Abstract
We repurposed existing genotypes in DNA biobanks across the Electronic Medical Records and Genomics network to perform a genome-wide association study for primary hypothyroidism, the most common thyroid disease. Electronic selection algorithms incorporating billing codes, laboratory values, text queries, and medication records identified 1317 cases and 5053 controls of European ancestry within five electronic medical records (EMRs); the algorithms' positive predictive values were 92.4% and 98.5% for cases and controls, respectively. Four single-nucleotide polymorphisms (SNPs) in linkage disequilibrium at 9q22 near FOXE1 were associated with hypothyroidism at genome-wide significance, the strongest being rs7850258 (odds ratio [OR] 0.74, p = 3.96 × 10−9). This association was replicated in a set of 263 cases and 1616 controls (OR = 0.60, p = 5.7 × 10−6). A phenome-wide association study (PheWAS) that was performed on this locus with 13,617 individuals and more than 200,000 patient-years of billing data identified associations with additional phenotypes: thyroiditis (OR = 0.58, p = 1.4 × 10−5), nodular (OR = 0.76, p = 3.1 × 10−5) and multinodular (OR = 0.69, p = 3.9 × 10−5) goiters, and thyrotoxicosis (OR = 0.76, p = 1.5 × 10−3), but not Graves disease (OR = 1.03, p = 0.82). Thyroid cancer, previously associated with this locus, was not significantly associated in the PheWAS (OR = 1.29, p = 0.09). The strongest association in the PheWAS was hypothyroidism (OR = 0.76, p = 2.7 × 10−13), which had an odds ratio that was nearly identical to that of the curated case-control population in the primary analysis, providing further validation of the PheWAS method. Our findings indicate that EMR-linked genomic data could allow discovery of genes associated with many diseases without additional genotyping cost.
Introduction
Since 2005, more than 900 genome-wide association studies (GWASs) have identified genomic variants associated with more than 200 diseases and traits.1 Although genotyping costs are decreasing, GWASs targeting a single disease or trait are expensive, and recruitment of the necessary number of patients can be challenging. Recently, electronic medical record (EMR)-linked DNA biobanks have allowed researchers to study the genetic basis of disease by using phenotypes derived solely from information contained within the EMR.2–8 Most EMRs contain a dense longitudinal record of a patient's health conditions, the evolution of those conditions, and responses to treatments. EMR-linked DNA biobanks might allow genetic analysis of many phenotypes with little to no incremental genotyping cost because patients often have more than one clinically apparent disease. In this study, we explored the hypothesis that genetic data from five previously conducted EMR-based GWASs could be reused in a study of a different phenotype of broad public health importance, primary hypothyroidism (PH).
PH is the most common thyroid disorder; it affects 1%–5% of the population,9 and up to 12% of the elderly express subclinical hypothyroidism.10 The majority of cases result from chronic lymphocytic thyroiditis, or Hashimoto thyroiditis (HT [MIM 140300]). Treatment involves thyroid hormone replacement and lifelong monitoring of serum hormone levels. A number of rare mutations have been shown to result in congenital hypothyroidism, which is readily detectible through newborn screening. These mutations can cause central hypothyroidism (e.g., TSHB [MIM 188540]) or primary hypothyroidism due to thyroid dysgenesis (e.g., TSHR [MIM 603372]), alterations in thyroid transcription factors (NKX2-1 [MIM 600635], FOXE1 [MIM 602617], and PAX8 [MIM 167415]), or dyshormonogenesis (NIS [MIM 601843], TG [MIM 188450], DUOX2 [MIM 606759], DUOXA2 [MIM 612772], SLC26A4 [MIM 605646], and DEHAL1[MIM 612025]).11–13 There is evidence of a genetic component in autoimmune thyroid disease, including PH;14,15 candidate-gene analysis and linkage studies16–19suggest that loci contributing to the pathogenesis of PH include CTLA4 (MIM 123890), PTPN22 (MIM 600716), and the thyroglobulin (TG) gene. A GWAS of levels of thyroid-stimulating hormone (TSH) in euthyroid individuals has identified associations at 1p36.13,20 PDE8B21,22(MIM 603390). and to a lesser degree, FOXE1.22,23
Implementation of EMRs can lead to a higher quality of care, reduced cost, and improved adherence to guidelines.24–26 More recently, investigators have used EMRs as a longitudinal resource for clinical and genomic research.27 Significant challenges inherent in EMR-based research include the accurate identification of cases and controls from a data source that was not designed for such a task. Methods of achieving high precision have been developed and include cohort-selection algorithms that identify subjects via a combination of billing codes, records of medication prescription, laboratory or report data, and use of informatics techniques such as natural language processing to search for biomedical concepts in unstructured clinical documentation.2,28,29 The portability of such algorithms from one EMR system to another has not been systematically evaluated and remains a potential obstacle to secondary use of EMR data,30 a goal for the recently released Meaningful Use criteria of the Health Information Technology for Economic and Clinical Health (HITECH) Act.
We sought to develop and implement a transportable EMR-based algorithm to identify hypothyroidism cases and controls. This study was performed in the Electronic Medical Records and Genomics (eMERGE) Network, a project that is sponsored by the National Human Genome Research Institute and which involves five institutions (Group Health Cooperative [GHC], Marshfield Clinic [MFC], Mayo Clinic [MC], Northwestern University [NU], and Vanderbilt University Medical Center [VU]) that each have DNA biorepositories linked to their EMRs.3 Using genotype data derived from five prior GWASs performed in five institutions, we conducted a GWAS to identify genetic variants associated with PH. We then performed a phenome-wide association study (PheWAS) on a single-nucleotide polymorphism (SNP) associated with PH to investigate the pleiotropy of this region. The PheWAS method uses custom case and control definitions to perform an unbiased scan of diseases represented in billing codes accrued in the medical record; we have previously demonstrated its validity through rediscovery of known associations.4
Subjects and Methods
eMERGE Network Sites
As of mid-2011, the eMERGE Network consisted of five institutions (noted above) that each had DNA biorepositories linked to their EMRs. Each EMR is linked to a DNA biobank. Details of these biobanks have been published elsewhere.31,32 Each site has investigated at least one primary phenotype (Table 1) by performing a GWAS after implementing and validating computer selection logic to identify cases and controls from available EMR data. After genotyping for these primary GWASs was complete, each site identified individuals with the network-wide phenotype PH from within their genotyped population.
Table 1.
Evaluation of Primary Hypothyroidism Algorithm at the Five eMERGE Sites
| Site | Primary Phenotype | Total Genotyped Subjects |
Primary Hypothyroidism |
|||
|---|---|---|---|---|---|---|
| Cases | Controls | Case PPV (%) | Control PPV (%) | |||
| Group Health | dementia | 2532 | 397 | 1,160 | 98 | 100 |
| Marshfield | cataracts | 4113 | 514 | 1,187 | 91 | 100 |
| Mayo Clinic | peripheral arterial disease | 3043 | 233 | 1,884 | 82 | 96 |
| Northwestern | type 2 diabetes | 1217 | 92 | 470 | 98 | 100 |
| Vanderbilt | normal cardiac conduction | 2712 | 81 | 352 | 98 | 100 |
| All sites | 13,617 | 1317 | 5053 | 92.4a | 98.5a | |
Genotype counts represent all subjects who were found by the hypothyroidism algorithms at each site and who were genotyped. Counts are limited to those classified as “white” in the electronic medical record of each site. PPV = positive predictive value.
Average weighted for number of samples contributed to the total.
Five different EMR systems are used across eMERGE; three sites (MFC, MC, and VU) use custom-developed EMR systems. NU and GHC use EpicCare (Epic Systems Corporation, Verona, WI). NU also uses Cerner PowerChart (Cerner Corporation, Kansas City, MO). Each eMERGE site maintains research data warehouses to make EMR data accessible for clinical and genetic research. Details of these systems have been published previously.3,31–33
Evaluation and Development of Phenotype-Selection Logic
The algorithm identifying cases and controls for PH was initially created and iteratively refined at VU and then deployed at the other four eMERGE sites. At VU, two physicians not associated with algorithm development reviewed algorithm results with access to the entire deidentified clinical records of individuals meeting selection criteria. The results of the manual classification were then used for improving the algorithms, and the procedure was iterated until the positive predictive value reached the predesignated target of ≥ 95% for a random selection of unreviewed algorithm-determined cases and controls.
The final algorithm required the presence of a thyroid replacement medication for at least three months and at least one International Classification of Disease, 9th edition code for hypothyroidism, or abnormal thyroid function study, defined as a study showing the laboratory value of thyroid stimulating hormone (TSH) to be greater than 5 μIU/ml or showing the laboratory value of abnormal free thyroxine (FT4) to be less than 0.5 ng/dl. Secondary causes of hypothyroidism (e.g., postsurgical hypothyroidism or medication-induced hypothyroidism) and other thyroid conditions (e.g., Graves disease [GRD; MIM 275000], pregnancy-related hypothyroidism, or thyroid cancer) were excluded by queries for medications, billing codes, and radiology tests (so that contrast exposure was excluded). Control subjects had at least one normal TSH value (defined as between 0.5 and 5 μIU/ml) and had no occurrences of hypothyroidism billing codes, abnormal thyroid function tests, or thyroid replacement medications. Both cases and controls excluded patients who had received medications potentially altering thyroid function (such medications include phenytoin, lithium, methimazole, and propylthiouracil). The complete description of the final algorithm is available online (see Web Resources). So that algorithm accuracy could be determined, a randomly selected subset of cases and controls was manually reviewed at each site by trained chart abstractors or physicians, each of whom was blinded to the algorithm's case or control determination and had full access to the EMR data. The review included a total of 300 cases and 300 controls. Reviewers were instructed to mark as false positives individuals designated as having subclinical hypothyroidism only and not overt hypothyroidism.
Genotyping
Genotyping was performed at the Center for Genotyping and Analysis at the Broad Institute (two sites) and the Center for Inherited Disease Research at Johns Hopkins University (three sites) with the Human660W-Quadv1_A BeadChip, consisting of 561,490 SNPs and 95,876 intensity-only probes on a total of 13,617 subjects of European American ancestry, as designated in the EMRs.
Data were cleaned with the quality-control (QC) pipeline developed by the eMERGE Genomics Working Group.34 This process includes evaluation of sample and marker call rate, gender mismatch and anomalies, duplicate and HapMap concordance, batch effects, Hardy-Weinberg equilibrium (HWE), sample relatedness, and population stratification (implemented with STRUCTURE35 and EIGENSTRAT36). Relatedness was determined on the basis of identity by descent (IBD) estimates generated from the genome-wide genotype data in PLINK. All study sites had individuals with an IBD estimate greater than 0.0625. We also identified two inter-site related pairs from Mayo and Marshfield. In all cases of suspected relatedness, one individual from each related pair and the child from identified trios were removed from the analysis.
After QC, 522,164 SNPs were used for analysis on the basis of the following QC criteria: SNP call rate > 99%, sample call rate > 99%, minor allele frequency > 0.01, 99.99% concordance rate in duplicates, unrelated samples only, and individuals of European descent only (determination of European descent was based on STRUCTURE analysis showing a >90% probability of being in the CEU [Utah residents with ancestry from northern and western Europe] cluster). We flagged all markers with HWE p < 1 × 10−4 for further evaluation after analysis with standard criteria.34 The QC and data analysis were performed with a combination of PLINK,37 PLATO,38 and the R statistical package.
Statistical Analysis
We evaluated the positive predictive value of case and control determination for the automated algorithm. The positive predictive value was calculated as the number of true positive cases (or controls), as determined by human expert review, divided by the total number of cases (or controls) selected by the algorithm.
Genetic analyses were limited to subjects of European American ancestry (as determined by genetic ancestry) given the relatively few individuals identified not to be of European descent (28 cases and 146 controls). Single-locus tests of association were performed via logistic regression in PLINK under the assumption of an additive genetic model. The model was adjusted for birth decade, sex, and site of ascertainment. Birth decade was chosen instead of age of diagnosis because the latter is usually not available within EMR records and the former also allows adjustment for possible differences in iodine supplementation over time. A second model included birth decade, sex, site of ascertainment, and the first principal component from EIGENSTRAT. The genomic inflation factor for data in both models was 1.00. Linkage disequilibrium was calculated and plotted with LocusZoom.39 Tests of heterogeneity were performed with METAL.40 Analyses involving 1000 genomes data were carried out with SNP Annotation and Proxy Search (SNAP) in the CEU population panel.41
In addition to an unmatched analysis, we performed a genome-wide association study of cases matched to controls.
In the matched analysis, controls were matched to cases by site, sex, genotypic ancestry designation, and birth decade. We used principal components one and two to match by genetic ancestry with EIGENSTRAT; these values were allowed to vary slightly between matches. To increase the number of controls matched to cases, additional controls were added if the birth decade was within one decade. For unrelated cases and controls, the control-to-case matching ratio was at least 3:1. Cases were stratified with their related controls, where appropriate, and matched with additional nonrelated controls, when necessary, so that a 3:1 control-to-case ratio was achieved. Conditional logistic regression was performed for each SNP under the assumption of an additive genetic model adjusted for birth decade and two principal components.
Replication
Variants reaching genome-wide significance (p < 5 × 10−8) in the discovery set were tested for association with PH in the Mayo Genome Consortia (MayoGC), which includes Mayo Clinic patients with both EMR and GWAS data derived from prior studies on Illumina HumanHap550, Human610-Quad, and Human660W-Quad platforms. Currently, the cohort includes 6508 patients from three studies, the eMERGE peripheral arterial disease cases and controls, case and control patients from a study of venous thromboembolism, and control patients from a study of pancreatic cancer.42,43 Participants in the venous thromboembolism study were genotyped with the Illumina Human660W-Quad platform. Participants in the pancreatic cancer study were genotyped with Illumina HumanHap550 and the Human 610-Quad chips. The PH algorithm was used for identification of cases and controls from the non-eMERGE subjects (n = 3110). The top associations in the initial GWAS were analyzed in the replication set via PLINK with logistic regression under the assumption of an additive genetic model adjusted for age and sex. MayoGC samples used for the replication analysis had distinct medical-record numbers from the primary analysis, and IBD analysis within PLINK was used for removal of any first-degree relatives (or twins or replicate samples) of those individuals used within the primary eMERGE analysis.
Phenome-wide Association Study
After defining a region of interest in GWAS, we used all individuals of European Ancestry in the eMERGE data set (N = 13,617) to investigate for possible pleiotropy due to this locus. To define diseases, we queried all International Classification of Disease (ICD), 9th edition, codes from the respective EMRs from the five eMERGE sites. The PheWAS software uses these ICD codes to classify each person as having one of 957 possible clinical phenotypes (typically representing diseases). Each phenotype is treated independently such that one person can have both the general diagnosis of hypothyroidism as well as the specific diagnosis of Hashimoto's thyroiditis, presumably as the cause of their hypothyroidism, if both are supported by the individual's billing record. For each disease, the PheWAS code defines relevant control groups for each disease or finding, such that patients with related diseases do not serve as controls for that disease (e.g., a patient with psoriatic arthritis cannot serve as a control for an analysis of rheumatoid arthritis). Analysis of each phenotype then proceeds using a pairwise analysis of all case and control groups for each SNP. We have observed that positive predictive values increase when codes are present more than once in the EMR, and here we required each case to have at least two ICD codes in a PheWAS case group. We also did not analyze phenotypes occurring in less than 20 patients (a prevalence of 0.15% in the data set). Controls were excluded if they had any ICD codes in the PheWAS control exclusion ranges. After generation of case and control groups for each of the PheWAS phenotypes, association analyses were performed with PLINK using logistic regression adjusted for age, sex, and the first three principal component analyses as calculated by EIGENSTRAT. To determine if PheWAS-detected disease associations were the result of co-occurrence with hypothyroidism or a result of pleiotropy, we then performed a PheWAS adjusted for diagnosis of hypothyroidism (defined by PheWAS code) as well as age, sex, and the first three principal components.
Results
Identification of subjects with Primary Hypothyroidism
Table 1 presents the positive predictive value of the PH phenotype algorithm at each of the five sites. A total of 13,617 subjects of European American ancestry were genotyped for one of five primary eMERGE GWAS. The PH algorithm identified 1,317 cases and 5,053 controls from these samples. The remaining 7,247 individuals with GWAS data across eMERGE were excluded because either they met an exclusion criterion (e.g., on medications such as amiodarone) or there was insufficient evidence that they qualified as a control (e.g., lacking a TSH laboratory test or insufficient visit data). The average positive predictive value, weighted by site sample size, was 92.4% for cases and 98.5% for controls. Positive predictive values exceeded 90% for controls at all sites and for cases in four of five sites. Cases were predominantly women (73%) with median birth decade in the 1930s (Table 2). All patient populations averaged more than a decade of data within their EMRs. Figure S1 presents a schematic of the algorithm.
Table 2.
Characteristics of Hypothyroidism Cases and Controls
|
Primary Analysis |
Replication Set |
||||
|---|---|---|---|---|---|
| Cases (n = 1317) | Controls (n = 5053) | Non-TSH Controls (n = 5632) | Cases (n = 263) | Controls (n = 1616) | |
| Median birth decade | 1930 | 1930 | 1940 | 1930 | 1940 |
| Age (yr)a | 68.7 ± 14.0 | 60.7 ± 12.5 | 52.9 ± 14.6 | 58.9 ± 14.2 | 53.9 ± 13.5 |
| Female (%) | 73.0 | 48.3 | 43.8 | 78.7 | 43.9 |
| Follow-up (yr)b | 19.3 ± 9.4 | 15.5 ± 9.5 | 13.0 ± 11.0 | 11.9 ± 4.9 | 10.1 ± 5.6 |
Plus-minus values are mean ± SD.
Age is calculated as the first age matching case definition (e.g., a qualifying billing code, laboratory value, or medication) or, for controls, first billing code.
Follow-up entries were calculated as the number of years the patients was observed by billing codes.
Genome-wide Analysis for Primary Hypothyroidism
The analysis identified four SNPs associated with hypothyroidism at genome-wide significance after adjustment for birth decade, sex, and study site: rs7850258, rs965513, rs925489, and rs10759944 (Figure 1, Table 3, Table S1). The minor alleles of all four SNPs are underrepresented among cases compared with controls at p < 8.2x10−9 and odds ratio (OR) 0.74 (95% confidence interval 0.67-0.82). Analysis including the first principal component did not alter the results. The four SNPs located on chromosome 9 are in strong pair-wise linkage disequilibrium (r2 > 0.98) with one another and 58-71 kb from the nearest gene, FOXE1 (Figure 2). Analysis of the pilot low coverage CEU data from 1000 genomes found the four FOXE1 SNPs were not in linkage disequilibrium (r2 > 0.80) with any coding region variants (Figure S2).
Figure 1.
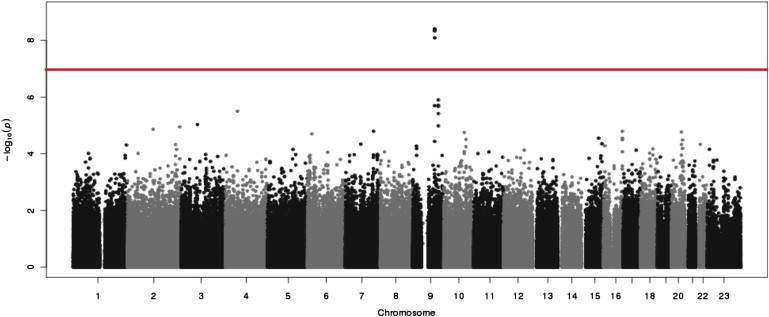
Genome-wide Association Analysis of Individuals with Primary Hypothyroidism versus Controls
SNP tests of association (logistic regression) under the assumption of an additive genetic model adjusted for sex, birth decade, and study site; the tests incorporated 522,164 SNPs. The red horizontal line indicates p = 5 × 10−8, the threshold for genome-wide significance.
Table 3.
SNPs Associated with Hypothyroidism at p < 10−6 with Either Primary Controls or Non-TSH Controls
| SNP | Chromosome | Position | Minor Allele | Nearest Gene |
Minor Allele Frequency |
Cases versus Controls |
Cases versus Non-TSH Controls |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Controls | Non-TSH Controls | OR (95% CI) | p Value | OR (95% CI) | p Value | |||||
| rs7850258 | 9 | 99588834 | A | FOXE1 | 0.285 | 0.348 | 0.3452 | 0.74 (0.67–0.82) | 3.96 × 10−9 | 0.74 (0.67–0.82) | 8.17 × 10−9 |
| rs965513 | 9 | 99595930 | A | FOXE1 | 0.286 | 0.348 | 0.3456 | 0.74 (0.67–0.82) | 4.19 × 10−9 | 0.74 (0.67–0.82) | 1.15 × 10−8 |
| rs925489 | 9 | 99586421 | C | FOXE1 | 0.286 | 0.348 | 0.3452 | 0.74 (0.67–0.82) | 4.68 × 10−9 | 0.74 (0.67–0.82) | 1.17 × 10−8 |
| rs10759944 | 9 | 99596793 | A | FOXE1 | 0.286 | 0.347 | 0.3445 | 0.75 (0.68-0.83) | 8.19 × 10−9 | 0.74 (0.67–0.82) | 1.50 × 10−8 |
| rs4979402 | 9 | 116262496 | G | DFNB31 | 0.288 | 0.247 | 0.2518 | 1.29 (1.16–1.42) | 1.23 × 10−6 | 1.17 (1.05–1.30) | 0.0035 |
| rs4979397 | 9 | 116259709 | T | DFNB31 | 0.286 | 0.246 | 0.2500 | 1.28 (1.16–1.42) | 1.91 × 10−6 | 1.17 (1.06–1.31) | 0.0033 |
| rs1877432 | 9 | 99583701 | A | 9q22.3; near FOXE1 | 0.437 | 0.382 | 0.3885 | 1.25 (1.14–1.37) | 1.99 × 10−6 | 1.28 (1.16–1.41) | 4.29 × 10−7 |
| rs1408528 | 9 | 116260594 | C | DFNB31 | 0.286 | 0.246 | 0.2507 | 1.28 (1.16–1.42) | 2.07 × 10−6 | 1.17 (1.05–1.30) | 0.0039 |
| rs17827152 | 4 | 55173443 | A | 4q12; closest to KIT | 0.263 | 0.219 | 0.2348 | 1.28 (1.15–1.42) | 3.20 × 10−6 | 1.16 (1.04–1.29) | 0.0084 |
| rs1535971 | 9 | 116269221 | T | DFNB31 | 0.299 | 0.260 | 0.2629 | 1.27 (1.15-1.40) | 3.65 × 10−6 | 1.17 (1.05–1.30) | 0.0035 |
| rs17043990 | 3 | 72963160 | C | SHQ1 | 0.0091 | 0.0024 | 0.0050 | 3.71 (2.08–6.60) | 8.34 × 10−6 | 2.47 (1.44–4.25) | 0.0011 |
| rs4733792 | 8 | 128909458 | T | PVT1 | 0.4445 | 0.4077 | 0.3884 | 1.18 (1.08,1.30) | 2.35 × 10−4 | 1.26 (1.14–1.39) | 2.40x10−6 |
| rs4733789 | 8 | 128903585 | C | PVT1 | 0.4461 | 0.4112 | 0.391 | 1.17 (1.07, 1.28) | 5.51 × 10−4 | 1.26 (1.14–1.38) | 3.01x10−6 |
| rs17280788 | X | 9638573 | T | TBL1X | 0.2612 | 0.2382 | 0.2133 | 1.14 (1.02–1.28) | 0.019 | 1.35 (1.20–1.52) | 9.94x10−7 |
| rs1791303 | 18 | 55606206 | T | CCBE1 | 0.3651 | 0.4068 | 0.4111 | 0.83 (0.75, 0.91) | 6.70 × 10−4 | 0.79 (0.72–0.87) | 2.13x10−6 |
| rs4940904 | 18 | 55606041 | T | CCBE1 | 0.3662 | 0.4071 | 0.4112 | 0.83 (0.76,0.91) | 1.10 × 10−4 | 0.79 (0.72–0.88) | 2.93x10−6 |
Analyses performed with logistic regression adjusted for birth decade, sex, and site of ascertainment. DFNB31 SNPs are in the gene. FOXE1 SNPs are from 58–71 kb to the gene. The DIRAS2 SNP is in the gene. SHQ1 (MIM 602322) SNPs are in the gene.
Figure 2.
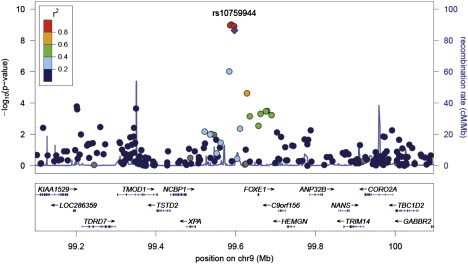
Regional Associations Near FOXE1
To further investigate these genome-wide significant results, we examined the tests of association within each of the five sites. All four SNPs near FOXE1 were associated with PH at p < 0.03 with similar effect sizes and direction (OR 0.60-0.81, Table 4). The associations with PH were strongest within the Marshfield Clinic Personalized Medicine Research Project set, which had the largest case sample size (n = 514) among the study sites. There was no evidence of heterogeneity across the study sites for these four SNPs (p = 0.9).
Table 4.
Association of SNPs near FOXE1 with Hypothyroidism in Each eMERGE Site and Replication Set
| SNP |
eMERGE GWAS Sites |
Replication Set |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
All Sites (n = 1317/5053)a |
GHC (n = 233/1884) |
Marshfield (n = 514/1187) |
Mayo (n = 233/1884) |
Northwestern (n = 92/470) |
Vanderbilt (n = 81/352) |
n = 263/1616 |
||||||||
| Odds Ratio (95% CI) | p Value | Odds ratio (95% CI) | p Value | Odds Ratio (95% CI) | p Value | Odds Ratio (95% CI) | p Value | Odds Ratio (95% CI) | P-Value | Odds Ratio (95% CI) | p Value | Odds Ratio (95% CI) | p Value | |
| rs7850258 | 0.74 (0.67–0.82) | 3.93 × 10−9 | 0.81 (0.68–0.96) | 0.02 | 0.74 (0.62–0.88) | 6.24 × 10−4 | 0.78 (0.63–0.97) | 0.02 | 0.64 (0.44–0.92) | 0.02 | 0.60 (0.40–0.91) | 0.02 | 0.60 (0.48–0.74) | 5.7 × 10−6 |
| rs965513 | 0.74 (0.67–.082) | 4.1510−9 | 0.81 (0.68–0.96) | 0.02 | 0.74 (0.62–0.88) | 6.24 × 10−4 | 0.77 (0.62–0.96) | 0.02 | 0.64 (0.44–0.92) | 0.02 | 0.60 (0.40–0.91) | 0.02 | 0.59 (0.47–0.74) | 5.8 × 10−6 |
| rs925489 | 0.74 (0.67–.082) | 4.64 × 10−9 | 0.81 (0.67–0.95) | 0.01 | 0.75 (0.63–0.89) | 8.71 × 10−4 | 0.78 (0.62–0.96) | 0.02 | 0.64 (0.44–0.92) | 0.02 | 0.60 (0.40–0.91) | 0.02 | 0.59 (0.46–0.74) | 1.1 × 10−5 |
| rs10759944 | 0.75 (0.68–.083) | 8.13 × 10−9 | 0.81 (0.68–0.97) | 0.02 | 0.75 (0.63–0.89) | 9.11 × 10−4 | 0.78 (0.63–0.97) | 0.02 | 0.64 (0.44–0.92) | 0.02 | 0.60 (0.40–0.91) | 0.02 | 0.60 (0.47–0.75) | 7.6 × 10−6 |
Counts represent number of cases/number of controls.
The replication population contained 263 cases and 1616 controls (Table 2). Analysis of the four genome-wide significant variants near FOXE1 replicated the most significant findings in the discovery data set with p < 1.1x10−5 (Table 4).
To assess the possibility of ascertainment bias, i.e., the hypothesis that the physician ordering of a TSH laboratory test may bias toward patients with other autoimmune diseases, we also compared the PH cases to patients who had no evidence of thyroid disease or thyroid-active medications, but did not have TSH testing performed (“non-TSH controls”; n = 5,643 of the 7,247 patients excluded from the primary analysis). GWAS, adjusted for age and sex, of PH cases to non-TSH controls revealed results consistent in direction and magnitude to those of our primary analysis. Results are presented in Table 3 and Figure S3. The top four associated SNPs were identical between both analyses and also reached genome-wide significance (rs7850258, OR = 0.74, p = 8.2x10−9). These data argue against a role for ascertainment bias.
In addition to the genome-wide significant association identified for SNPs along chromosome 9 near FOXE1, we also identified several SNPs in the same chromosome associated with PH near genome-wide significance. Specifically, SNPs rs4979402, rs4979397, rs1408528, and rs1535971 are associated with PH at p < 3.7x10−6 (Table 3) and are located in DFNB31 [MIM 607928]. DFNB31 is ∼16.6Mb from FOXE1 and not in linkage disequilibrium with the FOXE1 variants identified in this study (r2 < 3x10−4); thus, these associations potentially represent an independent locus associated with PH. However, these regions were not as strongly associated with hypothyroidism when compared with the non-TSH control group. The analysis to non-TSH controls identified additional areas of possible significance near PVT1 [MIM 165140] (rs4733792), near TBL1X [MIM 300196] (rs17280788), and within CCBE1 [MIM 612753] (rs1791303) (Table 3). A genome-wide analysis conditioned on rs925489 demonstrated that each of the loci in Table 3 are independent of the FOXE1 variants with similar P values and effects as those presented in Table 3.
The matched GWAS used 1314 cases and 3150 controls. Of the 4464 subjects, 4185 (94%) were unrelated. The results of the matched analysis were essentially identical to the primary analysis except that significance levels tended to be slightly lower (p < 2.6x10−8 for the top association, Figures S4 and S5, Tables S2–S4), likely due to reduced sample size.
Phenome-wide association study of rs965513
Once associations with PH were established, we conducted a PheWAS on rs965513 near FOXE1 to investigate for other phenotypes potentially associated with this locus. We selected rs965513 because it was strongly associated with PH in our analysis and had also previously been associated with an increased risk of thyroid cancer in individuals of European descent.23,44 For this analysis, we combined all self-identified European American individuals from the five eMERGE sites (n = 13,617). Subjects had a total of 5,964,605 billing codes accrued over a total of 217,619 patient-years. A total of 866 phenotypes from more than 20 cases were analyzed, yielding a Bonferroni correction of p = 0.05/866 = 5.8 × 10−5; this correction is probably overly stringent given that many phenotypes are related to each other. Figure 3 and Table 5 show phenotypes associated with rs965513, which included acquired hypothyroidism (OR = 0.76, p = 2.7 × 10−13), thyroiditis (OR = 0.58, p = 1.4 × 10−5), nodular (OR = 0.76, p = 3.1 × 10−5), and multinodular (OR = 0.69, p = 3.9 × 10−5) goiters, nutritional deficiency anemias (OR = 1.41, p = 3.7 × 10−5), and thyrotoxicosis (OR = 0.76, p = 1.5 × 10−3). The locus was also associated with chronic lymphocytic thyroiditis (OR = 0.58, p = 2.5 × 10−4), a subset of thyroiditis, the most common cause of PH. However, Graves disease (an autoimmune cause of hyperthyroidism and one of the major etiologies of thyrotoxicosis) was not associated with this locus (OR = 1.03, p = 0.82). Associations with thyroid cancer, the previously reported association, trended toward significance in this analysis (OR = 1.29, p = 0.09), although only 96 cases were present in this data set. However, rs965513 was weakly associated with other malignancies: lymphoid tumors, including lymphosarcoma and reticulosarcoma (n = 130, OR = 1.35, p = 0.02), salivary gland cancers (n = 22, OR = 1.97, p = 0.025), and eye cancers (n = 20, OR = 1.97, p = 0.03). The PheWAS analysis adjusted for hypothyroidism status demonstrated more-significant associations with nutritional-deficiency anemia, pernicious anemia, atrial flutter, and thyroid cancer. Association with thyroid-related phenotypes, such as goiters and thyroiditis, were weaker; many of these also result in PH. Indeed, 80% of the individuals with Hashimoto thyroiditis and 73% of those with thyroiditis also had diagnoses of PH.
Figure 3.
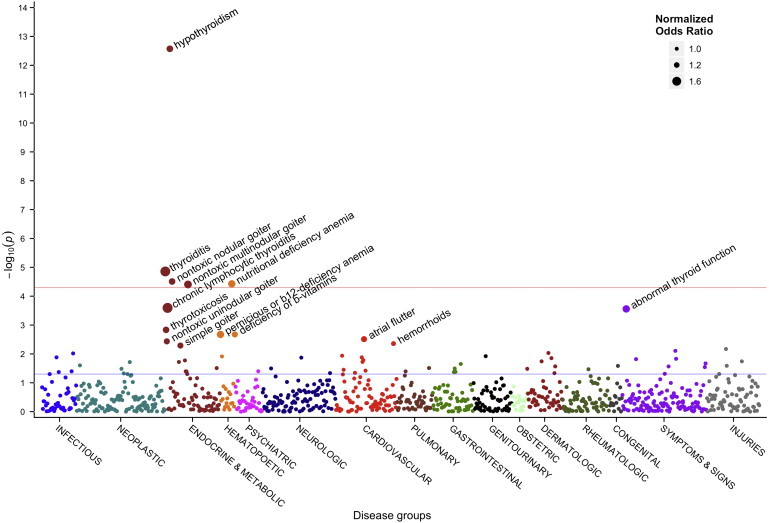
Manhattan Plot of Phenome-wide Association Study for rs965513
The PheWAS considered 866 phenotypes via single SNP tests of association (logistic regression) adjusted for age and sex. The red line indicates Bonferroni significance for these associations, p = 5.8 × 10−5. The blue line indicates p = 0.05. For labeled associations, the dot size varies by the magnitude of the odds ratio. For purposes of display, we normalized all odds ratios by taking the multiplicative absolute value such that all values were greater than 1 (e.g., an odds ratio of 0.5 becomes 2).
Table 5.
Phenotypes Associated with rs965513 Near FOXE1 in PheWAS Analysis
| Associated Phenotype | n |
Primary PheWAS |
PheWAS Adjusted for PH |
||
|---|---|---|---|---|---|
| Odds Ratio (95% CI) | p Value | Odds Ratio (95% CI) | p Value | ||
| Hypothyroidism | 2108 | 0.76 (0.70–0.81) | 2.7 × 10−13 | – | – |
| Thyroiditis | 185 | 0.58 (0.46–0.74) | 1.4 × 10−5 | 0.52 (0.33–0.82) | 4.8 × 10−3 |
| Nontoxic nodular goiter | 605 | 0.76 (0.67–0.86) | 3.1 × 10−5 | 0.78 (0.66–0.93) | 4.0 × 10−3 |
| Nutritional deficiency anemiaa | 332 | 1.41 (1.20–1.66) | 3.7 × 10−5 | 1.44 (1.23–1.70) | 9.7 × 10−6 |
| Nontoxic multinodular goiter | 319 | 0.69 (0.57–0.82) | 3.9 × 10−5 | 0.75 (0.59–0.96) | 0.02 |
| Hashimoto's thyroiditis | 127 | 0.58 (0.43–0.77) | 2.5 × 10−4 | 0.51 (0.27–0.97) | 0.04 |
| Abnormal thyroid function studies | 295 | 0.71 (0.59–0.85) | 2.8 × 10−4 | 0.77 (0.64–0.93) | 7.0 × 10−3 |
| Thyrotoxicosis | 354 | 0.76 (0.64–0.90) | 1.5 × 10−3 | 0.71 (0.53–0.95) | 0.02 |
| Deficiency of B-vitamins | 393 | 1.26 (1.09–1.46) | 2.1 × 10−3 | 1.30 (1.13–1.51) | 3.9 × 10−4 |
| Pernicious or B12-deficiency anemia | 182 | 1.40 (1.13–1.74) | 2.1 × 10−3 | 1.45 (1.17–1.80) | 7.7 × 10−4 |
| Atrial flutter | 486 | 1.25 (1.08–1.45) | 3.1 × 10−3 | 1.29 (1.12–1.50) | 6.9 × 10−4 |
| Hemorrhoids | 2916 | 0.90 (0.84–0.97) | 4.4 × 10−3 | 0.91 (0.85–0.98) | 0.01 |
| Simple goiter | 389 | 0.80 (0.68–0.93) | 5.1 × 10−3 | 0.94 (0.77–1.16) | 0.57 |
| Thyroid cancer | 96 | 1.29 (0.96–1.72) | 0.09 | 1.55 (1.16–2.09) | 3.5 × 10−3 |
| Iatrogenic hypothyroidismb | 197 | 0.85 (0.69–1.06) | 0.15 | – | – |
| Benign thyroid neoplasm | 55 | 1.23 (0.84–1.81) | 0.29 | 1.36 (0.93–1.99) | 0.11 |
| Graves disease | 106 | 1.03 (0.78–1.38) | 0.82 | 0.86 (0.44–1.67) | 0.65 |
All associations with p < 5 × 10−3, as well as other thyroid-related phenotypes, are shown. Bonferroni significance for these associations are p = 0.05/866 = 5.8 × 10−5.
Includes B12, folate, and protein-deficiency anemias as well as other unspecified nutritional anemias. It excludes iron-deficiency anemia.
Includes hypothyroidism resulting from surgery or radioiodine ablation.
Discussion
We demonstrate that one can repurpose previously genotyped samples linked to longitudinal EMR-derived data to identify variants associated with a disease unrelated to the original study hypotheses. In this study, we found that 9q22 variants near FOXE1, also known as thyroid transcription factor 2, are associated with PH in a European-American population. This association was observed independently with consistent effect size and direction in each of the five sites from which samples were derived, and it was independently verified in a sixth EMR-derived population. Importantly, neither the primary analysis nor the replication set required new genotyping. Furthermore, the phenotype algorithm developed at a single site proved portable to four other institutions and used only patient information derived as a byproduct of normal clinical care. PheWAS of this region suggests that these variants near FOXE1 are specifically associated with Hashimoto thyroiditis and might be associated with other thyroid conditions as well. These results support a growing body of evidence that DNA-linked EMRs might represent a powerful and cost-effective platform for future genomic investigation of many phenotypes.
FOXE1 is an intronless gene. These variants near FOXE1 have been previously associated with increased risk for both papillary and follicular thyroid cancer.23 Of note, the rs965513 minor allele has been shown to confer risk of thyroid cancer in this and prior studies,23 whereas the major allele confers risk to PH, as demonstrated in this study. Some retrospective studies have suggested an association between thyroiditis and thyroid cancer, although the primary process is unclear.45,46 In addition, higher TSH levels have been associated with an increased incidence of thyroid cancer and advanced-stage disease.47 Mutations in the coding region of FOXE1 have also been associated with a rare form of syndromic congenital hypothyroidism (Bamforth-Lazarus syndrome: hypothyroidism, cleft palate, choanal atresia, spiky hair [MIM 241850]),11 and cleft lip with or without cleft palate and isolated cleft palate).48,49 These observational data from humans are consistent with model systems showing that mice null for Foxe1 also exhibit cleft palate and thyroid abnormalities.50 Moreover, prior research has demonstrated that the FOXE1 variants identified in this study are also associated with lower serum TSH levels in euthyroid individuals.23,51 FOXE1 also has a polymorphic polyalanine stretch; variations in the length of this polyalanine stretch have been shown to result in variable transcription activity and risk of congenital hypothyroidism resulting from thyroid dysgenesis.52,53
FOXE1 is expressed during embryologic development and plays an important role in morphogenesis of the thyroid gland.54 Targeted disruption of Foxe1 in mice leads to a sublingual thyroid or thyroid agenesis, and patients with biallelic mutations are usually athyreotic.55 Later in development and in adults, the FOXE1 gene product binds to response elements in the promoter regions of the thyroglobulin (TG) and thyroid peroxidase (TPO [MIM 606765]) genes and helps regulate transcription of both proteins.56,57 The majority of PH patients exhibit autoantibodies to thyroglobulin or thyroid peroxidase,58 suggesting a possible link between a change in regulation of TPO production and the formation of autoantibodies.
The strongest association in the PheWAS analysis was hypothyroidism with an odds ratio nearly identical to that of the curated case-control population in the primary analysis, providing further validation of the PheWAS method. PheWAS also supports an association with thyroiditis, of which the most common form is Hashimoto thyroiditis. The relatively more significant odds ratio for those individuals with thyroiditis (0.58 versus 0.74) suggests that the underlying pathology for FOXE1-related risk of PH may be thyroiditis, especially when one considers that the underlying cause of most PH is Hashimoto thyroiditis. Although other variants, such as CTLA4 and PTPN22, that confer risk for PH have been associated with Graves disease,19 rs965513 was not. B-12-deficiency anemia occurs more commonly in individuals and family members with PH;59 however, the PheWAS associations of rs965513 with B-12-deficiency anemia and PH were inversely related, despite the fact that patients with hypothyroidism were more likely to have B-12-deficiency anemia in the eMERGE population (OR = 4.8). Thus, these results suggest that other genetic or clinical factors might mediate the clinical and familial associations of PH with B-12-deficiency anemia.
The association of FOXE1 variants with at least two structures derived from the embryonal foregut suggests that disorders of other foregut-derived adult organs might also display a genetic association with this locus. These organs include the pharynx; mouth and palate; thymus gland and related lymphoid tissue; esophagus, stomach, hepatic cells, and bile cannaliculi; gall bladder and common bile duct; pancreatic acinar and island cells; and upper duodenum.60 The association between these variants and thyroid cancer as well as, possibly, several cancers of lymphoid and salivary glands suggests that FOXE1 plays a role in the development and differentiation of these tissues. Further investigation of the PheWAS findings highlighted in this analysis will require future study with carefully defined phenotypes.
Previous genetics studies of PH have included family-based linkage and association studies.19 These studies have implicated several chromosomal regions61 and candidate genes,62 but none have localized a gene linked to PH. Interestingly, the HLA region, which is a candidate region in light of the autoimmune basis of PH, has not been consistently linked to autoimmune thyroid disease63 and was not associated with PH in this data set (Table S5 in the Supplemental Data available online). The lack of consistent findings across prior studies could be due to a variety of factors, such as insufficient statistical power, unaccounted-for environmental influences, genetic heterogeneity, and phenotypic heterogeneity. Indeed, case definitions differed greatly across studies. Some studies included both Graves disease and autoimmune hypothyroidism in their case definition, whereas others did not. The present study excluded Graves disease from PH case definition, and the fact that Graves disease was not associated with rs965513 by the PheWAS analysis supports this exclusion. Previous candidate-gene studies have shown associations with hypothyroidism and PTPN22 rs2476601;17 this association was observed in this analysis (controls: p = 5.0 × 10−4, OR = 1.29; non-TSH controls: p = 4.3 × 10−4, OR = 1.30), albeit not at genome-wide significance. CTLA4 rs3087243 has also been associated with hypothyroidism,16 but no associations with CTLA4 were detected here; however, this SNP and others in strong LD with it are not assayed on the Illumina 660-W BeadChip.
The results of this study demonstrate the successful reuse of EMR-linked genotype data as a discovery resource. This approach could support investigation of many diseases because individuals commonly experience many clinically important health conditions over the course of their lifetime. As the number of EMR-linked DNA biobanks increase, the possibility for numerous in silico genetic analyses also increases. The EMR contains a longitudinal record of disease, response to treatment, and evolution of care—all possible targets for future genomic and pharmacogenomic association studies. Moreover, as EMRs accrue additional phenotypic information through more widespread adoption and use in routine clinical care, the utility of such resources will grow.
A rate-limiting step toward EMR-based genetic association studies is identification of relevant case and control populations. Often, this involves either manual review of patient charts or creation of phenotype algorithms that incorporate a combination of techniques such as queries of billing code data, laboratory values, and natural language processing to interrogate data found in clinical notes. The phenotype selection logic developed at one site was successfully deployed with similar performance at five different sites with different EMR systems. One site, the Mayo Clinic, noted a lower positive predictive value for PH case identification. Algorithm classification errors largely resulted from exclusionary events predating electronic records (e.g., thyroidectomy in the 1940s) or from events occurring at outside medical facilities (e.g., military personnel treated at the VA). More advanced application of natural language processing might improve the ability to detect such non-coded data. Developing libraries of phenotype selection algorithms is a goal of the eMERGE network; a repository of phenotype selection algorithms is available online (see Web Resources).
Several limitations of this study deserve comment. Application of phenotype selection logic requires robust EMR systems capable of efficient data querying so that cohorts of patients matching specific criteria can be identified. Although our specific phenotype selection logic performed well for PH at these five sites, similar efforts will be needed if new selection logics uniquely suited for the study of other traits are to be developed. Our ascertainment and evaluation of phenotype characterization for cases and controls was limited to information available in the medical record; thus, some of our patients might have secondary causes for hypothyroidism, but these might not have been recognized by their treating physicians. Given that PH accounts for the vast majority of hypothyroid cases, we suspect that this misclassification bias is low. Although we asked reviewers validating our algorithm to exclude patients having only subclinical hypothyroidism from our analysis, it is possible that some of these patients had subclinical hypothyroidism instead of overt PH. However, patients with subclinical hypothyroidism often develop overt PH, especially in the absence of secondary causes (e.g., radiation exposure) specifically excluded in this study.64,65 Although we did not have formal iodine measurement available in our patients, the United States has supplemented dietary iodine intake through iodinated salt since 1920, making iodine-related hypothyroidism rare.66,67 Finally, our replication set was about a decade younger than the discovery set. However, because all control subjects had normal TSH values and absence of thyroid replacement medications in their medication lists, we believe that false-negative controls were rare.
In summary, this study demonstrates several notable findings. First, the study identifies and replicates variants near FOXE1 as genetic risk factors for PH. Second, it quantifies the portability of automated selection logic applied across five health centers with heterogeneous data models, EMR systems, and local practice patterns. Third, this study demonstrates the utility of EMR data for finding multiple clinically relevant phenotypes, based on the co-occurrence of multiple diseases in individuals. Finally, this study shows the potential for discovery of previously unknown genetic associations through the reuse of existing genomic data linked to clinical findings recorded in EMRs.
Acknowledgments
The eMERGE Network was initiated and funded by the National Human Genome Research Institute (NHGRI) through the following grants: U01-HG-004610 (Group Health Cooperative); U01-HG-004608 (Marshfield Clinic); U01-HG-004599 (Mayo Clinic); U01-HG-004609 (Northwestern University); and U01-HG-004603 (Vanderbilt University, also serving as the Coordinating Center). The National Institute of General Medical Sciences (NIGMS) provided additional funding to the eMERGE Network through these grants. The Mayo Genome Consortia are supported by the Mayo Foundation, Mayo Clinic Genome-wide Association Study of Venous Thromboembolism (HG-004735) from the NHGRI (GENEVA Consortium), and the Mayo Clinic SPORE in Pancreatic Cancer (P50CA102701) from the National Cancer Institute.
Supplemental Data
Web Resources
The URLs for the date presented herein are as follows:
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org
eMERGE Library of Phenotype Algorithms, http://gwas.org/index.php/Library_of_Phenotype_Algorithms
PheWAS software, http://knowledgemap.mc.vanderbilt.edu/research/content/phewas
References
- 1.Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., Manolio T.A. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ritchie M.D., Denny J.C., Crawford D.C., Ramirez A.H., Weiner J.B., Pulley J.M., Basford M.A., Brown-Gentry K., Balser J.R., Masys D.R. Robust replication of genotype-phenotype associations across multiple diseases in an electronic medical record. Am. J. Hum. Genet. 2010;86:560–572. doi: 10.1016/j.ajhg.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCarty C.A., Chisholm R.L., Chute C.G., Kullo I.J., Jarvik G.P., Larson E.B., Li R., Masys D.R., Ritchie M.D., Roden D.M., eMERGE Team The eMERGE Network: A consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med. Genomics. 2011;4:13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Denny J.C., Ritchie M.D., Basford M.A., Pulley J.M., Bastarache L., Brown-Gentry K., Wang D., Masys D.R., Roden D.M., Crawford D.C. PheWAS: Demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26:1205–1210. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kurreeman F., Liao K., Chibnik L., Hickey B., Stahl E., Gainer V., Li G., Bry L., Mahan S., Ardlie K. Genetic basis of autoantibody positive and negative rheumatoid arthritis risk in a multi-ethnic cohort derived from electronic health records. Am. J. Hum. Genet. 2011;88:57–69. doi: 10.1016/j.ajhg.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kullo I.J., Ding K., Jouni H., Smith C.Y., Chute C.G. A genome-wide association study of red blood cell traits using the electronic medical record. PLoS ONE. 2010;5:e13011. doi: 10.1371/journal.pone.0013011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denny J.C., Ritchie M.D., Crawford D.C., Schildcrout J.S., Ramirez A.H., Pulley J.M., Basford M.A., Masys D.R., Haines J.L., Roden D.M. Identification of genomic predictors of atrioventricular conduction: Using electronic medical records as a tool for genome science. Circulation. 2010;122:2016–2021. doi: 10.1161/CIRCULATIONAHA.110.948828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kullo I.J., Ding K., Shameer K., McCarty C.A., Jarvik G.P., Denny J.C., Ritchie M.D., Ye Z., Crosslin D.R., Chisholm R.L. Complement receptor 1 gene variants are associated with erythrocyte sedimentation rate. Am. J. Hum. Genet. 2011;89:131–138. doi: 10.1016/j.ajhg.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vanderpump M., Tunbridge W. The epidemiology of thyroid disease. In: Braverman L.E., Utiger R.D., editors. The Thyroid. Ninth Edition. Lippincott-Raven Publishers; Philadelphia, PA: 1996. pp. 474–482. [Google Scholar]
- 10.Aoki Y., Belin R.M., Clickner R., Jeffries R., Phillips L., Mahaffey K.R. Serum TSH and total T4 in the United States population and their association with participant characteristics: National Health and Nutrition Examination Survey (NHANES 1999-2002) Thyroid. 2007;17:1211–1223. doi: 10.1089/thy.2006.0235. [DOI] [PubMed] [Google Scholar]
- 11.Park S.M., Chatterjee V.K. Genetics of congenital hypothyroidism. J. Med. Genet. 2005;42:379–389. doi: 10.1136/jmg.2004.024158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Vliet G. Development of the thyroid gland: lessons from congenitally hypothyroid mice and men. Clin. Genet. 2003;63:445–455. doi: 10.1034/j.1399-0004.2003.00107.x. [DOI] [PubMed] [Google Scholar]
- 13.Kopp P.A. Reduce, recycle, reuse—Iodotyrosine deiodinase in thyroid iodide metabolism. N. Engl. J. Med. 2008;358:1856–1859. doi: 10.1056/NEJMe0802188. [DOI] [PubMed] [Google Scholar]
- 14.Brix T.H., Kyvik K.O., Hegedüs L. A population-based study of chronic autoimmune hypothyroidism in Danish twins. J. Clin. Endocrinol. Metab. 2000;85:536–539. doi: 10.1210/jcem.85.2.6385. [DOI] [PubMed] [Google Scholar]
- 15.Hansen P.S., Brix T.H., Iachine I., Kyvik K.O., Hegedüs L. The relative importance of genetic and environmental effects for the early stages of thyroid autoimmunity: a study of healthy Danish twins. Eur. J. Endocrinol. 2006;154:29–38. doi: 10.1530/eje.1.02060. [DOI] [PubMed] [Google Scholar]
- 16.Ueda H., Howson J.M.M., Esposito L., Heward J., Snook H., Chamberlain G., Rainbow D.B., Hunter K.M.D., Smith A.N., Di Genova G. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature. 2003;423:506–511. doi: 10.1038/nature01621. [DOI] [PubMed] [Google Scholar]
- 17.Criswell L.A., Pfeiffer K.A., Lum R.F., Gonzales B., Novitzke J., Kern M., Moser K.L., Begovich A.B., Carlton V.E.H., Li W. Analysis of families in the multiple autoimmune disease genetics consortium (MADGC) collection: the PTPN22 620W allele associates with multiple autoimmune phenotypes. Am. J. Hum. Genet. 2005;76:561–571. doi: 10.1086/429096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stefan M., Jacobson E.M., Huber A.K., Greenberg D.A., Li C.W., Skrabanek L., Conception E., Fadlalla M., Ho K., Tomer Y. Novel variant of the thyroglobulin promoter triggers thyroid autoimmunity through an epigenetic interferon α-modulated mechanism. J. Biol. Chem. 2011;286:31168–31179. doi: 10.1074/jbc.M111.247510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomer Y. Genetic susceptibility to autoimmune thyroid disease: Past, present, and future. Thyroid. 2010;20:715–725. doi: 10.1089/thy.2010.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Panicker V., Wilson S.G., Walsh J.P., Richards J.B., Brown S.J., Beilby J.P., Bremner A.P., Surdulescu G.L., Qweitin E., Gillham-Nasenya I. A locus on chromosome 1p36 is associated with thyrotropin and thyroid function as identified by genome-wide association study. Am. J. Hum. Genet. 2010;87:430–435. doi: 10.1016/j.ajhg.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arnaud-Lopez L., Usala G., Ceresini G., Mitchell B.D., Pilia M.G., Piras M.G., Sestu N., Maschio A., Busonero F., Albai G. Phosphodiesterase 8B gene variants are associated with serum TSH levels and thyroid function. Am. J. Hum. Genet. 2008;82:1270–1280. doi: 10.1016/j.ajhg.2008.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Medici M., van der Deure W.M., Verbiest M., Vermeulen S.H., Hansen P.S., Kiemeney L.A., Hermus A.R.M.M., Breteler M.M., Hofman A., Hegedüs L. A large-scale association analysis of 68 thyroid hormone pathway genes with serum TSH and FT4 levels. Eur. J. Endocrinol. 2011;164:781–788. doi: 10.1530/EJE-10-1130. [DOI] [PubMed] [Google Scholar]
- 23.Gudmundsson J., Sulem P., Gudbjartsson D.F., Jonasson J.G., Sigurdsson A., Bergthorsson J.T., He H., Blondal T., Geller F., Jakobsdottir M. Common variants on 9q22.33 and 14q13.3 predispose to thyroid cancer in European populations. Nat. Genet. 2009;41:460–464. doi: 10.1038/ng.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ammenwerth E., Schnell-Inderst P., Machan C., Siebert U. The effect of electronic prescribing on medication errors and adverse drug events: a systematic review. J. Am. Med. Inform. Assoc. 2008;15:585–600. doi: 10.1197/jamia.M2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaushal R., Jha A.K., Franz C., Glaser J., Shetty K.D., Jaggi T., Middleton B., Kuperman G.J., Khorasani R., Tanasijevic M., Bates D.W., Brigham and Women's Hospital CPOE Working Group Return on investment for a computerized physician order entry system. J. Am. Med. Inform. Assoc. 2006;13:261–266. doi: 10.1197/jamia.M1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bates D.W., Leape L.L., Cullen D.J., Laird N., Petersen L.A., Teich J.M., Burdick E., Hickey M., Kleefield S., Shea B. Effect of computerized physician order entry and a team intervention on prevention of serious medication errors. JAMA. 1998;280:1311–1316. doi: 10.1001/jama.280.15.1311. [DOI] [PubMed] [Google Scholar]
- 27.Kohane I.S. Using electronic health records to drive discovery in disease genomics. Nat. Rev. Genet. 2011;12:417–428. doi: 10.1038/nrg2999. [DOI] [PubMed] [Google Scholar]
- 28.Pacheco J.A., Avila P.C., Thompson J.A., Law M., Quraishi J.A., Greiman A.K., Just E.M., Kho A. A highly specific algorithm for identifying asthma cases and controls for genome-wide association studies. AMIA Annu. Symp. Proc. 2009;2009:497–501. [PMC free article] [PubMed] [Google Scholar]
- 29.Liao K.P., Cai T., Gainer V., Goryachev S., Zeng-treitler Q., Raychaudhuri S., Szolovits P., Churchill S., Murphy S., Kohane I. Electronic medical records for discovery research in rheumatoid arthritis. Arthritis Care Res. (Hoboken) 2010;62:1120–1127. doi: 10.1002/acr.20184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilke R.A., Xu H., Denny J.C., Roden D.M., Krauss R.M., McCarty C.A., Davis R.L., Skaar T., Lamba J., Savova G. The emerging role of electronic medical records in pharmacogenomics. Clin. Pharmacol. Ther. 2011;89:379–386. doi: 10.1038/clpt.2010.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roden D.M., Pulley J.M., Basford M.A., Bernard G.R., Clayton E.W., Balser J.R., Masys D.R. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin. Pharmacol. Ther. 2008;84:362–369. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCarty C.A., Wilke R.A., Giampietro P.F., Wesbrook S.D., Caldwell M.D. Marshfield Clinic Personalized Medicine Research Project (PMRP): Design, methods and recruitment for a large population-based biobank. Personalized Medicine. 2005;2:49–79. doi: 10.1517/17410541.2.1.49. [DOI] [PubMed] [Google Scholar]
- 33.McCarty C.A., Nair A., Austin D.M., Giampietro P.F. Informed consent and subject motivation to participate in a large, population-based genomics study: the Marshfield Clinic Personalized Medicine Research Project. Community Genet. 2007;10:2–9. doi: 10.1159/000096274. [DOI] [PubMed] [Google Scholar]
- 34.Turner S., Armstrong L.L., Bradford Y., Carlson C.S., Crawford D.C., Crenshaw A.T., de Andrade M., Doheny K.F., Haines J.L., Hayes G. Quality control procedures for genome-wide association studies. Curr. Protoc. Hum. Genet. 2011;68:1.19.1–1.19.18. doi: 10.1002/0471142905.hg0119s68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pritchard J.K., Stephens M., Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 37.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grady B.J., Torstenson E., Dudek S.M., Giles J., Sexton D., Ritchie M.D. Finding unique filter sets in plato: A precursor to efficient interaction analysis in gwas data. Pac. Symp. Biocomput. 2010;2010:315–326. [PMC free article] [PubMed] [Google Scholar]
- 39.Pruim R.J., Welch R.P., Sanna S., Teslovich T.M., Chines P.S., Gliedt T.P., Boehnke M., Abecasis G.R., Willer C.J. LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Willer C.J., Li Y., Abecasis G.R. METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson A.D., Handsaker R.E., Pulit S.L., Nizzari M.M., O'Donnell C.J., de Bakker P.I.W. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petersen G.M., Amundadottir L., Fuchs C.S., Kraft P., Stolzenberg-Solomon R.Z., Jacobs K.B., Arslan A.A., Bueno-de-Mesquita H.B., Gallinger S., Gross M. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat. Genet. 2010;42:224–228. doi: 10.1038/ng.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amundadottir L., Kraft P., Stolzenberg-Solomon R.Z., Fuchs C.S., Petersen G.M., Arslan A.A., Bueno-de-Mesquita H.B., Gross M., Helzlsouer K., Jacobs E.J. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat. Genet. 2009;41:986–990. doi: 10.1038/ng.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takahashi M., Saenko V.A., Rogounovitch T.I., Kawaguchi T., Drozd V.M., Takigawa-Imamura H., Akulevich N.M., Ratanajaraya C., Mitsutake N., Takamura N. The FOXE1 locus is a major genetic determinant for radiation-related thyroid carcinoma in Chernobyl. Hum. Mol. Genet. 2010;19:2516–2523. doi: 10.1093/hmg/ddq123. [DOI] [PubMed] [Google Scholar]
- 45.Büyükaşık O., Hasdemir A.O., Yalçın E., Celep B., Sengül S., Yandakçı K., Tunç G., Küçükpınar T., Alkoy S., Cöl C. The association between thyroid malignancy and chronic lymphocytic thyroiditis: Should it alter the surgical approach? Endokrynol. Pol. 2011;62:303–308. [PubMed] [Google Scholar]
- 46.Segal K., Ben-Bassat M., Avraham A., Har-El G., Sidi J. Hashimoto's thyroiditis and carcinoma of the thyroid gland. Int. Surg. 1985;70:205–209. [PubMed] [Google Scholar]
- 47.Haymart M.R., Glinberg S.L., Liu J., Sippel R.S., Jaume J.C., Chen H. Higher serum TSH in thyroid cancer patients occurs independent of age and correlates with extrathyroidal extension. Clin. Endocrinol. (Oxf.) 2009;71:434–439. doi: 10.1111/j.1365-2265.2008.03489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marazita M.L., Lidral A.C., Murray J.C., Field L.L., Maher B.S., Goldstein McHenry T., Cooper M.E., Govil M., Daack-Hirsch S., Riley B. Genome scan, fine-mapping, and candidate gene analysis of non-syndromic cleft lip with or without cleft palate reveals phenotype-specific differences in linkage and association results. Hum. Hered. 2009;68:151–170. doi: 10.1159/000224636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moreno L.M., Mansilla M.A., Bullard S.A., Cooper M.E., Busch T.D., Machida J., Johnson M.K., Brauer D., Krahn K., Daack-Hirsch S. FOXE1 association with both isolated cleft lip with or without cleft palate, and isolated cleft palate. Hum. Mol. Genet. 2009;18:4879–4896. doi: 10.1093/hmg/ddp444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.De Felice M., Ovitt C., Biffali E., Rodriguez-Mallon A., Arra C., Anastassiadis K., Macchia P.E., Mattei M.G., Mariano A., Schöler H. A mouse model for hereditary thyroid dysgenesis and cleft palate. Nat. Genet. 1998;19:395–398. doi: 10.1038/1289. [DOI] [PubMed] [Google Scholar]
- 51.Lowe J.K., Maller J.B., Pe'er I., Neale B.M., Salit J., Kenny E.E., Shea J.L., Burkhardt R., Smith J.G., Ji W. Genome-wide association studies in an isolated founder population from the Pacific Island of Kosrae. PLoS Genet. 2009;5:e1000365. doi: 10.1371/journal.pgen.1000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Szczepanek E., Ruchala M., Szaflarski W., Budny B., Kilinska L., Jaroniec M., Niedziela M., Zabel M., Sowinski J. FOXE1 polyalanine tract length polymorphism in patients with thyroid hemiagenesis and subjects with normal thyroid. Horm. Res. Paediatr. 2011;75:329–334. doi: 10.1159/000322874. [DOI] [PubMed] [Google Scholar]
- 53.Carré A., Castanet M., Sura-Trueba S., Szinnai G., Van Vliet G., Trochet D., Amiel J., Léger J., Czernichow P., Scotet V., Polak M. Polymorphic length of FOXE1 alanine stretch: Evidence for genetic susceptibility to thyroid dysgenesis. Hum. Genet. 2007;122:467–476. doi: 10.1007/s00439-007-0420-5. [DOI] [PubMed] [Google Scholar]
- 54.Fagman H., Nilsson M. Morphogenetics of early thyroid development. J. Mol. Endocrinol. 2011;46:R33–R42. doi: 10.1677/jme-10-0084. [DOI] [PubMed] [Google Scholar]
- 55.Clifton-Bligh R.J., Wentworth J.M., Heinz P., Crisp M.S., John R., Lazarus J.H., Ludgate M., Chatterjee V.K. Mutation of the gene encoding human TTF-2 associated with thyroid agenesis, cleft palate and choanal atresia. Nat. Genet. 1998;19:399–401. doi: 10.1038/1294. [DOI] [PubMed] [Google Scholar]
- 56.Zannini M., Avantaggiato V., Biffali E., Arnone M.I., Sato K., Pischetola M., Taylor B.A., Phillips S.J., Simeone A., Di Lauro R. TTF-2, a new forkhead protein, shows a temporal expression in the developing thyroid which is consistent with a role in controlling the onset of differentiation. EMBO J. 1997;16:3185–3197. doi: 10.1093/emboj/16.11.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ortiz L., Aza-Blanc P., Zannini M., Cato A.C., Santisteban P. The interaction between the forkhead thyroid transcription factor TTF-2 and the constitutive factor CTF/NF-1 is required for efficient hormonal regulation of the thyroperoxidase gene transcription. J. Biol. Chem. 1999;274:15213–15221. doi: 10.1074/jbc.274.21.15213. [DOI] [PubMed] [Google Scholar]
- 58.Dayan C.M., Daniels G.H. Chronic autoimmune thyroiditis. N. Engl. J. Med. 1996;335:99–107. doi: 10.1056/NEJM199607113350206. [DOI] [PubMed] [Google Scholar]
- 59.Boelaert K., Newby P.R., Simmonds M.J., Holder R.L., Carr-Smith J.D., Heward J.M., Manji N., Allahabadia A., Armitage M., Chatterjee K.V. Prevalence and relative risk of other autoimmune diseases in subjects with autoimmune thyroid disease. Am. J. Med. 2010;123:183.e1–183.e9. doi: 10.1016/j.amjmed.2009.06.030. [DOI] [PubMed] [Google Scholar]
- 60.Martini F.H., Timmons M.J., Tallitsch R.B. Seventh Edition. Benjamin Cummings; San Francisco: 2011. Human Anatomy. [Google Scholar]
- 61.Allen E.M., Hsueh W.-C., Sabra M.M., Pollin T.I., Ladenson P.W., Silver K.D., Mitchell B.D., Shuldiner A.R. A genome-wide scan for autoimmune thyroiditis in the Old Order Amish: Replication of genetic linkage on chromosome 5q11.2-q14.3. J. Clin. Endocrinol. Metab. 2003;88:1292–1296. doi: 10.1210/jc.2002-020634. [DOI] [PubMed] [Google Scholar]
- 62.Hadj Kacem H., Rebai A., Kaffel N., Masmoudi S., Abid M., Ayadi H. PDS is a new susceptibility gene to autoimmune thyroid diseases: association and linkage study. J. Clin. Endocrinol. Metab. 2003;88:2274–2280. doi: 10.1210/jc.2002-021460. [DOI] [PubMed] [Google Scholar]
- 63.Ban Y., Davies T.F., Greenberg D.A., Concepcion E.S., Tomer Y. The influence of human leucocyte antigen (HLA) genes on autoimmune thyroid disease (AITD): Results of studies in HLA-DR3 positive AITD families. Clin. Endocrinol. (Oxf.) 2002;57:81–88. doi: 10.1046/j.1365-2265.2002.01566.x. [DOI] [PubMed] [Google Scholar]
- 64.Huber G., Staub J.-J., Meier C., Mitrache C., Guglielmetti M., Huber P., Braverman L.E. Prospective study of the spontaneous course of subclinical hypothyroidism: prognostic value of thyrotropin, thyroid reserve, and thyroid antibodies. J. Clin. Endocrinol. Metab. 2002;87:3221–3226. doi: 10.1210/jcem.87.7.8678. [DOI] [PubMed] [Google Scholar]
- 65.Kabadi U.M. ‘Subclinical hypothyroidism’. Natural course of the syndrome during a prolonged follow-up study. Arch. Intern. Med. 1993;153:957–961. doi: 10.1001/archinte.153.8.957. [DOI] [PubMed] [Google Scholar]
- 66.Pearce E.N. U.S. iodine nutrition: Where do we stand? Thyroid. 2008;18:1143–1145. doi: 10.1089/thy.2008.1554. [DOI] [PubMed] [Google Scholar]
- 67.Caldwell K.L., Makhmudov A., Ely E., Jones R.L., Wang R.Y. Iodine status of the U.S. population, National Health and Nutrition Examination Survey, 2005–2006 and 2007–2008. Thyroid. 2011;21:419–427. doi: 10.1089/thy.2010.0077. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


