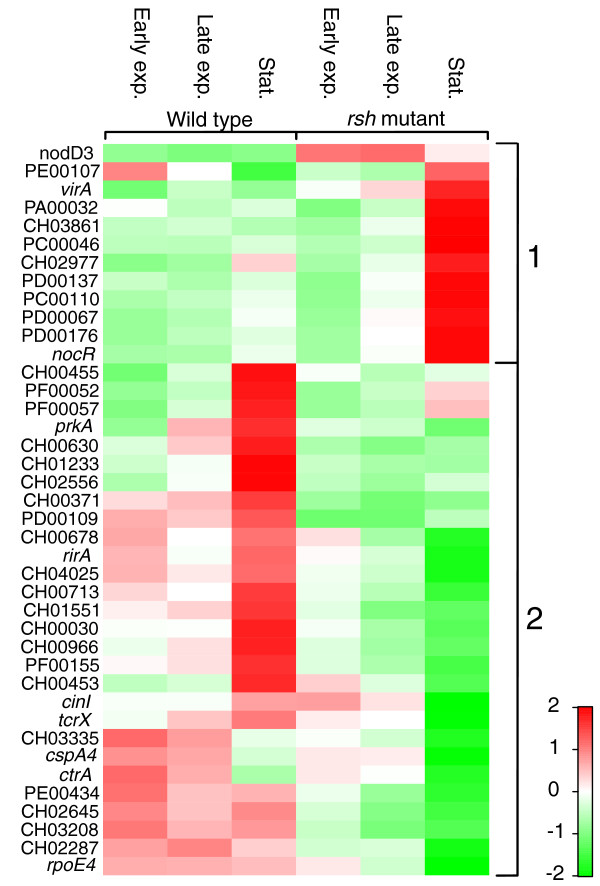
By clustering the differentially expressed genes of these two categories, we identified two main groups (Figure 5). The first group contains genes that are under negative (p)ppGpp control during primarily the stationary phase and include the LysR transcriptional regulators nocR and nodD3, the two-component sensor kinase virA and two diguanylate cyclases, PD00137 and PE00107. The second group contains genes that are under positive (p)ppGpp control during primarily the stationary phase, encoding among others the transcriptional regulators RirA and BolA-like CH02287, the CarD-like regulator CH04025, the two-component response regulators CH02556 and CH03335, and the N-acyl-L-homoserine lactone (AHL) synthase CinI.
Figure 5.
Clustering of differentially expressed signal transduction and transcription-related genes. The heat map visualizes the expression profiles of all differentially expressed genes belonging to the transcription category and signal transduction category in the wild type and rsh mutant during stationary phase. The expression values in each row were standardized by subtraction of the mean and division by the standard deviation and hierarchically clustered. Expression values are reflected by red-green coloring as indicated. Genes showing similar expression patterns are grouped as follows: group 1, genes under negative (p)ppGpp control during stationary phase; group 2, genes under positive (p)ppGpp control during stationary phase. Exp., exponential; Stat., stationary.
Several of these transcriptional regulators have previously been shown to play a role in the adaptation to adverse conditions in other species and can partly explain the pleiotropic stress phenotype of the rsh mutant. In E. coli, BolA controls expression of a number of cell wall proteins, is partially responsible for the coccoid morphology of stationary phase cells and is also expressed in a (p)ppGpp-dependent manner, being under control of RpoS [5,18,33]. Reduced BolA levels may therefore contribute to the altered morphology of the R. etli rsh mutant. Furthermore, expression of the global iron-responsive regulator RirA that controls the synthesis of heme, FeS-clusters and bacterioferritin in rhizobia, is under positive (p)ppGpp control as well [37,38]. Accordingly, expression of bacterioferritin (bfr) was positively upregulated in the R. etli wild type, suggesting that (p)ppGpp contributes to iron homeostasis. Conversely, a lack of iron may cause an increase in the level of (p)ppGpp in order to regulate iron homeostasis in the cell as reported in E. coli and B. subtilis [39,40]. Since iron plays a crucial role in the oxidative stress response, incomplete iron sequestration may also contribute to the increased oxidative stress sensitivity of the rsh mutant [30,41]. Other regulators under positive (p)ppGpp control include two members of the cold shock protein family (CspA3, CspA4), a putative member of the UspA family (CH01233), the SOS response regulator LexA and the two-component regulator TcrX. TcrX is orthologous to PhyR of Methylobacterium extorquens, which regulates many stress response genes and was shown to play a role in the osmotic stress response in R. etli as well [42,43].
Sigma factors
R. etli CFN42 possesses 23 sigma factors that determine the promoter specificity of the RNAP holoenzyme by binding to the core enzyme. Therefore, differential expression and/or activity of sigma factors can redirect global gene expression. During exponential growth, transcription is largely under control of the housekeeping sigma factor σ70 as its binding affinity for RNAP and intracellular concentration are much higher compared to the other sigma factors. These alternative sigma factors have specific regulons and will redirect transcription upon unfavorable conditions. Bacteria like R. etli that have a complex lifestyle or encounter diverse environmental conditions usually display an increased number of sigma factors [5,44].
Upon transition to stationary phase, the reversible switch to a less σ70-dominated expression in E. coli is accomplished not solely by (p)ppGpp but also by DksA and the anti-σ70 factor Rsd. In R. etli, expression of dksA is reduced over eight-fold in stationary phase compared to early exponential phase in a (p)ppGpp-independent manner. The role of DksA in α-proteobacteria is so far unknown. Furthermore, no Rsd homolog is found in R. etli or other other α-proteobacteria. R. etli may compensate for the lack of a specific anti-σ70 factor, as we observed a (p)ppGpp-independent drop in expression of the housekeeping sigma factor sigA to below the detection limit while expression in E. coli of σ70 remains constant during stationary phase.
Of all the alternative sigma factors, only the extracytoplasmic function (ECF) sigma factor PF00052 was upregulated at least two-fold during stationary phase compared to early exponential phase in the wild type. The ECF sigma factor rpoE4 is expressed at the same level during all conditions in the wild type, but dropped below the expression threshold during stationary phase in the rsh mutant. Consequently, two ECF sigma factors, PF00052 and rpoE4, were upregulated over two-fold in the wild type compared to the rsh mutant during stationary phase (Figure 5). In E. coli, only the level of the stationary phase sigma factor rpoS (σS) increases with (p)ppGpp concentration and plays a crucial role as global regulator in the (p)ppGpp-dependent stress response [45,46]. In contrast, ε- and α-proteobacteria, including R. etli, lack such a stationary phase sigma factor and, so far, it is unclear which system takes over this function.
Our data suggest that both PF00052 and RpoE4 may be important sigma factors in R. etli adaptation to stationary phase and possibly fulfill a role similar to RpoS in E. coli. First, both sigma factors are expressed during stationary phase. Second, both sigma factors are the most highly upregulated (p)ppGpp-dependent alternative sigma factors during stationary phase. Third, both share considerable sequence similarity and were recently classified in a group of proposed general stress response sigma factors that is exclusively found in α-proteobacteria [47]. Fourth, it was recently shown in R. etli that RpoE4 regulates gene expression in response to several stress conditions including oxidative, saline and osmotic stress. Fifth, we found that PF00052 is also involved in the (p)ppGpp-dependent stress response and is in part functionally redundant with RpoE4 (see below).
Transcriptome analysis of an R. etli rpoE4 mutant and overexpression strain revealed 98 genes to be regulated by this sigma factor [42]. Since transcription of rpoE4 is (p)ppGpp-dependent, we investigated to what extent the reported RpoE4 regulon is (p)ppGpp-dependent. In total, 60 of the 98 genes belonging to the reported regulon are differentially expressed in our data (Additional file 4). Of these genes, 82% are (p)ppGpp-dependent and 92% are up- or downregulated during stationary phase compared to early exponential phase in the wild type. Upon rpoE4 overexpression, 74% of the reported upregulated genes were found to be (p)ppGpp-dependent.
Considering the RpoE4-regulated genes, all 16 genes predicted to encode proteins associated with cell envelope biogenesis are also (p)ppGpp-dependent. Similarly, E. coli's sole ECF sigma factor, σE, regulates many genes involved in the biogenesis and stress response of the cell envelope [48,49]. Other RpoE4 and (p)ppGpp-dependent genes include a putative Mn-catalase (CH00462), a putative pyridoxine-phosphate oxidase (CH03474), an alpha-glucoside ABC transporter (algE), and a CarD-like transcriptional regulator (CH04025). The latter is a crucial regulator in Mycobacterium tuberculosis that is upregulated in response to oxidative stress, DNA damage and starvation [48,49]. The above suggests that the pleiotropic stress phenotype of the R. etli rsh mutant can be explained, at least in part, by downregulation of (p)ppGpp-dependent sigma factors that play a crucial role in orchestrating the stress response.
Non-coding RNAs
Our data indicate that (p)ppGpp controls expression of many protein-coding genes. In addition, we identified 33 alarmone-dependent ncRNAs expressed during stationary phase. Of these, 28 were positively regulated by (p)ppGpp, including one glycine riboswitch, 17 novel ncRNAs, 4 previously identified but uncharacterized ncRNAs, and the 6 well characterized ncRNAs (6S RNA, tmRNA, signal recognition particle 4.5S RNA, RNase P, and ctRNA of plasmids p42d and p42e) (Additional file 5). Only five ncRNAs, all novel, were negatively regulated by (p)ppGpp.
So far, no ncRNAs have been reported to be (p)ppGpp-dependent in any organism. However, in recent years, an increasing number of ncRNAs have been found to be regulated by alternative sigma factors in E. coli, Salmonella enterica serovar Typhimurium, L. monocytogenes, B. subtilis and S. coelicolor [50-53]. Therefore, the (p)ppGpp-dependent ncRNAs of R. etli could be regulated by alternative sigma factors as well. Additionally, ncRNAs can also regulate sigma factors, as is the case for σS in E. coli whose translation is regulated by DsrA and RprA [50-53].
The level of 6S RNA was almost 14-fold lower in the alarmone-deficient mutant during stationary phase in R. etli. This is unlike in E. coli, where 6S RNA is not under (p)ppGpp control either in vitro or in vivo [54,55]. However, 6S RNA transcription appears to be complexly regulated in E. coli as several stress regulators, such as Fis, H-NS, Lrp and StpA, were shown to be inhibitors under in vitro conditions [55]. Recently, transcriptional analysis of a 6S RNA-deficient mutant showed 273 genes to be differentially expressed during stationary phase. Surprisingly, loss of 6S RNA in E. coli also resulted in an increase of the basal (p)ppGpp level mediated by an altered activity of SpoT and not RelA [56]. Therefore, 6S RNA is clearly embedded in stationary phase adaptation, although its association with (p)ppGpp in R. etli and E. coli may differ.
Expression of bacterial RNase P and tmRNA was almost 13- and 6-fold downregulated, respectively, in the R. etli rsh mutant compared to the wild type. Although the synthesis and processing of tRNA is expected to be downregulated during growth arrest, 38% of the tRNAs were upregulated in the wild type during stationary phase compared to early exponential phase and 56% of the tRNAs were upregulated in an alarmone-dependent manner. The upregulation of bacterial RNase P during stationary phase in an alarmone-dependent manner is in line with the unexpected upregulation of several tRNAs as RNase P is required to process the 5' end of precursor tRNAs. Expression of tmRNA is also upregulated in R. etli. This alarmone-dependence of tmRNA expression has not been reported in E. coli, although a lack of 6S RNA results in a three-fold higher expression of the SmpB protein, which acts together with tmRNA [56]. However, the expression level of tmRNA was not reported. Interestingly, the 6S RNA mutation is compensated for by an increase of the basal (p)ppGpp level, which indicates that the tmRNA/SmpB system might be alarmone-dependent in E. coli also. Still, (p)ppGpp is not needed for mRNA cleavage in the A site of the ribosome by tmRNA [57]. In contrast, both tmRNA and smpB of Streptococcus pyogenes were shown to be upregulated in a relA-independent amino acid starvation response [58].
Translational apparatus
In addition to inducing general stress and nutrient scavenging regulons, the accumulation of (p)ppGpp upon growth arrest in E. coli is characterized by a stringent downregulation of expression of the translational apparatus as a mechanism to fine-tune the metabolically expensive process of protein synthesis according to the growth state of the cell [9,59]. As expected, during the stationary phase all 56 genes encoding ribosomal proteins were downregulated in the R. etli wild type compared to the exponential phase. However, nearly all (53 out of 56) of these were downregulated in the rsh mutant as well. Although this (p)ppGpp-independent downregulation is in conflict with the established E. coli paradigm of the stringent response, a similar response was described in a rel mutant of Corynebacterium glutamicum upon addition of serine hydroxamate [9,34,59]. Therefore, the difference in transcriptional regulation of ribosomal protein expression during growth arrest suggests that the stringent response in R. etli may deviate from the classical model in E. coli.
Other genes encoding parts of the translational machinery that were positively regulated by (p)ppGpp in R. etli include the homolog of E. coli yhbH (CH00406) and two EF-Tu elongation factors (tufA, tufB). In E. coli, YhbH is involved in the temporary storage or dimerization of ribosomes during stationary phase. This process was shown to contribute to the survival of E. coli [28]. In accordance with our data, the YhbH ortholog of B. subtilis (yvyD) is also under positive (p)ppGpp control [60]. However, in contrast to the positive (p)ppGpp-dependent regulation of tufA and tufB in R. etli, TE-Tu factors in E. coli and B. subtilis were previously shown to be under negative control of (p)ppGpp [8,60]. Interestingly, translation factors such as TE-Tu are GTPases that can bind (p)ppGpp and associate with the ribosome, indicating that they may have a downstream role in (p)ppGpp-dependent gene regulation [13].
Post-translational modification, repair and recombination
The (p)ppGpp-dependent stress adaptation during stationary phase involves 20 genes belonging to the post-translational modification category, of which 15 were positively regulated. These include several components of the ATP-dependent Clp protease system, such as clpX, clpP2, clpP3, clpA and clpS, as well as the ATP-dependent proteases lon and ftsH [61]. These proteases allow cells to cope with misfolded or denatured proteins, the abundance of which increases during stress conditions, such as heat stress, in order to prevent protein aggregation and to enable recycling of amino acids [5]. A similar (p)ppGpp-dependent regulation was observed for clpA in E. coli as well as clpP1 and clpC in C. glutamicum [33,34]. Thus, the (p)ppGpp-dependent increase of tmRNA in R. etli correlates with the increase in proteases as the Clp system and Lon are needed to degrade tmRNA-tagged polypeptides in E. coli [62].
Proteases and chaperones are also involved in regulating transcriptional regulators and other growth-phase regulated proteins, such as RpoS, Dps and GlnA in E. coli [63]. Therefore, by controlling proteolysis, the alarmone (p)ppGpp mediates the cellular reprogramming of R. etli at the post-transcriptional level as well. Other positively controlled genes include the probable serine protease CH01273, the small heat shock protein PF00472 as well as genes required to cope with oxidative stress, such as osmC and grlA.
Rather unexpectedly, very few genes of the repair and recombination category were under positive stringent control. However, several IS-related genes were negatively controlled by (p)ppGpp, including 47 transposases, one resolvase, and one integrase. Therefore, these data suggest that the alarmone may assist in repressing insertional activity and mobility of IS-related elements.
Other processes
The impact of (p)ppGpp as a global regulator of transcription is further illustrated by its control of genes involved in diverse cellular processes. In E. coli, the alarmone plays a central role in restructuring metabolism upon nutrient starvation and growth arrest, thereby increasing the range of active metabolic pathways and nutrient scavenging potential [33]. In R. etli, the alarmone likely has a similar role in metabolism as differential gene expression was detected for 22 genes involved in amino acid metabolism, 41 genes in carbohydrate metabolism, 9 in lipid metabolism and 22 in energy production.
(p)ppGpp was shown in E. coli to induce amino acid biosynthesis pathways depending on the availability of limiting amino acids. However, compared to exponential phase, no clear upregulation during stationary phase of one or more specific amino acid pathways was found in the R. etli wild type. Only a few genes involved in amino acid metabolism were positively controlled by (p)ppGpp (phhA, cysE1, glnA2, trpE). In addition, 13 amino acid synthesis genes (trpF, trpA, hisB, asnB, aroQ1, aroF, ilvI, aatA, lysC, argG2, tyrA, leuD, asd) along with the P-II regulator glnB, which regulates glutamine synthetase in response to nitrogen levels, were downregulated in a (p)ppGpp-independent manner during stationary phase compared to exponential phase. Therefore, during stationary phase, amino acid biosynthesis in R. etli is downregulated rather than upregulated as in E. coli.
Several genes encoding key enzymes of carbohydrate metabolism were induced by (p)ppGpp, including the transaldolase tal of the pentose pathway, glgC involved in starch and sucrose metabolism, the glycolytic gene fbaB and the gene encoding trehalose-6-phophatase, otsB. These genes were also shown to be under positive control by (p)ppGpp in E. coli upon amino acid starvation [33]. FbaB is a fructose-bisphosphate aldolase whose reaction product can exert feedback control on the glycolytic flux and is also required for ribosome recycling during carbon starvation [6]. Moreover, OtsB produces the disaccharide trehalose from trehalose-6-phosphate, which is produced by OtsA using UDP-glucose and glucose-6-phosphate. Not only is trehalose an energy and carbon source, it also stabilizes and protects proteins and membranes from dehydration, oxidation and cold [64]. Recently, it was shown that all three trehalose synthesis pathways known to date are present in S. meliloti. However, only the OtsA pathway is important for osmo-inducible trehalose synthesis [65]. In R. etli, overexpressing otsA improves symbiotic efficiency and drought tolerance of its host P. vulgaris [66]. During stationary phase, otsA and otsB have a different expression pattern; otsB is induced by (p)ppGpp while otsA is constitutively expressed in the wild type but under negative (p)ppGpp control upon growth arrest. It is possible that this (p)ppGpp-dependent regulation of trehalose synthesis contributes to the previously observed increased sensitivity of the rsh mutant to osmotic stress [30].
The link between the stringent response and the available carbon sources remains unclear. In E. coli, the (p)ppGpp synthetase/hydrolase SpoT interacts with acyl carrier proteins (ACPs) of fatty acid metabolism [67]. R. etli contains four acyl carrier proteins, of which only two (acpP, acpXL) were expressed during growth and downregulated upon growth arrest independently of (p)ppGpp. In contrast to E. coli, no clear (p)ppGpp-dependent regulation of lipid metabolism genes was observed. Also, most of the nucleotide biosynthesis genes are downregulated during stationary phase compared to exponential phase in the wild type, reflecting the decreased need for nucleotides. Only six nucleotide biosynthesis genes were found to be under control of (p)ppGpp in R. etli. This is in accordance with the observed (p)ppGpp-independent downregulation of ribosomal proteins.
As well as regulating R. etli's biosynthetic potential, (p)ppGpp also controls its transport capacity during stationary phase. Twenty-one genes related to ABC transporters were under positive (p)ppGpp control, such as potF, dppA, proX, aglK, and gguB, while 12 were repressed. Most of these transporters allow for the uptake of amino acids, peptides and monosaccharides. In addition, two secretion-associated genes (secB and pilA) were upregulated and seven genes involved in type IV secretion were downregulated (virB1a, 2a, D4, B6a, B8a, B8d, B10). Interestingly, the pilin subunit pilA was the most highly expressed protein-encoding gene in R. etli during stationary phase.
Energy production drops during the stationary phase as more than 25 genes predicted to be involved in oxidative phosphorylation were downregulated compared to exponential phase in the wild type. In contrast to E. coli, 76% of the differentially expressed genes that belong to the energy production category, 95 in total, are not under (p)ppGpp control [33]. During free-living growth, R. etli uses cytochrome aa3 terminal oxidases, encoded by ctaCDGE, coxPONM and CH00981-CH00985 [31,68]. The ctaCDGE terminal oxidase was downregulated during stationary phase in both the wild type and the rsh mutant. On the other hand, the coxPONM alternative terminal oxidase was upregulated during stationary phase in the wild type but not in the rsh mutant. The third probable terminal oxidase was not expressed. Therefore, the alternative terminal oxidase coxPONM is likely to play an important role during (p)ppGpp-dependent stationary phase adaptation.
In addition to the decrease in energy production, flagellum synthesis and motility is also downregulated during stationary phase, reflecting that it is a highly energy demanding process. In E. coli, flagellar genes are under positive (p)ppGpp control [18,19,69]. Similarly in R. etli, (p)ppGpp positively regulates flagellar gene expression. However, this regulation occurs primarily during the exponential phase instead of the stationary phase, as 25 of the 35 flagellar genes were expressed above threshold during the exponential phase compared to 10 during the stationary phase. Of the latter, three flagellar hook-related genes (flgD, flgE, flgL) and two flagellin synthesis regulators (flaF, flbT) were upregulated in the wild type compared to the rsh mutant [70]. FlgD forms a scaffold on which the hook subunit FlgE polymerizes on the envelope-embedded rod to form the flexible hook structure. FlgL is a junction protein connecting the rigid flagellar filament. In short, (p)ppGpp regulates several crucial flagellar genes in R. etli.
The effect of (p)ppGpp on global gene expression during early exponential phase
By comparing the expression data of the wild type and rsh mutant during early exponential growth, we identified 203 differentially expressed genes, of which 59 were under positive stringent control and 144 under negative stringent control. This is surprising as transcription during the exponential phase of a (p)ppGpp-deficient mutant and the wild type is generally thought to be very similar. The alarmone is considered to be a stationary phase or growth arrest-specific messenger that switches the cellular metabolism to a non-growing state. During favorable growth conditions, (p)ppGpp is produced at a low basal level and rapidly accumulates in response to growth-perturbing conditions. Furthermore, during the exponential phase, a (p)ppGpp-deficient mutant of E. coli is phenotypically very similar to the wild type, although a decreased growth phase-independent thermotolerance has been reported [71]. However, to the best of our knowledge, to this date no detailed comparison of global transcription during exponential growth has been described for a wild type and relA spoT mutant of E. coli, which serves as the stringent response model organism. Still, it was recently shown that almost 300 genes were differentially expressed in an rpoS mutant of E. coli during exponential growth, even though RpoS is known as the stationary phase sigma factor [72]. This would suggest that a difference in expression during logarithmic growth in a (p)ppGpp-deficient mutant could be expected as (p)ppGpp regulates the expression and activity of RpoS in E. coli. Moreover, a major difference in expression during growth in the absence of (p)ppGpp was also previously observed in M. tuberculosis and C. glutamicum [34,73]. Our data are in agreement with these reports, showing that the low basal level of (p)ppGpp is functionally relevant during active growth. This additional function is also in agreement with the observed increase in sensitivity to several acute and chronic stresses of a R. etli rsh mutant during exponential growth [30].
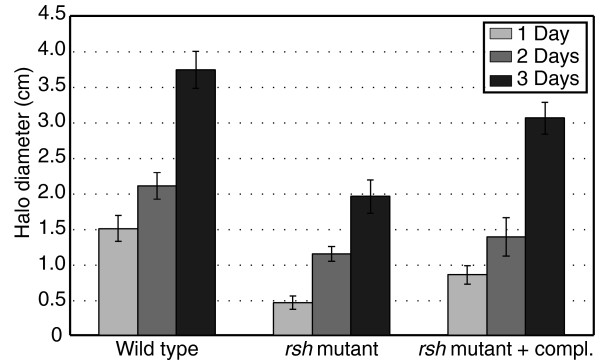
A comparison of the (p)ppGpp-dependent genes during early exponential phase and stationary phase showed that only 50 genes were differentially expressed in both states. Of this fraction, only half of the genes showed similar positive or negative control during both phases. This suggests that the function of (p)ppGpp differs during active growth and growth arrest, possibly through involvement of other regulators. To further understand the impact and role of the alarmone during exponential growth, we again grouped the up- and downregulated genes in functional categories (Figure 4b). As 71% of these genes were under negative (p)ppGpp regulation, the alarmone plays a primarily repressing role during logarithmic growth, in contrast to the observed predominantly inducing role upon growth arrest. For example, the alarmone induces 19 transporters during stationary phase while it represses 12 during early exponential phase. Other genes under negative (p)ppGpp control include 10 conjugal transfer proteins, 5 IS-related transposases and 26 ribosomal proteins. In contrast, nine motility genes were upregulated by (p)ppGpp, such as three of the four basal-body rod proteins (flgBCG), one of the three flagellar switch proteins that interact with the chemotaxis system (fliN) and three chemotaxis proteins (motA, cheW5, cheY1). Therefore, (p)ppGpp has a similarly inducing role on flagellar genes during growth as observed upon growth arrest. A swimming test on 0.2% agar plates corroborates this observation (Figure 6). The (p)ppGpp-deficient mutant showed reduced swimming activity compared to the wild type, a phenotype that could be partially complemented by providing the rsh gene in trans. Hence, the alarmone is required for optimal motility, as was also previously reported for E. coli [19,69].
Figure 6.
Swimming motility test. Swimming halo diameter observed on 0.2% agar TY plates on three consecutive days for wild type, rsh mutant and complemented rsh mutant. The mean values and standard deviation of five biological replicates are shown. The differences over time are statistically significant between the different strains (P < 0.001).
Remarkably, in addition to growth-related genes, many post-translational modification genes were under negative stringent control in the rsh mutant. These comprise numerous chaperones, including the three major ones, tig, dnaK and groEL-groES, involved in folding of new proteins as well as in proper assembly of unfolded proteins and refolding of misfolded proteins generated under stress conditions [74]. DnaK is also involved in chromosomal DNA replication and is part of the osmotic stress response, in addition to osmC [74]. Other upregulated heat shock proteins in the rsh mutant include four peptidases (hslV, lon, traF, htpX2) and one protease (ftsH). Although exponentially growing cells are considered to be less stressed, this increased expression of many heat shock proteins in actively growing cells in the absence of (p)ppGpp might indicate a defect or disruption in protein homeostasis, rather then merely an increase in translational activity. Therefore, this stress response during growth is in accordance with increased stress sensitivity of the rsh mutant as observed previously [30].
Functional analysis of three (p)ppGpp-dependent regulators
To gain insight into the (p)ppGpp-controlled adaptation of R. etli to the stationary phase and diverse stresses as previously reported [30], we selected three different types of previously uncharacterized regulators based on their strongly (p)ppGpp-dependent expression during stationary phase and belonging to the group of transcriptional regulators (ecfG2, phrR) or signal transduction (prkA). The microarray expression patterns of these regulators were confirmed using RT-qPCR (Figure 3). A phenotypical analysis was performed on the corresponding knockout strains to determine the regulators' contribution to the (p)ppGpp-regulated stress response.
Extracytoplasmic function sigma factor PF00052 or EcfG2
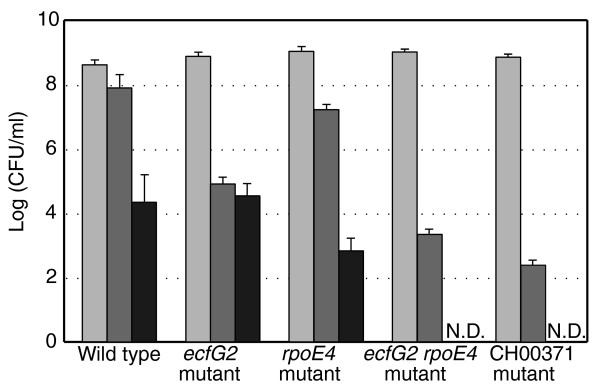
R. etli PF00052 is the most highly upregulated alternative sigma factor during stationary phase in the wild type and upregulated over two-fold compared to the rsh mutant. Analysis of the PF00052 knockout mutant showed decreased survival following oxidative stress, approximately three orders of magnitude lower compared to the wild type (Figure 7). In accordance, the rsh mutant also displays an increased oxidative stress phenotype [30]. In contrast to the rsh mutant, however, the PF00052 mutant does not show a decreased viability after osmotic or heat shock.
Figure 7.
Stress survival of (p)ppGpp-dependent regulator mutants. Survival of the wild type, ecfG2 mutant, rpoE4 mutant, ecfG2-rpoE4 mutant and CH00371 mutant was determined by plating on TY medium after stress treatment and is shown as the mean log(colony forming units (CFU)/ml) of three biological replicates with error bars corresponding to standard deviations. Light gray bars represent control samples incubated at 30°C for the same time period as test samples. Dark gray bars represent samples incubated for 30 minutes in the presence of 10 mM H2O2 at 30°C. Black bars represent samples incubated for 30 minutes at 45°C. N.D., no colonies detected.
Recently, a comprehensive phylogenetic analysis classified the ECF sigma factor family into 43 groups supported by domain architecture, genomic context conservation and potential targets [47]. R. etli contains 18 ECF sigma factors, of which three (rpoE4, sigK and PE00004), in addition to PF00052, were expressed during growth arrest as well. RpoE4 and PF00052 are 42% identical and both belong to theECF15/EcfG group, which exclusively contains α-proteobacterial ECFs, such as EcfG1 of Methylobacterium extorquens, RpoE2 of S. meliloti and SigT of C. crescentus. This group of proposed general stress response sigma factors is characterized by a conserved genomic context that encodes an EcfG-like sigma factor, a cytoplasmic NepR-like anti-sigma factor, a PhyR-like response regulator and a sensor histidine kinase. When the latter perceives a signal, it phosphorylates the regulator, which in turn binds to the anti-sigma factor, thereby releasing the sigma factor and initiating a signal transduction cascade [47]. In case of R. etli rpoE4, an anti-sigma factor (CH03274) and a sensor/regulator (tcrXY or CH03275-CH03276) are found upstream of rpoE4, and tcrX is transcribed in a strongly (p)ppGpp-dependent way (Additional file 3). Though PF00052 is also a member of the EcfG group, a similar genomic context was not recognized. Several other α-proteobacteria also have two EcfG representatives, one that is present in the conserved genomic context while the other is not - for example, S. meliloti 1021 and Agrobacterium tumefaciens str. C58 [47]. A revised ECF nomenclature has been proposed [47] and has recently been adopted for EcfG-like ECF sigma factors in M. extorquens [43] and Bradyrhizobium japonicum [75]. Accordingly, we will hereafter refer to PF00052 as EcfG2.
Due to the high similarity between RpoE4 and EcfG2, we also analyzed the stress sensitivity of an rpoE4 mutant. This revealed a decrease in survival of one order of magnitude upon oxidative and heat stress compared to the wild type (Figure 7). Hence, the rpoE4 mutant exhibits an oxidative stress phenotype less severe than the ecfG2 mutant but, in contrast to the latter, a significant heat stress phenotype. This suggests that RpoE4 and EcfG2 control expression of, at least in part, non-overlapping sets of target genes. To determine if both sigma factors function completely independently in these stress responses, an rpoE4-ecfG2 double mutant was constructed. After heat or oxidative stress treatment, the double mutant showed higher sensitivity than either of the single mutants separately (Figure 7). Moreover, survival of the double mutant is even lower than expected based upon the respective phenotypes of the single mutants, indicating a synergistic effect. Hence, we conclude that both sigma factors have partially overlapping functions in the (p)ppGpp-mediated stress response of R. etli to the tested stress conditions.
R. etli has two heat shock sigma factors that may contribute to the observed heat stress phenotype. RpoH1 was shown to be the main heat shock sigma factor, though a more complete response requires RpoH2 [76]. The promoter region of both sigma factors contains an EcfG-like binding site.
Recently, expression of R. etli rpoH2 was shown to be under positive control of RpoE4, while expression of rpoH1 is not [42]. To determine whether either or both rpoH sigma factors are regulated by EcfG2, their expression was analyzed by qPCR. The expression level of rpoH1 was not altered in the ecfG2 mutant compared to the wild type, while the transcription level of rpoH2 was reduced by approximately 25%. In the rpoE4-ecfG2 double mutant, expression of rpoH2 was reduced over 100-fold, confirming its RpoE4-dependency (data not shown). In contrast, the level of rpoH1 was upregulated over 2.5-fold in the double mutant. This increase is more likely a way to compensate for the impaired heat shock response, rather than a negative control of its expression.
Partial redundancy of EcfG-like sigma factors may occur in other α-proteobacteria as well. In S. meliloti, two EcfG-like proteins are encoded by rpoE2 and rpoE5. RpoE2 was previously reported to regulate many stress response genes during stationary phase. In agreement with our observations, an S. meliloti rpoE2 mutant showed increased sensitivity to H2O2 during stationary phase and rpoE5 is upregulated under heat stress [77,78].
Putative transcriptional regulator CH00371 or PhrR
CH00371 encodes a putative DNA-binding transcriptional regulator of unknown function belonging to the xenobiotic response element family. Expression of this gene is under positive (p)ppGpp-control during all growth phases, although most pronounced upon growth arrest. CH00371 is 80% identical to PhrR, a putative repressor protein of S. meliloti. This regulator was shown to be induced by low pH, hence designated PhrR for pH-regulated [79]. In order to investigate whether CH00371 plays a role in the acid stress response, growth of the R. etli wild type and CH00371 mutant was examined at pH levels ranging from pH 3.5 to 10. No growth difference was observed, either at acidic or basic pH (data not shown). In addition to low pH, oxidative stress agents and heat shock at neutral pH induce phrR in S. meliloti as well. Therefore, survival of the CH00371 mutant was determined under oxidative stress and after heat shock. Compared to the wild type, survival decreased by over four orders of magnitude upon oxidative shock following exposure to hydrogen peroxide (Figure 7). No survival was observed after heat treatment. Moreover, a plate assay demonstrated growth inhibition of a CH00371 mutant on medium containing H2O2 but not in the presence of the superoxide generators menadione and paraquat, nor of the organic hydroperoxide producer cumene hydroperoxide (data not shown).
In order to identify downstream elements in the regulatory cascade mediated by CH00371 during R. etli growth arrest, we carried out qPCR expression analysis of a selection of genes presumed to be associated with the oxidative and heat stress responses (Additional file 6). These candidate target genes were selected based on a literature search and sequence analysis. Several of these genes were downregulated. recA, a key regulator of the SOS response involved in DNA repair, and osmC, an osmotically induced peroxidase, were 70% and 30% downregulated in the CH00371 mutant compared to the wild type, respectively. The superoxide dismutase sodC was downregulated by 20%. Surprisingly, expression of katG was not significantly altered despite the oxidative stress phenotype. However, the expression level of katG was very low. This may indicate that KatG primarily exerts its function upon induction by oxidative shock. In addition, oxidative homeostasis is likely impaired in the CH00371 mutant as 14 genes related to oxidative stress resistance, such as gshB, sufBCD and cysK, showed increased expression of over 25% compared to the wild type. Interestingly, four genes of the EcfG2-RpoE4-regulon (CH00600, CH01778, CH01802 and CH02172) were also downregulated over 25%, possibly contributing to the observed heat stress phenotype.
Furthermore, the expression level of CH00371 was not altered in the ecfG2 mutant and ecfG2 rpoE4 double mutant compared to the wild type, nor vice versa (data not shown). Hence, these novel regulators exert their role in the observed stress phenotypes of the respective mutants independently of each other.
Putative serine kinase CH02817 or PrkA
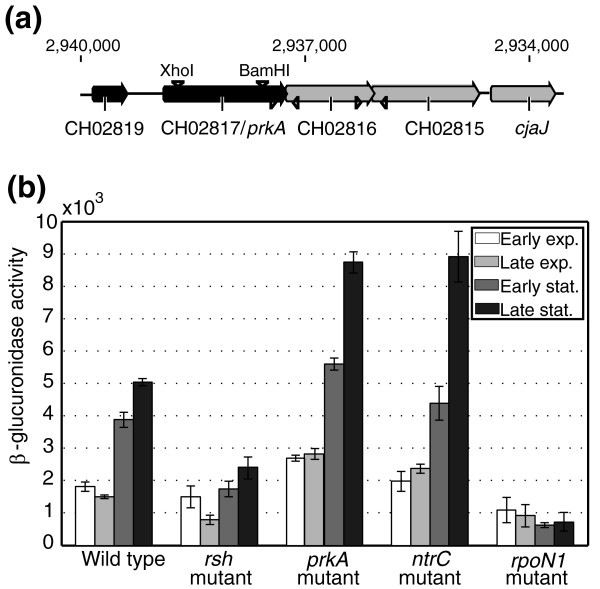
CH02817 or prkA is upregulated over 27-fold in the wild type compared to the rsh mutant during stationary phase, making it the most strongly (p)ppGpp-induced gene detected in our array. This was confirmed by RT-qPCR (Figure 3). PrkA belongs to the PrkA family of serine protein kinases and is highly conserved, with homologs in many eubacteria and archae suggesting a conserved function. E. coli and B. subtilis PrkAs were shown to phosphorylate serine residues of proteins [80,81] and display 66% and 34% identity with the R. etli ortholog, respectively. Protein phosphorylation usually changes the function of the target by modulating its activity, its localization or interaction with other proteins, thereby converting extracellular signals into cellular responses, such as adaptation of the central metabolism, production of secondary metabolites and pathogenicity [82]. Although the specific regulatory function of PrkA remains unknown, the B. subtilis ortholog was shown to be an important inner spore coat protein under control of the developmental sigma factor σE [83]. Furthermore, prkA is part of a highly conserved gene cluster together with the two downstream genes CH02816 and CH02815. This suggests that prkA is likely the first gene of a three-gene operon in R. etli (Figure 8a), which we confirmed by RT-PCR (data not shown). The second operon gene encodes a protein containing a von Willebrand Factor type A domain and the third encodes a SpoVR-like protein. So far, these genes have not been functionally characterized.
Figure 8.
prkA genomic context and expression analysis. (a) R. etli prkA is the first gene of a three-gene operon. Open reading frames are represented by right facing arrows, genomic coordinates are indicated above. Restriction sites for the deletion insertion of the prkA mutant are depicted by downwards facing triangles, and primer sites for RT-PCR used to determine prkA operon structure are depicted by left and right facing triangles. (b) Expression of prkA-gusA transcriptional reporter fusion was monitored in different R. etli mutant backgrounds during growth in AMS succinate. The strains were the wild type R. etli CFN42, rsh mutant, prkA mutant, ntrC mutant and rpoN1 mutant. Expression levels are shown in Miller units and are the means of three biological replicates with error bars representing the standard deviation. Exp., exponential; Stat., stationary.
To determine the function of prkA in R. etli, we constructed a non-polar deletion mutant. A phenotypical analysis of this mutant, including survival during stationary phase, osmotic stress, oxidative stress and heat stress, revealed no clear stress phenotype. Therefore, PrkA does not seem to play a crucial role in the stress response of R. etli. This is rather unexpected given the high level of (p)ppGpp-dependency and strong induction under stress conditions in other organisms. In E. coli and S. Typhimurium, the prkA homolog, annotated as yeaG, was also shown to be highly upregulated by (p)ppGpp upon entry into stationary phase as part of the RpoS regulon [81]. In addition, yeaG is upregulated by Lrp during stationary phase in E. coli [45,46]. Other stress conditions were also reported to induce yeaG expression, such as acid and osmotic stress in E. coli and sublethal concentrations of polymyxin in S. Typhimurium [6].
Additionally, PrkA was postulated to be involved in nitrogen metabolism or the nitrogen starvation response in E. coli based on its potential association with NtrCB and GlnP [46,84]. To examine the transcriptional regulation of prkA in R. etli, we monitored expression of a transcriptional prkA-gusA promoter fusion in various genetic backgrounds (Figure 8b). β-Glucuronidase activity was measured during exponential and stationary phase under the same conditions used for microarray sampling, confirming that the expression of prkA is highly upregulated upon growth arrest in the wild type and positively controlled by (p)ppGpp. Moreover, increased expression of prkA was observed in a prkA mutant, indicating that prkA is negatively autoregulated. Expression of prkA was also shown to be regulated by NtrC and RpoN1. NtrC is a transcriptional regulator involved in nitrogen assimilation and growth in nitrogen-limited conditions, as well as a member of the σN-dependent activator family [85]. RpoN1 codes for the main σN operating under free-living growth conditions in R. etli [86]. In the rpoN1 mutant background, prkA showed an even stronger downregulation than in the rsh mutant during stationary phase, showing prkA transcription to be strongly σN-dependent. Because NtrC is a common activator of σN-dependent genes, a similar downregulation of prkA in the ntrC mutant was expected. However, no downregulation of prkA was observed during growth and early stationary phase in the ntrC mutant. Instead, prkA was highly upregulated during late stationary phase compared to the wild type, suggesting that prkA is under negative control of NtrC.
To further analyze the role of PrkA in cellular metabolism, we compared growth of the wild type and prkA mutant on 384 different nitrogen sources using glucose as the sole carbon source. Even though transcriptional control of prkA expression by rpoN1 suggests an involvement for PrkA in nitrogen metabolism, no growth defects were detected. Therefore, the specific function of this highly conserved protein in the (p)ppGpp regulon remains to be identified.
Conclusions
Analysis of growth phase-specific gene expression of the R. etli wild type and rsh mutant has provided insight into the (p)ppGpp regulon of R. etli, providing the first genome-wide view of the stringent response in an α-proteobacterium. Our results indicate that (p)ppGpp functions as a global regulator, with primarily an inducing role, in the adaptation to a non-growing lifestyle as shown by the extensive differential expression of genes belonging to all functional categories. Moreover, we showed both similarities and differences to its role in E. coli and other bacteria, reflecting the merit of investigating a well-studied regulatory response in species more distantly related to typical model organisms. Surprisingly, even though (p)ppGpp is considered to be a growth-arrest specific messenger, we identified a significant number of (p)ppGpp-dependent genes during early exponential phase as well, suggesting functional relevance of the low basal level of (p)ppGpp during active growth in R. etli. Additionally, the genome-wide transcriptome analysis of a strain deficient in a global regulator, and exhibiting a pleiotropic phenotype, enabled us to identify diverse regulators that control genes associated with a subset of stress phenotypes. The phenotypic analysis of three novel downstream regulators during stationary phase, that is, ecfG2, CH00371, and prkA, allowed us to obtain additional insight into the intricate regulatory role of this stress alarmone (Figure 9). Added detail to the complex picture of (p)ppGpp-dependent regulation of gene expression in R. etli was further provided by identifying several up- and downstream elements in the signal transduction cascades of these regulators. We conclude that (p)ppGpp is situated high up in the hierarchy of cellular gene regulation of R. etli, orchestrating its adaptation to growth stage or extracellular conditions through specific downstream regulators to control expression of a variety of target genes.
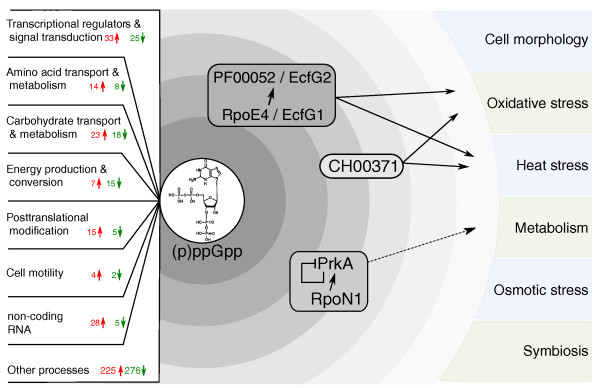
Figure 9.
The (p)ppGpp regulon of R. etli. The extensive impact of (p)ppGpp on gene expression of R. etli is illustrated by the number of up- and downregulated genes grouped according to functional categories. The remaining categories are combined as 'Other processes'. The rsh mutant is unable to synthesize (p)ppGpp and has a pleiotropic phenotype, such as an altered morphology, increased stress sensitivity and impaired symbiosis. As a global regulator, the regulon of (p)ppGpp is multilayered. Further insight into the (p)ppGpp-dependent stress response was obtained by the identification and subsequent characterization of three different regulators that are under strong positive regulation of (p)ppGpp during stationary phase. EcfG2/PF00052 and RpoE4, both ECF sigma factors, are partly functionally redundant for survival under heat stress and oxidative stress. The transcription factor CH00371 is also involved in survival during both heat and oxidative stress. PrkA, a serine kinase, likely plays a role in the (p)ppGpp-dependent adaptation of the cellular metabolism. Its transcription is positively controlled by RpoN1 and negatively autoregulated.
Materials and methods
Bacterial strains and growth conditions
The bacterial strains and plasmids used for this work are listed in Additional file 7. R. etli CFN42 strains were cultured in minimal AMS or complex TY medium at 30°C when used for RNA isolation or stress tests, respectively [29,87]. AMS medium was supplemented with 10 mM NH4Cl and 10 mM succinate unless otherwise indicated. E. coli strains were grown at 37°C in LB medium. In order to study gene expression during different growth phases in AMS medium, samples were taken based on optical density (OD) readings of OD600 = 0.3, OD600 = 0.7, and 6 hours after reaching the maximum optical density (OD)600, representing early exponential, late exponential and stationary phase, respectively [32]. Antibiotics were supplied at the following final concentrations (in μg-1 ml): ampicillin, 100; gentamicin, 50; kanamycin, 25; spectinomycin, 50; nalidixic acid, 15; neomycin, 60; and tetracycline, 1 (E. coli) or 0.1 (R. etli).
Mutant construction
The R. etli CFN42 rsh mutant was constructed by insertion of an SpR cassette, obtained from pHP45ΩSp, as described previously for R. etli CNPAF512 [29]. The ecfG2 and CH00371 mutants were constructed by first amplifying a 2.5-kb and a 1.9-kb fragment, respectively, using Pfx DNA polymerase and primers (CACCGCGGCCGCGGGTTT AAGGGGATAAATT and ACTGGCGGCCGCAAG GGCCGATCGAGATCCAC in the case of ecfG2; CACCGCGGCCGCAGCTGC AGGATCTTATGGGAATA and ACTGGCGGCCGCCGACGACCAGATCCTGAT CGC in the case of CH00371) that carried NotI recognition sites at their 5' ends (shown in italics). These fragments were subsequently cloned into pUC18Not, and a KmR cassette flanked by transcription termination signals, obtained from pHP45ΩKm, was inserted in the HindIII and EcoRV site of CH00371 and ecfG2, respectively. From these plasmids, the corresponding NotI fragments were cloned into the suicide plasmid pJQ200uc1. For construction of the rpoE4-ecfG2 double mutant, an ecfG2::ΩSp suicide construct was obtained as described for the ecfG2 mutant above, replacing the KmR cassette with a SpR cassette.
The non-polar prkA mutant was constructed by amplifying a 3.6-kb fragment using Pfx DNA polymerase and primers (CACCGTTAACTCGACAGGAAAAGGTAG AGC and CACCGTTAACTACTCG TCAAGAAGGAGGCT) that carried HpaI recognition sites at their 5' ends (shown in italics). This fragment was cloned into pCR4Blunt-TOPO (Invitrogen, Carlsbad, CA, USA). A fragment of 1.2 kb was removed from prkA by digesting with BamHI and XhoI (Figure 8a) and the construct was ligated after blunting, creating a deletion in prkA. An HpaI fragment was removed form this construct and cloned into the SmaI site of pJQ200uc1.
Finally, these suicide constructs were used for site-directed mutagenesis of the respective genes following triparental conjugation as described previously [88]. The obtained mutants were verified by Southern blot hybridization.
RNA isolation and cDNA synthesis for microarray detection
RNA was isolated as described previously [32]. Briefly, the RNA content of bacterial cultures was stabilized using a phenol:ethanol solution. Pellets were frozen in liquid nitrogen and stored at -80°C. Total RNA was extracted using the TRIzol Plus RNA Purification kit (Invitrogen). DNA contamination was removed by TURBO DNase (Ambion, Austin, TX, USA)) and afterwards checked by PCR (45 cycles). To increase RNA yields and account for experimental variation, RNA from six different cultures was pooled. RNA integrity was analyzed using Experion RNA StdSens Chips (Biorad, Hercules, CA, USA) before and after precipitation. All samples had an RNA Quality Indicator value of 10. RNA quantity and purity was assessed using the NanoDrop ND-1000. The A260/A280 ratio and A260/A230 ratio of all samples were ≥2.
cDNA was synthesized using random decamers (Ambion) and the SuperScript Double-Stranded cDNA Synthesis Kit (Invitrogen) according to the manufacturer's protocol.
High-density microarray design and data preprocessing
A whole-genome tiling array covering the entire R. etli genome sequence was used (see GEO GPL9409) and the data were analyzed as described previously [32]. Samples were hybridized and scanned by NimbleGen. The data were deposited in the NCBI Gene Expression Omnibus (GEO) and can be accessed through accession numbers [GEO:GSE23961], [GEO:GSM462173], [GEO:GSM462178], [GEO:GSM462180], [GEO:GSM590285], [GEO:GSM590286] and [GEO:GSM590287].
Differentially expressed genes were identified based on a standard deviation cutoff. These genes were considered induced or repressed if the absolute expression ratio was ≥2 (log2 ≥1). This threshold is cogent since most regulatory responses in nature appear to function using low level changes as a kind of energy saving solution [89]. Hierarchical clustering was performed using the software package R.
RT-(q)PCR
Expression levels were determined by RT-qPCR using SYBR Green, as described previously [32]. In short, primers were designed using Primer Express 3.0. Pooled total RNA (2 μg) of each growth condition (early/late exponential phase, stationary phase) was reverse transcribed to single-stranded cDNA using the SuperScript VILO cDNA Synthesis Kit according to the manufacturer's instructions (Invitrogen). DNA contamination of the RNA samples was checked by PCR (45 cycles) before RT. cDNA (40 ng) was used in each reaction. All reactions were performed in triplicate.
The microarray data were validated by determining the expression levels of 14 representative genes: flaCh1, potF, rpsH, flgB, rplR, otsA, aglE, a serine tRNA (CH01348), and genes encoding a chaperonin GroEL (CH00828), the sigma 54 modulation protein (CH00406), a permease protein of the Nod factor ABC transporter family (PD00277), the serine protein kinase prkA/CH02817, the transcriptional regulator CH00371 and the ECF sigma factor ecfG2/PF00052. The log2 ratios of the array data were compared to the log2 ratios of the qPCR results. 16S rRNA was not used as a reference gene as the level of mRNA/rRNA fluctuates during growth and the expression of rRNA is controlled by (p)ppGpp. New reference genes were identified using the geNorm algorithm in order to normalize the qPCR data [90]. Based on the microarray data, five genes were chosen that were relatively stable across all samples and assumed not to be co-regulated. These candidate reference genes were greA, cinRa, tatA, and genes encoding a zinc binding protein (CH00586) and a hypothetical protein (CH01579). 16S rRNA was included for comparison. Using geNorm, we determined repBa2 and tatA to be the most stable reference genes as they have the lowest gene expression stability values M (Figure S3a in Additional file 8). Consequently, a gene expression normalization factor could be calculated for each sample using the most stable genes. By plotting the pairwise variation V between two sequential normalization factors containing an increasing number of genes, we determined that the three best reference genes were an optimal number of reference genes for normalization (Figure S3b in Additional file 8). Although V2/3, the pairwise variation between the normalization factors calculated by the two and three most stable genes, is strictly higher then the proposed 0.15 cutoff value of Vandesompele et al. [90], the difference is very small and the pairwise variation decreases only slightly by taking an additional fourth reference gene. Therefore, the normalization factor would not significantly change if more internal control genes were to be included. Also, the degree of resolution does not require a fourth reference gene.
RT-PCR was performed on cDNA samples (40 ng) of stationary phase to determine the operon organization of prkA. The primers were designed accordingly (Additional file 6). Taq DNA polymerase was used in the PCR reactions (35 cycles).
Construction of prkA-gusA promoter fusion and β-glucuronidase assay
The prkA-gusA reporter fusion was constructed by first amplifying the 400 bp upstream of prkA by PCR using Pfx DNA polymerase and primers (ACTG AAGCTTTCTGCGGTTCGCCTATCGCA and ACTGTCTAGAAGCGCCGGAAG CGTATGATC) that carried a HindIII and XbaI recognition site at their 5' end (shown in italics), respectively. This promoter fragment was cloned into pFAJ1703 after digestion with HindIII and XbaI, thereby flanking the promoterless 5' end of gusA. Quantitative analysis of GusA activity was carried out as described previously [29].
Stress and stationary phase survival
To study stress survival, wild-type and mutant cells from a freshly grown culture on a MM79 agar plate were resuspended in 10 mM MgSO4 at an OD600 of approximately 0.5. For each regulator, two independently constructed mutants were analyzed in order to exclude the involvement of secondary mutations. To test heat stress survival, 1 ml of each sample was incubated at 45°C for 30 minutes. In case of oxidative stress, 0.1 ml of 100 mM H2O2 was added to 0.9 ml of each sample for 30 minutes or 1 hour while for osmotic stress 0.5 ml of 5 M NaCl was added to 0.5 ml of sample. Samples were plated on TY agar containing nalidixic acid using the Eddy Jet spiral plater (IUL Instruments, Barcelona, Spain). Control samples were incubated without the stress agent at 30°C and the colony forming units (CFU) were determined at the same time point as the stressed samples. The total number of CFU per ml was determined after 3 days of incubation at 30°C using the Flash and Go automated colony counter (IUL Instruments). All experiments were repeated at least two times using three independent biological replicates.
To assess long-term survival, pellets of overnight cultures of wild-type and mutant strains were washed and resuspended in 10 mM MgSO4 at an OD600 of 0.5. A volume of 100 ml of AMS medium (10 mM NH4Cl and succinate) was inoculated with 1 ml of cell suspension and incubated at 30°C for 2 weeks. Samples of 1 ml were removed at the indicated time points and subjected to viable cell counts as described above.
Swimming test
To study swimming activity, TY plates containing 0.2% agar were spot inoculated with cultures in exponential phase and incubated at 30°C in a closed container as described previously [91]. Each strain was tested five-fold in two independent experiments. The swimming halo diameter was measured after one, two and three days.
Growth analysis
Biolog Phenotype Microarray panels PM3/6/7/8 were used to test growth on nitrogen sources and PM10 was used to test pH susceptibility (Biolog, Hayward, CA, USA). AMS medium was inoculated (1:1,000) with overnight cultures of R. etli strains, washed and the OD600 was corrected to approximately 0.5. No NH4Cl was added in case of PM3/6/7/8. The Biolog redox indicator dye Mix A was added to the medium (1:100). The microplates were loaded with 100 μl in each well and incubated for 7 days at 30°C. Dye reduction was monitored every 12 h by measuring the OD570 using a Synergy Mx Microplate Reader (BioTek, Winooski, VT, USA).
Abbreviations
CFU: colony forming units; ECF: extracytoplasmic function; IS: insertion sequence; ncRNA: non-coding RNA; OD: optical density; ppGpp: guanosine tetraphosphate; pppGpp: guanosine pentaphosphate; RNAP: RNA polymerase; rrn: ribosomal RNA; RT-qPCR: reverse transcription-quantitative polymerase chain reaction.
Authors' contributions
MV carried out the experiments and bioinformatics analysis. MV, MF, KB, and JM conceived the study and contributed to the interpretation of the data. LC, KE, and KM performed and contributed to the microarray data normalization and processing. MV, MF and JM were involved in drafting the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Figure S1. Growth curve of R. etli CFN42 in AMS medium. (a) Optical density (OD) readings during growth of the wild type and rsh mutant shown in green and red, respectively. The arrows indicate the time points of sampling. (b) Colony forming units (CFU) during growth of the wild type and rsh mutant.
Figure S2. MA plots comparing transcriptome data. Scatter plots of the microarray data that plot the distribution of the log2 intensity ratio (M-value) versus the log2 average intensity (A-value). Differentially expressed genes that are upregulated or downregulated are shown in red or green, respectively. The number of genes with a growth phase or (p)ppGpp-dependent expression profile are indicated by histogram bars at the right of the MA plot. (a) Wild type compared to rsh mutant in stationary phase. (b) Wild type compared to rsh mutant in exponential phase.
Table S1. The differentially expressed genes during stationary phase and exponential phase in the wild type compared to the rsh mutant.
Table S2. The RpoE4-regulated genes according to Martinez-Salazar et al. (2009) that were found to be alarmone-dependent in this study [42].
Table S3. The alarmone-dependent ncRNAs.
Table S4. The RT-qPCR fold changes compared to array fold changes and qPCR primers.
Table S5. The bacterial strains and plasmids used in this study.
Figure S3. RT-qPCR identification of stable endogenous genes. (a) Determining the most stable reference genes using the average expression stability value M of the remaining reference genes during a stepwise exclusion of the least stable internal control gene. The genes are ranked according to increasing expression stability. At the left are the least stable genes and at the right are the most stable ones. (b) Determining the optimal number of reference genes using the pairwise variation V between two sequential normalization factors containing an increasing number of genes with 0.15 as a proposed cutoff value by Vandesompele et al. [90].
Contributor Information
Maarten Vercruysse, Email: maarten.vercruysse@biw.kuleuven.be.
Maarten Fauvart, Email: maarten.fauvart@biw.kuleuven.be.
Ann Jans, Email: ann.jans@biw.kuleuven.be.
Serge Beullens, Email: serge.beulens@biw.kuleuven.be.
Kristien Braeken, Email: kristien.braeken@gmail.com.
Lore Cloots, Email: lore.cloots@biw.kuleuven.be.
Kristof Engelen, Email: kristof.engelen@biw.kuleuven.be.
Kathleen Marchal, Email: kathleen.marchal@biw.kuleuven.be.
Jan Michiels, Email: jan.michiels@biw.kuleuven.be.
Acknowledgements
MV is indebted to the Institute for the Promotion of Innovation through Science and Technology in Flanders (IWT-Flanders). This work was supported by grants from the Research Council of the KU Leuven (GOA/011/2008) and from the Fund for Scientific Research-Flanders (G.0637.06 and G.0412.10).
References
- Fauvart M, Michiels J. Rhizobial secreted proteins as determinants of host specificity in the rhizobium-legume symbiosis. FEMS Microbiol Lett. 2008;285:1–9. doi: 10.1111/j.1574-6968.2008.01254.x. [DOI] [PubMed] [Google Scholar]
- Gibson KE, Kobayashi H, Walker GC. Molecular determinants of a symbiotic chronic infection. Annu Rev Genet. 2008;42:413–441. doi: 10.1146/annurev.genet.42.110807.091427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson-Boivin C, Giraud E, Perret X, Batut J. Establishing nitrogen-fixing symbiosis with legumes: how many rhizobium recipes? Trends Microbiol. 2009;17:458–466. doi: 10.1016/j.tim.2009.07.004. [DOI] [PubMed] [Google Scholar]
- Downie JA. The roles of extracellular proteins, polysaccharides and signals in the interactions of rhizobia with legume roots. FEMS Microbiol Rev. 2010;34:150–170. doi: 10.1111/j.1574-6976.2009.00205.x. [DOI] [PubMed] [Google Scholar]
- Navarro Llorens JM, Tormo A, Martinez-Garcia E. Stationary phase in gram-negative bacteria. FEMS Microbiol Rev. 2010;34:476–495. doi: 10.1111/j.1574-6976.2010.00213.x. [DOI] [PubMed] [Google Scholar]
- Tani TH, Khodursky A, Blumenthal RM, Brown PO, Matthews RG. Adaptation to famine: a family of stationary-phase genes revealed by microarray analysis. Proc Natl Acad Sci USA. 2002;99:13471–13476. doi: 10.1073/pnas.212510999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potrykus K, Cashel M. (p)ppGpp: Still Magical? (*). Annu Rev Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- Chang DE, Smalley DJ, Conway T. Gene expression profiling of Escherichia coli growth transitions: an expanded stringent response model. Mol Microbiol. 2002;45:289–306. doi: 10.1046/j.1365-2958.2002.03001.x. [DOI] [PubMed] [Google Scholar]
- Traxler MF, Chang DE, Conway T. Guanosine 3',5'-bispyrophosphate coordinates global gene expression during glucose-lactose diauxie in Escherichia coli. Proc Natl Acad Sci USA. 2006;103:2374–2379. doi: 10.1073/pnas.0510995103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braeken K, Moris M, Daniels R, Vanderleyden J, Michiels J. New horizons for (p)ppGpp in bacterial and plant physiology. Trends Microbiol. 2006;14:45–54. doi: 10.1016/j.tim.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Wolz C, Geiger T, Goerke C. The synthesis and function of the alarmone (p)ppGpp in firmicutes. Int J Med Microbiol. 2010;300:142–147. doi: 10.1016/j.ijmm.2009.08.017. [DOI] [PubMed] [Google Scholar]
- Magnusson LU, Farewell A, Nystrom T. ppGpp: a global regulator in Escherichia coli. Trends Microbiol. 2005;13:236–242. doi: 10.1016/j.tim.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Srivatsan A, Wang JD. Control of bacterial transcription, translation and replication by (p)ppGpp. Curr Opin Microbiol. 2008;11:100–105. doi: 10.1016/j.mib.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Barker MM, Gaal T, Josaitis CA, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J Mol Biol. 2001;305:673–688. doi: 10.1006/jmbi.2000.4327. [DOI] [PubMed] [Google Scholar]
- Gummesson B, Magnusson LU, Lovmar M, Kvint K, Persson O, Ballesteros M, Farewell A, Nystrom T. Increased RNA polymerase availability directs resources towards growth at the expense of maintenance. EMBO J. 2009;28:2209–2219. doi: 10.1038/emboj.2009.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jishage M, Kvint K, Shingler V, Nystrom T. Regulation of sigma factor competition by the alarmone ppGpp. Genes Dev. 2002;16:1260–1270. doi: 10.1101/gad.227902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo LM, Johansson LU, Solera D, Skarfstad E, Shingler V. The guanosine tetraphosphate (ppGpp) alarmone, DksA and promoter affinity for RNA polymerase in regulation of sigma-dependent transcription. Mol Microbiol. 2006;60:749–764. doi: 10.1111/j.1365-2958.2006.05129.x. [DOI] [PubMed] [Google Scholar]
- Durfee T, Hansen AM, Zhi H, Blattner FR, Jin DJ. Transcription profiling of the stringent response in Escherichia coli. J Bacteriol. 2008;190:1084–1096. doi: 10.1128/JB.01092-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aberg A, Fernandez-Vazquez J, Cabrer-Panes JD, Sanchez A, Balsalobre C. Similar and divergent effects of ppGpp and DksA deficiencies on transcription in Escherichia coli. J Bacteriol. 2009;191:3226–3236. doi: 10.1128/JB.01410-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalebroux ZD, Yagi BF, Sahr T, Buchrieser C, Swanson MS. Distinct roles of ppGpp and DksA in Legionella pneumophila differentiation. Mol Microbiol. 2010;76:200–219. doi: 10.1111/j.1365-2958.2010.07094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean RE, Ireland PM, Jordan JE, Titball RW, Oyston PC. RelA regulates virulence and intracellular survival of Francisella novicida. Microbiology. 2009;155:4104–4113. doi: 10.1099/mic.0.031021-0. [DOI] [PubMed] [Google Scholar]
- Yan X, Zhao C, Budin-Verneuil A, Hartke A, Rince A, Gilmore MS, Auffray Y, Pichereau V. The (p)ppGpp synthetase RelA contributes to stress adaptation and virulence in Enterococcus faecalis V583. Microbiology. 2009;155:3226–3237. doi: 10.1099/mic.0.026146-0. [DOI] [PubMed] [Google Scholar]
- Kazmierczak KM, Wayne KJ, Rechtsteiner A, Winkler ME. Roles of rel in stringent response, global regulation and virulence of serotype 2 Streptococcus pneumoniae D39. Mol Microbiol. 2009;72:590–611. doi: 10.1111/j.1365-2958.2009.06669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalebroux ZD, Svensson SL, Gaynor EC, Swanson MS. ppGpp conjures bacterial virulence. Microbiol Mol Biol Rev. 2010;74:171–199. doi: 10.1128/MMBR.00046-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviv M, Giladi H, Schreiber G, Oppenheim AB, Glaser G. Expression of the genes coding for the Escherichia coli integration host factor are controlled by growth phase, rpoS, ppGpp and by autoregulation. Mol Microbiol. 1994;14:1021–1031. doi: 10.1111/j.1365-2958.1994.tb01336.x. [DOI] [PubMed] [Google Scholar]
- Lemke JJ, Durfee T, Gourse RL. DksA and ppGpp directly regulate transcription of the Escherichia coli flagellar cascade. Mol Microbiol. 2009;74:1368–1379. doi: 10.1111/j.1365-2958.2009.06939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells DH, Long SR. The Sinorhizobium meliloti stringent response affects multiple aspects of symbiosis. Mol Microbiol. 2002;43:1115–1127. doi: 10.1046/j.1365-2958.2002.02826.x. [DOI] [PubMed] [Google Scholar]
- Calderon-Flores A, Du Pont G, Huerta-Saquero A, Merchant-Larios H, Servin-Gonzalez L, Duran S. The stringent response is required for amino acid and nitrate utilization, nod factor regulation, nodulation, and nitrogen fixation in Rhizobium etli. J Bacteriol. 2005;187:5075–5083. doi: 10.1128/JB.187.15.5075-5083.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moris M, Braeken K, Schoeters E, Verreth C, Beullens S, Vanderleyden J, Michiels J. Effective symbiosis between Rhizobium etli and Phaseolus vulgaris requires the alarmone ppGpp. J Bacteriol. 2005;187:5460–5469. doi: 10.1128/JB.187.15.5460-5469.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braeken K, Fauvart M, Vercruysse M, Beullens S, Lambrichts I, Michiels J. Pleiotropic effects of a rel mutation on stress survival of Rhizobium etli CNPAF512. BMC Microbiol. 2008;8:219. doi: 10.1186/1471-2180-8-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez V, Santamaria RI, Bustos P, Hernandez-Gonzalez I, Medrano-Soto A, Moreno-Hagelsieb G, Janga SC, Ramirez MA, Jimenez-Jacinto V, Collado-Vides J, Davila G. The partitioned Rhizobium etli genome: genetic and metabolic redundancy in seven interacting replicons. Proc Natl Acad Sci USA. 2006;103:3834–3839. doi: 10.1073/pnas.0508502103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercruysse M, Fauvart M, Cloots L, Engelen K, Thijs IM, Marchal K, Michiels J. Genome-wide detection of predicted non-coding RNAs in Rhizobium etli expressed during free-living and host-associated growth using a high-resolution tiling array. BMC Genomics. 2010;11:53. doi: 10.1186/1471-2164-11-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traxler MF, Summers SM, Nguyen HT, Zacharia VM, Hightower GA, Smith JT, Conway T. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol. 2008;68:1128–1148. doi: 10.1111/j.1365-2958.2008.06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockmann-Gretza O, Kalinowski J. Global gene expression during stringent response in Corynebacterium glutamicum in presence and absence of the rel gene encoding (p)ppGpp synthase. BMC Genomics. 2006;7:230. doi: 10.1186/1471-2164-7-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesketh A, Chen WJ, Ryding J, Chang S, Bibb M. The global role of ppGpp synthesis in morphological differentiation and antibiotic production in Streptomyces coelicolor A3(2). Genome Biol. 2007;8:R161. doi: 10.1186/gb-2007-8-8-r161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Antonio A, Collado-Vides J. Identifying global regulators in transcriptional regulatory networks in bacteria. Curr Opin Microbiol. 2003;6:482–489. doi: 10.1016/j.mib.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Todd JD, Sawers G, Rodionov DA, Johnston AW. The Rhizobium leguminosarum regulator IrrA affects the transcription of a wide range of genes in response to Fe availability. Mol Genet Genomics. 2006;275:564–577. doi: 10.1007/s00438-006-0115-y. [DOI] [PubMed] [Google Scholar]
- Singleton C, White GF, Todd JD, Marritt SJ, Cheesman MR, Johnston AW, Le Brun NE. Heme-responsive DNA binding by the global iron regulator Irr from Rhizobium leguminosarum. J Biol Chem. 2010;285:16023–16031. doi: 10.1074/jbc.M109.067215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinella D, Albrecht C, Cashel M, D'Ari R. Iron limitation induces SpoT-dependent accumulation of ppGpp in Escherichia coli. Mol Microbiol. 2005;56:958–970. doi: 10.1111/j.1365-2958.2005.04601.x. [DOI] [PubMed] [Google Scholar]
- Miethke M, Westers H, Blom EJ, Kuipers OP, Marahiel MA. Iron starvation triggers the stringent response and induces amino acid biosynthesis for bacillibactin production in Bacillus subtilis. J Bacteriol. 2006;188:8655–8657. doi: 10.1128/JB.01049-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrecht B, Heusdens C, Beullens S, Verreth C, Mulkers E, Proost P, Vanderleyden J, Michiels J. Defence of Rhizobium etli bacteroids against oxidative stress involves a complexly regulated atypical 2-Cys peroxiredoxin. Mol Microbiol. 2005;55:1207–1221. doi: 10.1111/j.1365-2958.2005.04457.x. [DOI] [PubMed] [Google Scholar]
- Martinez-Salazar JM, Salazar E, Encarnacion S, Ramirez-Romero MA, Rivera J. Role of the extracytoplasmic function sigma factor RpoE4 in oxidative and osmotic stress responses in Rhizobium etli. J Bacteriol. 2009;191:4122–4132. doi: 10.1128/JB.01626-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francez-Charlot A, Frunzke J, Reichen C, Ebneter JZ, Gourion B, Vorholt JA. Sigma factor mimicry involved in regulation of general stress response. Proc Natl Acad Sci USA. 2009;106:3467–3472. doi: 10.1073/pnas.0810291106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balleza E, Lopez-Bojorquez LN, Martinez-Antonio A, Resendis-Antonio O, Lozada-Chavez I, Balderas-Martinez YI, Encarnacion S, Collado-Vides J. Regulation by transcription factors in bacteria: beyond description. FEMS Microbiol Rev. 2009;33:133–151. doi: 10.1111/j.1574-6976.2008.00145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibanez-Ruiz M, Robbe-Saule V, Hermant D, Labrude S, Norel F. Identification of RpoS (sigma(S))-regulated genes in Salmonella enterica serovar typhimurium. J Bacteriol. 2000;182:5749–5756. doi: 10.1128/JB.182.20.5749-5756.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H, Polen T, Heuveling J, Wendisch VF, Hengge R. Genome-wide analysis of the general stress response network in Escherichia coli: sigmaS-dependent genes, promoters, and sigma factor selectivity. J Bacteriol. 2005;187:1591–1603. doi: 10.1128/JB.187.5.1591-1603.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staron A, Sofia HJ, Dietrich S, Ulrich LE, Liesegang H, Mascher T. The third pillar of bacterial signal transduction: classification of the extracytoplasmic function (ECF) sigma factor protein family. Mol Microbiol. 2009;74:557–581. doi: 10.1111/j.1365-2958.2009.06870.x. [DOI] [PubMed] [Google Scholar]
- Kanehara K, Ito K, Akiyama Y. YaeL (EcfE) activates the sigma(E) pathway of stress response through a site-2 cleavage of anti-sigma(E), RseA. Genes Dev. 2002;16:2147–2155. doi: 10.1101/gad.1002302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden JD, Ades SE. The extracytoplasmic stress factor, sigmaE, is required to maintain cell envelope integrity in Escherichia coli. PLoS One. 2008;3:e1573. doi: 10.1371/journal.pone.0001573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K, Pfeiffer V, Mika F, Lucchini S, Hinton JC, Vogel J. SigmaE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol Microbiol. 2006;62:1674–1688. doi: 10.1111/j.1365-2958.2006.05524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guisbert E, Rhodius VA, Ahuja N, Witkin E, Gross CA. Hfq modulates the sigmaE-mediated envelope stress response and the sigma32-mediated cytoplasmic stress response in Escherichia coli. J Bacteriol. 2007;189:1963–1973. doi: 10.1128/JB.01243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiercz JP, Hindra, Bobek J, Haiser HJ, Di Berardo C, Tjaden B, Elliot MA. Small non-coding RNAs in Streptomyces coelicolor. Nucleic Acids Res. 2008;36:7240–7251. doi: 10.1093/nar/gkn898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S, Kakeshita H, Nakamura K. Novel small RNA-encoding genes in the intergenic regions of Bacillus subtilis. Gene. 2009;428:2–8. doi: 10.1016/j.gene.2008.09.024. [DOI] [PubMed] [Google Scholar]
- Hsu LM, Zagorski J, Wang Z, Fournier MJ. Escherichia coli 6S RNA gene is part of a dual-function transcription unit. J Bacteriol. 1985;161:1162–1170. doi: 10.1128/jb.161.3.1162-1170.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neusser T, Gildehaus N, Wurm R, Wagner R. Studies on the expression of 6S RNA from E. coli: involvement of regulators important for stress and growth adaptation. Biol Chem. 2008;389:285–297. doi: 10.1515/BC.2008.023. [DOI] [PubMed] [Google Scholar]
- Neusser T, Polen T, Geissen R, Wagner R. Depletion of the non-coding regulatory 6S RNA in E. coli causes a surprising reduction in the expression of the translation machinery. BMC Genomics. 2010;11:165. doi: 10.1186/1471-2164-11-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza-Sanchez F, Gin JG, Hayes CS. Amino acid starvation and colicin D treatment induce A-site mRNA cleavage in Escherichia coli. J Mol Biol. 2008;378:505–519. doi: 10.1016/j.jmb.2008.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner K, Malke H. relA-Independent amino acid starvation response network of Streptococcus pyogenes. J Bacteriol. 2001;183:7354–7364. doi: 10.1128/JB.183.24.7354-7364.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul BJ, Ross W, Gaal T, Gourse RL. rRNA transcription in Escherichia coli. Annu Rev Genet. 2004;38:749–770. doi: 10.1146/annurev.genet.38.072902.091347. [DOI] [PubMed] [Google Scholar]
- Eymann C, Homuth G, Scharf C, Hecker M. Bacillus subtilis functional genomics: global characterization of the stringent response by proteome and transcriptome analysis. J Bacteriol. 2002;184:2500–2520. doi: 10.1128/JB.184.9.2500-2520.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera A, Aris A, Carrio M, Gonzalez-Montalban N, Villaverde A. Lon and ClpP proteases participate in the physiological disintegration of bacterial inclusion bodies. J Biotechnol. 2005;119:163–171. doi: 10.1016/j.jbiotec.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Keiler KC. Physiology of tmRNA: what gets tagged and why?. Curr Opin Microbiol. 2007;10:169–175. doi: 10.1016/j.mib.2007.03.014. [DOI] [PubMed] [Google Scholar]
- Weichart D, Querfurth N, Dreger M, Hengge-Aronis R. Global role for ClpP-containing proteases in stationary-phase adaptation of Escherichia coli. J Bacteriol. 2003;185:115–125. doi: 10.1128/JB.185.1.115-125.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbein AD, Pan YT, Pastuszak I, Carroll D. New insights on trehalose: a multifunctional molecule. Glycobiology. 2003;13:17R–27R. doi: 10.1093/glycob/cwg047. [DOI] [PubMed] [Google Scholar]
- Flechard M, Fontenelle C, Blanco C, Goude R, Ermel G, Trautwetter A. RpoE2 of Sinorhizobium meliloti is necessary for trehalose synthesis and growth in hyperosmotic media. Microbiology. 2010;156:1708–1718. doi: 10.1099/mic.0.034850-0. [DOI] [PubMed] [Google Scholar]
- Suarez R, Wong A, Ramirez M, Barraza A, Orozco Mdel C, Cevallos MA, Lara M, Hernandez G, Iturriaga G. Improvement of drought tolerance and grain yield in common bean by overexpressing trehalose-6-phosphate synthase in rhizobia. Mol Plant Microbe Interact. 2008;21:958–966. doi: 10.1094/MPMI-21-7-0958. [DOI] [PubMed] [Google Scholar]
- Battesti A, Bouveret E. Acyl carrier protein/SpoT interaction, the switch linking SpoT-dependent stress response to fatty acid metabolism. Mol Microbiol. 2006;62:1048–1063. doi: 10.1111/j.1365-2958.2006.05442.x. [DOI] [PubMed] [Google Scholar]
- Bott M, Preisig O, Hennecke H. Genes for a second terminal oxidase in Bradyrhizobium japonicum. Arch Microbiol. 1992;158:335–343. doi: 10.1007/BF00245362. [DOI] [PubMed] [Google Scholar]
- Magnusson LU, Gummesson B, Joksimovic P, Farewell A, Nystrom T. Identical, independent, and opposing roles of ppGpp and DksA in Escherichia coli. J Bacteriol. 2007;189:5193–5202. doi: 10.1128/JB.00330-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevance FF, Hughes KT. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol. 2008;6:455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Ishiguro EE. Temperature-sensitive growth and decreased thermotolerance associated with relA mutations in Escherichia coli. J Bacteriol. 2003;185:5765–5771. doi: 10.1128/JB.185.19.5765-5771.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong T, Kirchhof MG, Schellhorn HE. RpoS regulation of gene expression during exponential growth of Escherichia coli K12. Mol Genet Genomics. 2008;279:267–277. doi: 10.1007/s00438-007-0311-4. [DOI] [PubMed] [Google Scholar]
- Dahl JL, Kraus CN, Boshoff HI, Doan B, Foley K, Avarbock D, Kaplan G, Mizrahi V, Rubin H, Barry CE. The role of RelMtb-mediated adaptation to stationary phase in long-term persistence of Mycobacterium tuberculosis in mice. Proc Natl Acad Sci USA. 2003;100:10026–10031. doi: 10.1073/pnas.1631248100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A, Bukau B, Kramer G. Structure and function of the molecular chaperone Trigger Factor. Biochim Biophys Acta. 2010;1803:650–661. doi: 10.1016/j.bbamcr.2010.01.017. [DOI] [PubMed] [Google Scholar]
- Gourion B, Sulser S, Frunzke J, Francez-Charlot A, Stiefel P, Pessi G, Vorholt JA, Fischer HM. The PhyR-sigma(EcfG) signalling cascade is involved in stress response and symbiotic efficiency in Bradyrhizobium japonicum. Mol Microbiol. 2009;73:291–305. doi: 10.1111/j.1365-2958.2009.06769.x. [DOI] [PubMed] [Google Scholar]
- Martinez-Salazar JM, Sandoval-Calderon M, Guo X, Castillo-Ramirez S, Reyes A, Loza MG, Rivera J, Alvarado-Affantranger X, Sanchez F, Gonzalez V, Davila G, Ramirez-Romero MA. The Rhizobium etli RpoH1 and RpoH2 sigma factors are involved in different stress responses. Microbiology. 2009;155:386–397. doi: 10.1099/mic.0.021428-0. [DOI] [PubMed] [Google Scholar]
- Sauviac L, Philippe H, Phok K, Bruand C. An extracytoplasmic function sigma factor acts as a general stress response regulator in Sinorhizobium meliloti. J Bacteriol. 2007;189:4204–4216. doi: 10.1128/JB.00175-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flechard M, Fontenelle C, Trautwetter A, Ermel G, Blanco C. Sinorhizobium meliloti rpoE2 is necessary for H(2)O(2) stress resistance during the stationary growth phase. FEMS Microbiol Lett. 2009;290:25–31. doi: 10.1111/j.1574-6968.2008.01401.x. [DOI] [PubMed] [Google Scholar]
- Reeve WG, Tiwari RP, Wong CM, Dilworth MJ, Glenn AR. The transcriptional regulator gene phrR in Sinorhizobium meliloti WSM419 is regulated by low pH and other stresses. Microbiology. 1998;144:3335–3342. doi: 10.1099/00221287-144-12-3335. [DOI] [PubMed] [Google Scholar]
- Fischer C, Geourjon C, Bourson C, Deutscher J. Cloning and characterization of the Bacillus subtilis prkA gene encoding a novel serine protein kinase. Gene. 1996;168:55–60. doi: 10.1016/0378-1119(95)00758-X. [DOI] [PubMed] [Google Scholar]
- Tagourti J, Landoulsi A, Richarme G. Cloning, expression, purification and characterization of the stress kinase YeaG from Escherichia coli. Protein Expr Purif. 2008;59:79–85. doi: 10.1016/j.pep.2008.01.005. [DOI] [PubMed] [Google Scholar]
- Masters M, Blakely G, Coulson A, McLennan N, Yerko V, Acord J. Protein folding in Escherichia coli: the chaperonin GroE and its substrates. Res Microbiol. 2009;160:267–277. doi: 10.1016/j.resmic.2009.04.002. [DOI] [PubMed] [Google Scholar]
- Eichenberger P, Jensen ST, Conlon EM, van Ooij C, Silvaggi J, Gonzalez-Pastor JE, Fujita M, Ben-Yehuda S, Stragier P, Liu JS, Losick R. The sigmaE regulon and the identification of additional sporulation genes in Bacillus subtilis. J Mol Biol. 2003;327:945–972. doi: 10.1016/S0022-2836(03)00205-5. [DOI] [PubMed] [Google Scholar]
- Erickson KD, Detweiler CS. The Rcs phosphorelay system is specific to enteric pathogens/commensals and activates ydeI, a gene important for persistent Salmonella infection of mice. Mol Microbiol. 2006;62:883–894. doi: 10.1111/j.1365-2958.2006.05420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres Leon E, Ezkurdia I, Garcia B, Valencia A, Juan D. EcID. A database for the inference of functional interactions in E. coli. Nucleic Acids Res. 2009;37:D629–635. doi: 10.1093/nar/gkn853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michiels J, Moris M, Dombrecht B, Verreth C, Vanderleyden J. Differential regulation of Rhizobium etli rpoN2 gene expression during symbiosis and free-living growth. J Bacteriol. 1998;180:3620–3628. doi: 10.1128/jb.180.14.3620-3628.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michiels J, Vande Broek A, Vanderleyden J. Molecular cloning and nucleotide sequence of the Rhizobium phaseoli recA gene. Mol Gen Genet. 1991;228:486–490. doi: 10.1007/BF00260644. [DOI] [PubMed] [Google Scholar]
- D'Hooghe I, Michiels J, Vlassak K, Verreth C, Waelkens F, Vanderleyden J. Structural and functional analysis of the fixLJ genes of Rhizobium leguminosarum biovar phaseoli CNPAF512. Mol Gen Genet. 1995;249:117–126. doi: 10.1007/BF00290243. [DOI] [PubMed] [Google Scholar]
- Wren JD, Conway T. Meta-analysis of published transcriptional and translational fold changes reveals a preference for low-fold inductions. OMICS. 2006;10:15–27. doi: 10.1089/omi.2006.10.15. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braeken K, Daniels R, Vos K, Fauvart M, Bachaspatimayum D, Vanderleyden J, Michiels J. Genetic determinants of swarming in Rhizobium etli. Microb Ecol. 2008;55:54–64. doi: 10.1007/s00248-007-9250-1. [DOI] [PubMed] [Google Scholar]
- RhizoBase. http://genome.kazusa.or.jp/rhizobase/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Growth curve of R. etli CFN42 in AMS medium. (a) Optical density (OD) readings during growth of the wild type and rsh mutant shown in green and red, respectively. The arrows indicate the time points of sampling. (b) Colony forming units (CFU) during growth of the wild type and rsh mutant.
Figure S2. MA plots comparing transcriptome data. Scatter plots of the microarray data that plot the distribution of the log2 intensity ratio (M-value) versus the log2 average intensity (A-value). Differentially expressed genes that are upregulated or downregulated are shown in red or green, respectively. The number of genes with a growth phase or (p)ppGpp-dependent expression profile are indicated by histogram bars at the right of the MA plot. (a) Wild type compared to rsh mutant in stationary phase. (b) Wild type compared to rsh mutant in exponential phase.
Table S1. The differentially expressed genes during stationary phase and exponential phase in the wild type compared to the rsh mutant.
Table S2. The RpoE4-regulated genes according to Martinez-Salazar et al. (2009) that were found to be alarmone-dependent in this study [42].
Table S3. The alarmone-dependent ncRNAs.
Table S4. The RT-qPCR fold changes compared to array fold changes and qPCR primers.
Table S5. The bacterial strains and plasmids used in this study.
Figure S3. RT-qPCR identification of stable endogenous genes. (a) Determining the most stable reference genes using the average expression stability value M of the remaining reference genes during a stepwise exclusion of the least stable internal control gene. The genes are ranked according to increasing expression stability. At the left are the least stable genes and at the right are the most stable ones. (b) Determining the optimal number of reference genes using the pairwise variation V between two sequential normalization factors containing an increasing number of genes with 0.15 as a proposed cutoff value by Vandesompele et al. [90].