Abstract
Although genome-wide association studies have implicated many individual loci in complex diseases, identifying the exact causal alleles and the cell types within which they act remains greatly challenging. To ultimately understand disease mechanism, researchers must carefully conceive functional studies in relevant pathogenic cell types to demonstrate the cellular impact of disease-associated genetic variants. This challenge is highlighted in autoimmune diseases, such as rheumatoid arthritis, where any of a broad range of immunological cell types might potentially be impacted by genetic variation to cause disease. To this end, we developed a statistical approach to identify potentially pathogenic cell types in autoimmune diseases by using a gene-expression data set of 223 murine-sorted immune cells from the Immunological Genome Consortium. We found enrichment of transitional B cell genes in systemic lupus erythematosus (p = 5.9 × 10−6) and epithelial-associated stimulated dendritic cell genes in Crohn disease (p = 1.6 × 10−5). Finally, we demonstrated enrichment of CD4+ effector memory T cell genes within rheumatoid arthritis loci (p < 10−6). To further validate the role of CD4+ effector memory T cells within rheumatoid arthritis, we identified 436 loci that were not yet known to be associated with the disease but that had a statistically suggestive association in a recent genome-wide association study (GWAS) meta-analysis (pGWAS < 0.001). Even among these putative loci, we noted a significant enrichment for genes specifically expressed in CD4+ effector memory T cells (p = 1.25 × 10−4). These cell types are primary candidates for future functional studies to reveal the role of risk alleles in autoimmunity. Our approach has application in other phenotypes, outside of autoimmunity, where many loci have been discovered and high-quality cell-type-specific gene expression is available.
Introduction
Autoimmune diseases are complex traits with many scores of common variants throughout the genome that might subtly impact disease risk.1–4 But, using these loci to elucidate mechanisms from common variants has proven to be a challenging task, particularly because many of them do not directly alter coding sequences but potentially impact gene regulation modestly in a cell-specific manner.5 If the critical immune cell subsets were known for a given disease, then investigators could derive relevant cellular model systems for focused functional studies to understand pathogenic mechanisms. These studies might include broad genomics approaches, such as cell-type-specific expression quantitative trait loci (eQTL) screens to identify alleles that act to alter gene expression,6–8 or epigenetic screens to identify key active regulatory elements.9,10 Additionally, investigators could pursue focused mechanistic studies to understand the role of individual disease alleles within that tissue.
But for most autoimmune diseases the immune cell types specifically impacted by common risk variants are not defined. Past mechanistic studies in autoimmune model systems have often led to confusing results that might not easily translate to human disease. For example, separate influential studies in rheumatoid arthritis (RA [MIM 180300]) have implicated a wide range of pathogenic cell types, including B and T lymphocyte subsets,11 neutrophils,12 mast cells,13 macrophages,14 platelets,15 and synoviocytes.16,17 The importance of pursuing mechanistic studies in the appropriate cell type is highlighted by the fact that common variants can have conflicting functions in different closely related immune tissues. For example, a deletion of the promoter region of IRGM (MIM 608212), associated with Crohn disease (MIM 266600) might either increase or decrease allelic gene expression, depending on the tissue.18 Similarly an IL2RA (MIM 147730) autoimmune variant impacts different intermediate phenotypes even in closely related immune cells.19
Here, we hypothesize that predisposing autoimmune risk alleles impact a small number of pathogenic tissues or cell types. If this is the case, then the subset of genes with critical functions in those pathogenic cell types are likely to be within disease loci. However, in practice, a comprehensive and unbiased catalog of cell-type-specific gene function is simply not available. As an alternative, compendia of gene-expression data are available for many tissues. These compendia can serve as objective proxies for tissue-specific gene function. Practically, gene-expression profiles have been used to identify cell types of origin in malignancies.20,21 In addition, investigators commonly use gene-expression profiling of presumed pathogenic tissues to screen risk alleles and to prioritize genes for follow-up within complex trait loci. An orthogonal approach is to broadly consider a large collection of potential cell types and to then identify the single tissue that specifically expresses genes within loci that contain disease risk alleles. To our knowledge, no such systematic approach has yet been devised.
We developed a statistical method that, given a collection of disease-associated SNPs and a compendium of gene-expression profiles from a broad set of tissue types, scores tissues for enrichment of specifically expressed genes in linkage disequilibrium (LD) with the SNPs (see Figure 1 and Methods for details). For such a method to be effective, it is critical to use high-quality cell-specific expression data with minimal contamination and include replicates to reduce noise. To this end we use the Immunological Genome Project (ImmGen) data set assaying 223 mouse immune tissues individually double-sorted by FACS to ensure high purity and profiled in at least triplicate.22 Also, it is critical for the methodology to be robust to key confounders. Therefore, in our method we (1) use nonparametric-expression specificity scores to avoid confounding by the inherently skewed nature of expression levels, (2) correct for number of genes per SNP to avoid multiple-hypothesis testing biases, and (3) assess significance of disease-associated SNP sets by using matched SNP sets to avoid confounding by correlations in gene size and cell-specific expression,23 correlations in expression between proximate genes, and genomic biases in gene density and genetic variation across the genome.
Figure 1.
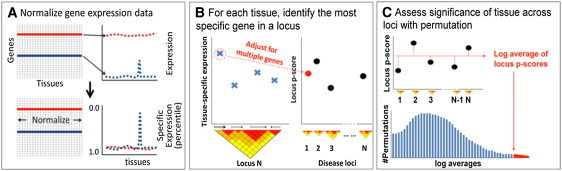
Statistical Approach
(A) Normalize gene expression data. We normalized the expression profile by dividing the expression value of each gene in each tissue by the Euclidean norm of the gene's expression across all tissues in order to emphasize tissue-specific expression. Scores were converted to nonparametric percentiles.
(B) For each tissue, identify the most specifically expressed gene in a locus. For each SNP, we first defined genes implicated by the SNP based on LD. For a specific tissue, we identified the most specifically expressed gene in the locus and then scored the SNP based on that gene's nonparametric specific expression score after adjusting for multiple genes within the locus.
(C) With permutations, assess the significance of each tissue across loci. We first calculated a score for the tissue by taking the average of the log adjusted-percentiles across loci from B. Then, we randomly selected matched SNP sets and scored them similarly. The proportion of random SNP sets with tissue scores exceeding that of the actual set of SNPs being tested was reported as the p value of the tissue.
Material and Methods
Summary of Statistical Method
First, after standard quality control and quantile normalization,24 we transform expression data into nonparametric “tissue-specific expression” scores for each gene (Figure 1A). In order to do this, we first divide raw expression values by the Euclidean norm of values for each gene across all tissue types. Then, for a given tissue, we order genes by normalized expression values and assign each gene a percentile. These uniformly distributed percentiles constitute nonparametric tissue-specific expression scores. Genes with low percentile scores for a tissue are highly specifically expressed in that tissue, whereas genes with high percentile scores are either not expressed in that tissue at all or ubiquitously expressed. Second, for a given tissue, we assign each disease-associated SNP a “locus p score” (Figure 1B). To do this, we first identify genes that are in LD with a disease-associated SNP by using standard methods.25,26 Then, we identify the single proximate gene most specifically expressed within that tissue. The SNP's locus p score for that tissue is determined to be the tissue-specific percentile score of that gene after correcting for multiple genes tested within that locus. These locus p scores should be roughly uniformly distributed under the null. Finally, we assign an overall significance score for each tissue by taking the average of the log of locus p scores across all disease-associated SNPs (see Figure 1C). Although an analytical p value can be calculated, to avoid realistic confounders that could inadvertently inflate theoretical p values, we calculate statistical significance scores by comparing the actual log average of locus p scores to that of random SNPs matched for total number of genes.
Gene-Expression Datasets
For this project we used two separate data sets. The Genomics Institute of the Novartis Research Foundation (GNF) tissue atlas consists of expression profiles of 79 human tissues and cells types including immune-cell types measured in duplicate (BioGPS, see Web Resources).27 The Immunological Genome Project (ImmGen) consists of expression profiles of 223 sorted cells from immunological tissues and blood obtained from mice.22 Each sample is sorted with at least three biological replicates.
Preprocessing and Normalizing Gene-Expression Datasets
For each data set, after applying standard quantile normalization,24 we averaged expression values from replicates for each probe set. To obtain the single most robust expression value for genes with multiple probe sets, we selected the single probe set within each gene transcript that had the highest minimal expression value across all tissues. The GNF data set then consisted of measurements on 17,581 unique genes in 79 tissue types. The ImmGen data set contained 21,968 unique Mus musculus genes. We used HomoloGene (March 2010) to map the M. musculus genes to 14,623 unique human homologs.
We then transformed both data sets into nonparametric tissue-specific expression scores for genes. First, we normalized the expression level of each gene to reflect the specificity of expression in each tissue type. To do so, for each gene in each tissue, we divided the raw expression value by the Euclidean norm of values across all tissues:
where Xi,j is the expression value of gene i at tissue j, and X′i,j is the specificity score. Thus, each gene and tissue received a score between 0 and 1, where a score of 1 means the gene is exclusively expressed in this tissue. Ubiquitously expressed genes have low normalized scores across tissue types.
Next, for a given tissue, we transformed these normalized scores, X′i,j, into nonparametric tissue-specificity percentile scores for each gene, Pi,j, where a low percentile represents high specificity relative to other genes for a given tissue and a high percentile represents low specificity.
Mapping SNPs to Genes
Disease-associated SNPs are linked to proximate genes in LD with a previously described approach.25,26 First, for each SNP, we defined genes implicated by the SNP by defining a disease region. To do so, we identified the furthest neighboring SNPs in LD with the SNP in the 3′ and 5′ directions (r2 > 0.5, CEU [Utah residents with Northern and Western European ancestry from the CEPH collection] HapMap). We then extended outward in each direction to the nearest recombination hotspot.28 This region would include the disease-associated SNP and all SNPs in LD. All genes that overlapped with this region were considered implicated by the SNP. If no genes were found in the region, we extended an additional 250 kb in each direction. If two SNPs contained overlapping genes, they were merged as one single locus.
Testing Tissue for Enrichment
Given our list of SNPs connected to genes and our nonparametric expression tissue-specificity percentiles, we scored the list of disease-associated SNPs for enrichment of genes specifically expressed in each individual tissue type.
To score each tissue j, we first identified the most specifically expressed gene near each SNP S in tissue j. We will refer to that gene as gS,j. We applied a Bonferroni correction to adjust the tissue-specificity percentile for testing of the multiple genes near each SNP:
where ns is the number of genes implicated by SNP S. The PS,j values are referred to in the main text as the locus p score. They should be roughly uniformly distributed. For each tissue, we scored for enrichment by summing the PS,j values of all SNPs:
Under the null, if PS,j scores were randomly distributed, then Tj should be distributed according to the gamma distribution:
where α is the shape parameter and is equal to the number of SNPs and β is the rate parameter and is set to 1. In this case, the p value for the tissue is calculated as:
However, analytical p values are not robust to realistic biological factors.
Significance Scores Are Based on Random SNP Sets
To estimate the significance in a more robust and unbiased manner, we calculated p values empirically by comparing observed Tj values to empirical values from random sets of SNPs. Given a set of disease-associated SNPs, we create a matched SNP set with exactly the same number of SNPs and approximately similar numbers of genes for each permutation. We drew random SNPs for permutation from a pool of 45,265 independent Hapmap SNPs that were “clumped” to insure minimal correlation.29 To create a matched SNP set with approximately similar gene numbers, for each disease-associated SNP that implicated fewer than 11 genes we selected a random SNP that implicated exactly the same number of genes and for SNPs that implicated more than 10 genes, we selected a random SNP that also implicated > 10 genes. To ensure a comparable number of genes, the total number of genes implicated by all random SNPs must be within 10% of that implicated by disease SNPs. We then scored each of matched SNP sets for enrichment of genes in tissue j and calculated Tj. The proportion of randomly selected matched SNP sets whose Tj is less than the Tj for the disease-associated SNPs set was reported as the p value.
To efficiently compute significance, we varied the number of random SNP sets that we evaluated for a tissue from 250 to 1,024,000. We started by evaluating each tissue with 250 SNP sets. For those tissues where at least 25 sets were observed to be more significant than the observed SNP set, we accepted the p value and did not evaluate for any more SNP sets. For those tissues for which fewer than 25 sets were more significant than the observed SNP set (ie p < 0.1), we doubled the number of SNP sets. The number of SNP sets was doubled until at least 25 events were more significant than the observed SNP set or until we reached 1,024,000 permutations. For p values > 2.5 × 10−5, this ensured a variance of <20%.
Assessing the Significance of Individual SNPs
For each SNP and tissue, we calculated an “empirical locus p value,” which assessed the degree to which an individual gene within a locus is contributing to enrichment of specific gene expression within a tissue. This value was calculated by comparing the locus p score for the actual disease-associated SNP, based on the most specifically expressed gene within a tissue, to that of the matched SNP in randomly selected SNP sets during the permutation process, as described above. The empirical locus p value was reported as the fraction of randomly selected matched SNPs with more extreme locus p scores than the actual locus p score.
Adjusting for Expression Profiles
In order to assess enrichment across tissues after accounting for the effect of tissues that have already been identified as significant from the data set, we have devised an adjusted analysis framework. Briefly, we used the X′ matrix of tissue specificity scores, then removed the component of each tissue expression profile that was correlated with the tissue that we are conditioning on.
Let the expression scores of the most significant tissue be vector v. We subtracted the components of v from another tissue's expression profile, u, in order to obtain a new profile, u', which is independent of v:
The new profile scores were used to recalculate tissue-specific percentiles P, which can then be reused with the same statistical framework as above.
Scoring Nominally Associated RA SNPs
In order to score RA SNPs not yet associated with RA, we used the p value results from a recently published meta-analysis of six genome-wide association study (GWAS) consisting of 5,539 autoantibody-positive RA cases and 20,169 controls of European descent. We selected all SNPs that had an association p value (pGWAS) of < 0.001. After excluding SNPs within the MHC region (ranging from 25.8–3.4 Mb on chromosome 6 in HG 18 coordinates), we grouped the resulting SNPs into independent loci. We grouped two SNPs within the same locus if they had r2 > 0.1 in HapMap or shared a common gene. For each locus, we selected the single SNP with the most significant association to RA. We excluded any of these SNPs that were in LD with a known RA-associated risk loci (r2 < 0.1) or implicated a gene that was also implicated by a known RA SNP. We tested these loci for enrichment of specifically expressed genes in each of the individual cell types in RA. Significance for each tissue was determined by selecting matched SNP sets as described above. Given the large number of SNPs, we allowed for the total gene number to be outside the ±10% criteria described above.
In order to calculate an overall association to CD4+ effector memory T cells, we averaged all four X′ specificity score profiles of each of the subsets of CD4+ effector memory T cells together to calculate significance of association and empirical locus p values.
Results
Statistical Properties and Robustness
We wanted to ensure that our statistical method was robust to realistic biological factors (e.g., neural tissues tend to express larger genes23) that can inadvertently inflate theoretical p values in certain cell types (see Figure S1, available online). Thus, we scored 10,000 sets of 20 random SNPs, each in LD with at least one gene, from a larger set of independent SNPs from the HapMap project. Applying our approach to assess gene-expression enrichment in both the 79 tissues from the GNF and to the 223 cell types from the ImmGen demonstrate appropriate type I error rate (see Figures S2A and SB). We also note that error rates are consistent across all cell types, and there is no evidence of inflation of significance scores at any given tissue. Furthermore, our method demonstrates little evidence of statistical inflation in 500 sets of 20 random SNPs in either of those two data sets (see Figures S2C and S2D).
As a positive control, we examined common variants from two phenotypes. First, we applied our method to 37 SNPs associated with serum low-density lipoprotein (LDL) cholesterol from a recent large genome-wide SNP association meta-analysis.30 We hypothesized that these genes would be most specifically enriched in the liver because the liver is the primary organ where LDL is regulated31 and known mutations impact hepatocyte cellular function.32,33 In aggregate, these SNPs implicated 165 genes in LD (see Table S1A). When we tested each of the 79 tissue expression profiles from GNF for specific expression of genes in LD with these SNPs, we did indeed observe that only the liver showed highly specific expression of genes in LD with cholesterol metabolism SNPs (p = 2.0 × 10−4, see Table S2 and Figure S3A). Other tissues that obtained nominal significance at p < 0.01, fetal liver (p = 0.0014) and the adipocyte (p = 0.0077), were no longer significant after adjusting for the liver-expression profile. This suggests that the other observed associations were the consequence of correlated expression (see Methods). In certain cases, loci harbored genes that were specifically expressed within the liver, and in these cases these genes were often compelling candidate genes (see Table S1A). Next, we applied our method to the 32 obesity-associated SNPs34 that in aggregate implicate 91 genes (see Table S1B). When we tested 79 tissues from the GNF for specific expression of genes within obesity loci, we observed that only the pituitary gland obtained nominal significance at p = 0.0032 (see Table S2 and Figure S3B). Although this was not statistically significant after accounting for 79 independent tests, we were encouraged that it emerged as the most significant tissue because pituitary dysfunction, from trauma or rare familial mutations, is a known cause of obesity.35,36 Furthermore, the authors of recent genome-wide genetic studies have speculated that obesity SNPs act on the hypothalamus-pituitary axis.34,37 Potentially, a more targeted expression data set of the brain with carefully dissected human tissues might have resulted in a more powerful analysis.
Although there is concern that multiple intercorrelated gene-expression profiles might compromise power, we found that even in extreme circumstances the power loss is minimal. Our nonparametric approach relies on the relative order of a gene's specific expression within a tissue, rather than the magnitude of its specificity. The addition of intercorrelated tissue profiles impacts the magnitude of specific expression scores for a tissue but has a minimal impact on the relative ordering of the genes themselves. To assess the robustness of our method to multiple intercorrelated expression profiles, we repeated our analysis of LDL SNPs after adding in 1, 10, 50, or 100, copies of the identical liver expression profile. In each case the liver showed the exact same highly specific expression of genes in LD with cholesterol metabolism SNPs (p = 2.0 × 10−4). As a second test we added 1, 10, 50, or 100 copies of modified liver expression profiles, where we permuted expression values of 50% of the genes independently; so that each added profile was correlated with but also had substantial differences from the original liver expression profile. In each case the liver showed highly specific expression of genes in LD with cholesterol metabolism SNPs (p ranging from 9.8 × 10−5 to 2.9 × 10−4). In instances where the correlative structure of the data is more complex, and power is impacted, dimensional reduction approaches to simplify the expression data are useful.38
Application to Autoimmune Disease
Convinced that this approach was statistically robust and could detect potentially pathogenic cell types, we applied it to autoimmune disease SNPs. We focused on three separate autoimmune diseases. For systemic lupus erythematosus (SLE [MIM 152700]), we identified 30 SNPs, implicating 27 independent loci with a total of 136 genes (see Table S3A).39–43 For Crohn disease, we identified 71 SNPs, implicating 69 independent loci with a total of 316 genes (see Table S3B).3 Finally, for RA, we identified 40 SNPs, in aggregate implicating 39 independent loci with a total of 132 genes (see Table S3C).44,45 Testing each of these three autoimmune disease SNP sets against the 79 GNF tissues implicated only immune tissues (in each case multiple tissues with p < 2 × 10−5, see Table S2 and Figure S4). But given the limited number of immunological tissues and the high degree of correlation between them, we could not pinpoint the causal immune-cell types. We speculated that the ImmGen data set could more clearly demonstrate the key immune cell types for each of the different diseases because it was collected to represent a very broad view of transcriptional profiles in mouse immune-cell types across many lineages, developmental stages, and target organs. It includes hematopoietic stem, myeloid and lymphoid cells, and both innate and adaptive immune cells.
When we tested SLE loci for enrichment of specifically expressed genes within the 223 expression profiles contained within ImmGen data set, the single most significant immune-cell type was transitional B cells (stage T3) collected from the spleen (p = 5.9 × 10−6, see Table 1, Figure 2A, and Table S4A). Strikingly, all of the other statistically significant associations were other B cell subsets, including other closely related splenic transitional B cell subsets (p < 2 × 10−4 = 0.05/223). All of the B cell associations are obviated (see Table 1 and Table S4A), when we repeated our analysis after adjusting for three splenic transitional B cell profiles (B.T1.Sp, B.T2.Sp, and B.T3.Sp). This strongly suggests that other observed B cell associations are the result of expression correlation with transitional B cells and not representative of independent effects. The implication of transitional B cells by associated loci is consistent with much of the known pathobiology of SLE, which has implicated B cells more broadly. The pathogenic nature of antibodies produced by B cells in lupus has been long established and is supported by mouse models46 and by the demonstration of the efficacious nature of B cell targeted therapies in SLE.47 These results implicating transitional B cells specifically offer a finer resolution on this commonly accepted hypothesis.
Table 1.
Summary of Autoimmune Disease Association to 22 ImmGen Tissues
| Immune Tissues |
SLE |
Crohn Disease |
Rheumatoid Arthritis |
|||
|---|---|---|---|---|---|---|
| Unconditional | Adjusting for Transitional B Subtypes | Unconditional | Adjusting for DC Subtypes | Unconditional | Adjusting for Subtypes of CD4+ Effector Memory T Cells | |
| B.T1.Sp | 0.00018 | – | 0.060 | 0.12 | 0.0032 | 0.0050 |
| B.T2.Sp | 0.000013 | – | 0.013 | 0.029 | 0.00041 | 0.0015 |
| B.T3.Sp | 0.0000059 | – | 0.030 | 0.036 | 0.00072 | 0.0055 |
| B.Fo.PC | 0.000013 | 0.23 | 0.06 | 0.04 | 0.0024 | 0.0010 |
| B.Fo.Sp | 0.000022 | 0.11 | 0.0071 | 0.014 | 0.00032 | 0.0015 |
| B.FrF.BM | 0.000023 | 0.82 | 0.031 | 0.021 | 0.0025 | 0.0025 |
| CD19Control | 0.000013 | 0.70 | 0.011 | 0.026 | 0.00099 | 0.0025 |
| T.4Mem.LN | 0.024 | 0.18 | 0.018 | 0.012 | <0.0000010 | – |
| T.4Mem.Sp | 0.080 | 0.27 | 0.000061 | 0.00075 | <0.0000010 | – |
| T.4Mem44h62l.LN | 0.021 | 0.05 | 0.00037 | 0.009 | <0.0000010 | – |
| T.4Mem44h62l.Sp | 0.062 | 0.14 | 0.00087 | 0.032 | <0.0000010 | – |
| T.4.Pa.BDC | 0.0073 | 0.019 | 0.00032 | 0.00025 | 0.0000059 | 0.017 |
| T.4.Sp.B16 | 0.0016 | 0.0045 | 0.010 | 0.0090 | 0.0000078 | 0.0085 |
| T.4int8+.Th | 0.05 | 0.33 | 0.00083 | 0.0015 | 0.00017 | 0.18 |
| T.4Mem.Tbet..Sp | 0.61 | 0.75 | 0.020 | 0.043 | 0.00021 | 0.70 |
| T.4Mem.Tbet+.Sp | 0.27 | 0.43 | 0.023 | 0.068 | 0.000012 | 0.46 |
| T.4SP69+.Th | 0.08 | 0.23 | 0.0094 | 0.013 | 0.00016 | 0.17 |
| T.8Mem.Tbet+.Sp | 0.27 | 0.58 | 0.041 | 0.10 | 0.00018 | 0.64 |
| DC.103-11b+.PolyIC.Lu | 0.013 | 0.026 | 0.000016 | - | 0.0094 | 0.021 |
| DC.103+11b-.PolyIC.Lu | 0.043 | 0.062 | 0.000069 | - | 0.13 | 0.090 |
| NKT.4+.Lv | 0.16 | 0.29 | 0.00038 | 0.019 | 0.00012 | 0.010 |
| NK.H-.MCMV1.Sp | 0.018 | 0.0061 | 0.00014 | 0.0073 | 0.076 | 0.33 |
Here, we list all 22 out of 223 tissues from ImmGen that obtained nominally significant association (p < 0.01) in at least one of the three autoimmune phenotypes tested. For each disease, we listed results of our analysis without any conditioning, as well as results after removing the contributions of the most significant tissues. For each tissue, we listed an association significance p value for each phenotype.
Figure 2.
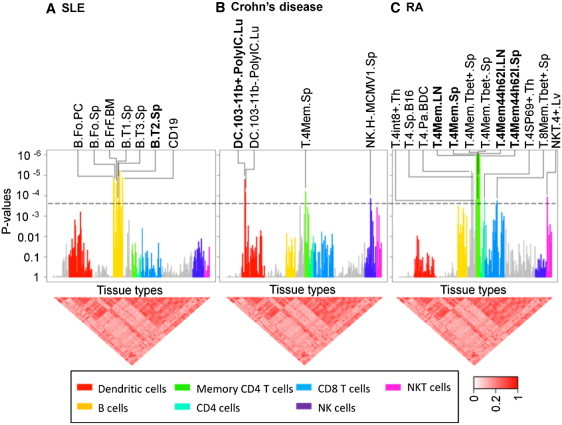
Application to Autoimmune Diseases
We evaluated SNPs associated with systemic lupus erythematous, rheumatoid arthritis, and Crohn disease for cell-specific gene enrichment in 223 murine immune-cell types. The Bonferroni-corrected p value is shown by a dotted line. In each case, we labeled cell types that are significant after multiple hypothesis testing (p < 2.2 × 10−4), and we bold the single most significant cell type. (A) In lupus, B cells, especially transitional B cells in the spleen (B.T2.Sp), showed significant enrichment of genes within disease loci. (B) In Crohn disease, epithelial-associated stimulated CD103- dendritic cells (DC.103-11b+PoluIC.Lu) achieved the highest statistical significance. (C) In rheumatoid arthritis, the four subsets of CD4+ effector memory T cells in both the spleen and lymph nodes (T.4Mem.LN, T.4Mem.Sp, T.4Mem44h62l.LN, and T.4Mem44h62l.Sp) showed the most significant gene enrichment (p < 10−6).
Some of the most significant loci might harbor compelling candidate genes (see Table S3A). For example, the rs13385731 locus (empirical locus p = 0.0017 for stage T3 transitional B cells) harbors the RAS pathway gene, RASGRP3 (MIM 609531), which has been shown to potentially play a role in downstream signaling from the B cell receptor.48 In other cases, we are able to identify specifically expressed genes that are not yet well characterized but might warrant further examination. For example, the rs6445975 SNP locus (empirical locus p = 1.6 × 10−5) contains PXK (MIM 611450), encoding a transcription factor whose role in immunology is not yet well characterized but is highly and specifically expressed in transitional B cells.
When we tested Crohn loci for enrichment of specifically expressed genes within the 223 cell types of the ImmGen data set, the single most significant cell type was an epithelial-associated stimulated dendritic cell subset (lung CD11b+ dendritic cells stimulated by polyinosinic:polycytydylic acid, p = 1.6 × 10−5, see Table 1, Figure 2B, and Table S4A). In Crohn disease, other cell types, including a single CD4+ memory T cell subset and natural killer cell subset demonstrate statistical significance after multiple hypothesis testing. Moreover, these effects are independent of the DC effect because their signals are maintained after adjusting for dendritic cell contributions (see Table 1 and Table S4A). Individual genes within loci and their relative significance are listed in Table S3B. Dendritic cells in the intestinal mucosa play a key role in mediating the intestinal inflammation associated with Crohn disease and have long been thought of as key mediators of disease activity.49,50 For example, NOD2 (MIM 605956) Crohn disease risk variants have been shown to disrupt autophagy in dendritic cells.51 The potential role of dendritic cells has been further highlighted in a mouse model where defective transforming growth factor (TGF)-beta activation can result in spontaneous colitis.52
When we tested RA loci for enrichment of specifically expressed genes within the 223 cell types of the ImmGen data set, we observed that each of the four subsets of CD4+ effector memory cells emerge as the most highly significant subset (p < 10−6 for all four subsets of CD4+ effector memory cells; see Table 1, Figure 2C, and Tables S2 and S4A). Strikingly, most of the other cell types achieving statistically significant association (but at a more modest level) are closely related CD4+ T cell subsets. Adjusting for the four profiles of CD4+ effector memory T cells obviates the significance of all of these cell types (see Table 1 and Table S4A), strongly suggesting that the associations found in these other T cell subsets are due to their high correlation in expression with CD4+ effector memory T cells.
Certain SNPs containing highly specifically expressed genes in CD4+ effector memory cells were particularly significant. In many cases these SNPs pointed to well-described candidate genes known to play key roles broadly in CD4+ T cell biology (see Table S3C). As examples, we note multiple genes that are specifically expressed in CD4+ effector memory cells: PTPN22 ([MIM 600716], rs2476601, empirical locus p = 0.056 for CD4+ memory T cells), CD2 ([MIM 186990], rs11586238, p = 0.040), PTPRC ([MIM 151460], rs10919563, p = 0.043), CD28 ([MIM 186760], rs1980422, p = 0.0045), IL2RA (rs2104286, p = 0.0010), and CTLA4 ([MIM 123890], rs3087243, p = 0.0010). The rs2104286 SNP has already been shown to correlate with surface expression of the protein product of IL2RA in CD4+ memory T cells19 and likely has CD4+ effector memory T cell function. However, in at least one instance, we identified a candidate gene that has not been specifically connected to T cell function. For example, the ANKRD55 (rs6859219, p = 0.017) has currently unknown biological function with respect to the immune system but is highly and specifically expressed in CD4+ effector memory cells.
To assess whether results were influenced by loci that overlap multiple diseases, we repeated our analyses for all three diseases excluding those loci that are implicated in more than one disease. This decreased the number of loci per disease substantially; the number of loci was reduced in SLE to 11 (from 27), in Crohn to 56 (from 69), and in RA to 23 (from 39). However, the pattern of tissue specific enrichment was not altered (see Table S4B).
Validating Enrichment of CD4+ Effector Memory T Cell Genes among RA Loci
In order to independently validate the role of CD4+ effector memory T cells in RA, we examined a second set of loci that were nominally associated to RA but not yet considered validated risk loci. Using a polygenic modeling approach, we have separately demonstrated that SNPs with nominal significance at a threshold of pGWAS < 0.001 in the latest RA GWAS meta-analysis are significantly associated with RA risk in aggregate in independent validation samples (E.S., S.R., R.P., unpublished data). We estimated that 5%–15% of the SNPs that define this polygenic signal represented true RA risk alleles (see Stahl et al. Table S2 for estimates), whereas the majority (>85%) of them represented statistical fluctuation. We hypothesized that if these SNPs were indeed enriched for true RA risk loci and if our result that CD4+ effector memory T cells are important for RA holds true, then the nominally associated SNPs should also be modestly enriched for genes specifically expressed in CD4+ effector memory T cells.
To test this hypothesis, we obtained the latest results of an RA GWAS meta-analysis and identified all SNPs with pGWAS < 0.001. To ensure independence, we combined SNPs in LD (r2 > 0.1) into individual loci and for each locus, we picked the single most significant SNP. To ensure that our results were independent of previously known RA loci, we removed all loci in LD with (r2 > 0.1) or sharing implicated genes with a known RA risk locus (see Table S3C). In the aggregate, we obtained a total of 436 loci implicating 1037 genes (see Table S5).
The most significant cell type was a subcutaneous lymph nodes CD4+ effector memory T cell subset (p = 8.2 × 10−5, T.4Mem44h62l.LN CD4+; see Figure 3A). This was the only cell type that obtained significance after correcting for multiple hypothesis testing. Indeed, each of the subsets of CD4+ effector memory T cells demonstrated at least nominally significant association at p < 0.008.
Figure 3.
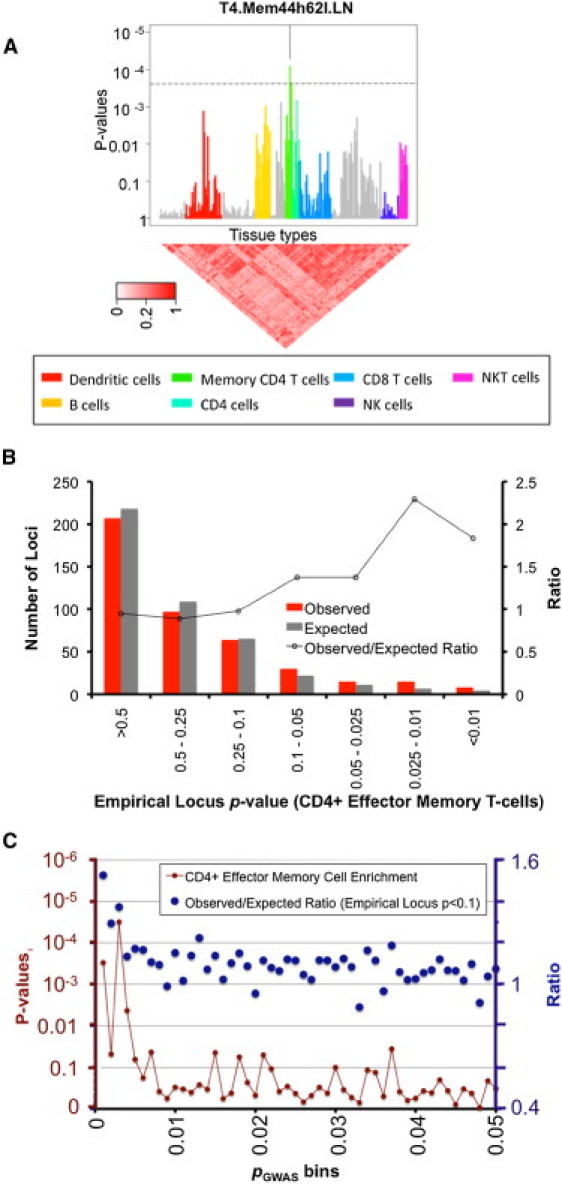
Validating Enrichment of Cell-Specific Expression in RA Loci
We evaluated 436 putative loci containing SNPs nominally associated to rheumatoid arthritis (pGWAS < 0.001) for cell-specific gene enrichment in 223 murine immune cell types. The Bonferroni-corrected p value is shown by a dotted line. (A) We labeled cell types that are significant after multiple hypothesis testing (p < 2.2 × 10−4). Only one of the four CD4+ effector memory cells (T.4Mem44h62l.LN) is significant. (B) We aggregated the expression specificity scores for the four different types of CD4+ effector memory T cells and calculated empirical locus p values for each of the 436 loci. These p values assessed the degree of specificity that the most highly specific CD4+ effector memory T cell genes in each locus achieved. In red, we plotted the histogram of these empirical locus p values, whereas in gray, we plotted the expected histogram of empirical locus p values. We plotted the ratio of those two values at each significance interval. We noted modest deflation at higher values (p > 0.5) and inflation at lower p values (p < 0.1). (C) We grouped loci by their association statistics (pGWAS) into 50 bins ranging from pGWAS < 0.001 (as in pane A and B) to 0.049 pGWAS < 0.05. Then, using aggregated specificity scores for types of CD4+ effector memory T cells, we evaluated these groups to see if they were enriched for specifically expressed genes. For each bin we plot the observed to expected ratio of loci with lower empirical locus p values (p < 0.1, blue, right axis), and the statistical significance of enrichment (red, left axis).
To identify the contribution of the individual loci toward the effector memory T cell enrichment, we averaged the specificity profiles of all four primary subsets of CD4+ effector memory cells together and again tested the aggregate effector memory T cell profile for association among these nominally associated loci. We again observed an association (p = 1.3 × 10−4). Looking at the individual loci and genes, we note that there are 68 loci that show specificity for populations of CD4+ effector memory T cells at a p < 0.1 level, whereas by chance alone we would expect only 43.6 (see Figure 3B). Based on these results, we might expect that as many as 25 true RA risk loci are embedded within this set. Of the loci tested, we list those with the most significant specific expression in CD4+ effector memory T cells (Table 2 and Table S5). We predict that subsequent ongoing genetic association studies for RA will eventually clarify which of these are true RA loci.
Table 2.
Nominally Associated RA Alleles near Genes Specifically Expressed in CD4+ Effector Memory Cells
| SNP | CHR | HG 18 Pos | Empirical Locus p (CD4+ Effector Memory T Cell) | Most Specifically Expressed Gene (Entrez ID, MIM) | Best Assoc RA p Value from GWAS |
|---|---|---|---|---|---|
| rs6683027 | chr1 | 204663522 | 0.0011 | CTSE (1510, 116890) | 4.26 × 10−4 |
| rs11867591 | chr17 | 62021413 | 0.0032 | PRKCA (5578, 176960) | 5.06 × 10−4 |
| rs2023628 | chr8 | 17091505 | 0.0043 | ZDHHC2 (51201, N/A) | 1.32 × 10−4 |
| rs10937694 | chr4 | 5979650 | 0.0054 | CRMP1 (1400, 602462) | 7.64 × 10−4 |
| rs7155603 | chr14 | 75030289 | 0.0084 | BATF (10538, 612476) | 1.53 × 10−5 |
| rs17215817 | chr8 | 131488842 | 0.0094 | DDEF1 (50807, 605953) | 8.22 × 10−5 |
| rs16898297 | chr8 | 101453401 | 0.0096 | RNF19A (25897, 607119) | 7.58 × 10−4 |
| rs7046901 | chr9 | 20236894 | 0.0097 | MLLT3 (4300, 159558) | 6.43 × 10−4 |
| rs10468137 | chr15 | 86012950 | 0.011 | NTRK3 (4916, 191316) | 7.32 × 10−4 |
| rs735684 | chr5 | 141465117 | 0.013 | NDFIP1 (80762, 612050) | 9.71 × 10−5 |
| rs6021275 | chr20 | 49588531 | 0.013 | NFATC2 (4773, 600490) | 6.30 × 10−4 |
| rs7579944 | chr2 | 30298530 | 0.014 | LBH (81606, 611763) | 1.08 × 10−4 |
| rs9907505 | chr17 | 73250509 | 0.017 | SEPT9 (10801, 604061) | 1.94 × 10−5 |
| rs2939931 | chr10 | 121626396 | 0.018 | INPP5F (22876, 609389) | 9.94 × 10−4 |
| rs9366347 | chr6 | 20474041 | 0.019 | MBOAT1 (154141, 611732) | 6.16 × 10−4 |
| rs1422673 | chr5 | 150419181 | 0.020 | TNIP1 (10318, 607714) | 9.51 × 10−5 |
Here, we listed nominally associated SNPs and their p values from RA GWAS (p < 0.001, first and last columns, respectively), their genomic coordinates (second and third column), a significance score suggesting enrichment for a single proximate gene for specific CD4+ effector memory T cells (fourth column), and the candidate gene with the most specific expression in CD4+ effector memory T cells (fifth column).
We assessed the degree of enrichment at more liberal GWAS significance thresholds. In order to do this, we grouped SNPs into 50 pGWAS bins, each of size 0.001, ranging from 0 to 0.05. Then for each group, we quantified the degree to which genes implicated by those SNPs were enriched for specific expression of CD4+ effector memory cells. We observed at least nominally significant enrichment for bins up to pGWAS < 0.005 with very little evidence of any enrichment at pGWAS > 0.02 (see Figure 3C).
Discussion
In the present study, we looked at gene expression data alone to ascertain the key cell types impacted by autoimmune loci. Previously, the potential value of using gene expression data, and other external information sources, in integrative analysis to understand relationships between disease-associated genes and to identify candidate genes for follow-up study has been demonstrated. For example, we have separately integrated protein-protein interaction data with expression data to identify specific pathways in disease.26 As another example, Prioritizer uses a large compendium of gene expression data along with a multitude of other data sources to identify likely candidate genes within loci.53 Chen et al.54 used a large-scale gene expression compendium to look for genes that vary most dramatically across Gene Expression Omnibus and to identify potential candidate genes.
Our approach is contingent on the quality and availability of a high-quality gene-expression database. A comprehensive data set containing all of the necessary human tissue types would be most ideal. Although the GNF data set is reasonably comprehensive, important immune cell types are not always present. On the other hand, ImmGen offers the highest quality and most comprehensive immunological data set that we are aware of. It does lack certain important derived cell types of potential interest. For example, derived helper T cell subgroups such as Th1, Th2, and Th17 cells are not individually profiled. One additional limitation of ImmGen is that it is based on mouse, and not human, tissues. Although the immune systems of the mouse and human are very similar in lineage and structure, there are also important differences. But the breadth of data collected for ImmGen would be impractical to obtain in human.
For each of the autoimmune diseases, we are able to identify very specific subsets of immune cells that could play a critical role in disease and that go well beyond broad immunological categories. For example, for RA we are able to not only establish that CD4+ T cells express genes within RA loci, but we are able to go beyond that and specifically implicate the very specific effector memory subset. All four of the subsets of CD4+ effector memory T cells achieve the greatest significance in this data set and adjusting for their effects obviates the other less significant observations. In this case, we validate our results by looking at independent SNP sets with more nominal disease association.
Intriguingly, we note for the autoimmune diseases that although a single cell type is most strongly associated, there is often evidence that more than one immune-cell type is involved. For example, for RA, there is a nominally significant cell type association for B cell subsets led by follicular B cells (B.Fo.Sp, p = 0.00032), stage 2 transitional B cells (B.T2.Sp, p = 0.00041), and nine other B cell subsets obtained p < 0.01. The loci driving the B cell subset association are distinct from those driving the association of CD4+ effector memory cells (see Figure S5). Thus, adjusting for CD4+ effector memory T cell profiles does not completely remove the B cell association signal. Similarly, for Crohn disease, after adjusting for the main effects of dendritic cells, there are remaining nominal signals in NK and CD4+ T cell subsets. Although these associations are not significant after accounting for the multiple hypotheses tested, they might suggest possible separate roles of other cell types in disease that might become more apparent as additional SNP discoveries accumulate. Because risk alleles across autoimmune diseases are known to overlap, diseases might be best understood by considering individual immune-cell types. Although the distribution of immune-cell types that are critical to particular diseases might vary, overlapping loci between different diseases might be explained by overlapping pathogenic cell types that might play a common role in the different diseases.
Acknowledgments
This work benefitted from data assembled by the ImmGen Consortium. This work was supported in part by the National Institutes of Health National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIH-NIAMS) Development Award (1K08AR055688) and by the Harvard Health Sciences and Technology Program. We thank Joel Hirschorn, Michael B. Brenner, David Altshuler, and Lude Franke for helpful discussions.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
BioGPS, http://biogps.gnf.org/
Immunological Genome Project, http://www.immgen.org/index_content.html
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org/
References
- 1.Barrett J.C., Clayton D.G., Concannon P., Akolkar B., Cooper J.D., Erlich H.A., Julier C., Morahan G., Nerup J., Nierras C., Type 1 Diabetes Genetics Consortium Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet. 2009;41:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raychaudhuri S. Recent advances in the genetics of rheumatoid arthritis. Curr. Opin. Rheumatol. 2010;22:109–118. doi: 10.1097/BOR.0b013e328336474d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franke A., McGovern D.P., Barrett J.C., Wang K., Radford-Smith G.L., Ahmad T., Lees C.W., Balschun T., Lee J., Roberts R. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat. Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Purcell S.M., Wray N.R., Stone J.L., Visscher P.M., O'Donovan M.C., Sullivan P.F., Sklar P., International Schizophrenia Consortium Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., Manolio T.A. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhong H., Beaulaurier J., Lum P.Y., Molony C., Yang X., Macneil D.J., Weingarth D.T., Zhang B., Greenawalt D., Dobrin R. Liver and adipose expression associated SNPs are enriched for association to type 2 diabetes. PLoS Genet. 2010;6:e1000932. doi: 10.1371/journal.pgen.1000932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dimas A.S., Deutsch S., Stranger B.E., Montgomery S.B., Borel C., Attar-Cohen H., Ingle C., Beazley C., Gutierrez Arcelus M., Sekowska M. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325:1246–1250. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Price A.L., Helgason A., Thorleifsson G., McCarroll S.A., Kong A., Stefansson K. Single-tissue and cross-tissue heritability of gene expression via identity-by-descent in related or unrelated individuals. PLoS Genet. 2011;7:e1001317. doi: 10.1371/journal.pgen.1001317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ernst J., Kheradpour P., Mikkelsen T.S., Shoresh N., Ward L.D., Epstein C.B., Zhang X., Wang L., Issner R., Coyne M. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heintzman N.D., Hon G.C., Hawkins R.D., Kheradpour P., Stark A., Harp L.F., Ye Z., Lee L.K., Stuart R.K., Ching C.W. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Firestein G.S. Evolving concepts of rheumatoid arthritis. Nature. 2003;423:356–361. doi: 10.1038/nature01661. [DOI] [PubMed] [Google Scholar]
- 12.Wipke B.T., Allen P.M. Essential role of neutrophils in the initiation and progression of a murine model of rheumatoid arthritis. J. Immunol. 2001;167:1601–1608. doi: 10.4049/jimmunol.167.3.1601. [DOI] [PubMed] [Google Scholar]
- 13.Lee D.M., Friend D.S., Gurish M.F., Benoist C., Mathis D., Brenner M.B. Mast cells: A cellular link between autoantibodies and inflammatory arthritis. Science. 2002;297:1689–1692. doi: 10.1126/science.1073176. [DOI] [PubMed] [Google Scholar]
- 14.Kinne R.W., Bräuer R., Stuhlmüller B., Palombo-Kinne E., Burmester G.R. Macrophages in rheumatoid arthritis. Arthritis Res. 2000;2:189–202. doi: 10.1186/ar86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boilard E., Nigrovic P.A., Larabee K., Watts G.F., Coblyn J.S., Weinblatt M.E., Massarotti E.M., Remold-O'Donnell E., Farndale R.W., Ware J. Platelets amplify inflammation in arthritis via collagen-dependent microparticle production. Science. 2010;327:580–583. doi: 10.1126/science.1181928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pap T., Müller-Ladner U., Gay R.E., Gay S. Fibroblast biology. Role of synovial fibroblasts in the pathogenesis of rheumatoid arthritis. Arthritis Res. 2000;2:361–367. doi: 10.1186/ar113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lefèvre S., Knedla A., Tennie C., Kampmann A., Wunrau C., Dinser R., Korb A., Schnäker E.M., Tarner I.H., Robbins P.D. Synovial fibroblasts spread rheumatoid arthritis to unaffected joints. Nat. Med. 2009;15:1414–1420. doi: 10.1038/nm.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCarroll S.A., Huett A., Kuballa P., Chilewski S.D., Landry A., Goyette P., Zody M.C., Hall J.L., Brant S.R., Cho J.H. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn's disease. Nat. Genet. 2008;40:1107–1112. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dendrou C.A., Plagnol V., Fung E., Yang J.H., Downes K., Cooper J.D., Nutland S., Coleman G., Himsworth M., Hardy M. Cell-specific protein phenotypes for the autoimmune locus IL2RA using a genotype-selectable human bioresource. Nat. Genet. 2009;41:1011–1015. doi: 10.1038/ng.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alizadeh A.A., Eisen M.B., Davis R.E., Ma C., Lossos I.S., Rosenwald A., Boldrick J.C., Sabet H., Tran T., Yu X. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 21.Golub T.R., Slonim D.K., Tamayo P., Huard C., Gaasenbeek M., Mesirov J.P., Coller H., Loh M.L., Downing J.R., Caligiuri M.A. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 22.Hyatt G., Melamed R., Park R., Seguritan R., Laplace C., Poirot L., Zucchelli S., Obst R., Matos M., Venanzi E. Gene expression microarrays: glimpses of the immunological genome. Nat. Immunol. 2006;7:686–691. doi: 10.1038/ni0706-686. [DOI] [PubMed] [Google Scholar]
- 23.Raychaudhuri S., Korn J.M., McCarroll S.A., Altshuler D., Sklar P., Purcell S., Daly M.J., International Schizophrenia Consortium Accurately assessing the risk of schizophrenia conferred by rare copy-number variation affecting genes with brain function. PLoS Genet. 2010;6:e1001097. doi: 10.1371/journal.pgen.1001097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 25.Raychaudhuri S., Plenge R.M., Rossin E.J., Ng A.C.Y., Purcell S.M., Sklar P., Scolnick E.M., Xavier R.J., Altshuler D., Daly M.J., International Schizophrenia Consortium Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 2009;5:e1000534. doi: 10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rossin E.J., Lage K., Raychaudhuri S., Xavier R.J., Tatar D., Benita Y., Cotsapas C., Daly M.J., International Inflammatory Bowel Disease Genetics Constortium Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 2011;7:e1001273. doi: 10.1371/journal.pgen.1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Su A.I., Wiltshire T., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Myers S., Bottolo L., Freeman C., McVean G., Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- 29.Lango Allen H., Estrada K., Lettre G., Berndt S.I., Weedon M.N., Rivadeneira F., Willer C.J., Jackson A.U., Vedantam S., Raychaudhuri S. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teslovich T.M., Musunuru K., Smith A.V., Edmondson A.C., Stylianou I.M., Koseki M., Pirruccello J.P., Ripatti S., Chasman D.I., Willer C.J. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stryer L. W.H. Freeman; New York: 2000. Biochemistry. [Google Scholar]
- 32.Hobbs H.H., Brown M.S., Goldstein J.L. Molecular genetics of the LDL receptor gene in familial hypercholesterolemia. Hum. Mutat. 1992;1:445–466. doi: 10.1002/humu.1380010602. [DOI] [PubMed] [Google Scholar]
- 33.Musunuru K., Strong A., Frank-Kamenetsky M., Lee N.E., Ahfeldt T., Sachs K.V., Li X., Li H., Kuperwasser N., Ruda V.M. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466:714–719. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Speliotes E.K., Willer C.J., Berndt S.I., Monda K.L., Thorleifsson G., Jackson A.U., Allen H.L., Lindgren C.M., Luan J., Magi R. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat. Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clément K., Vaisse C., Lahlou N., Cabrol S., Pelloux V., Cassuto D., Gourmelen M., Dina C., Chambaz J., Lacorte J.M. A mutation in the human leptin receptor gene causes obesity and pituitary dysfunction. Nature. 1998;392:398–401. doi: 10.1038/32911. [DOI] [PubMed] [Google Scholar]
- 36.Pinkney J., Wilding J., Williams G., MacFarlane I. Hypothalamic obesity in humans: what do we know and what can be done? Obes. Rev. 2002;3:27–34. doi: 10.1046/j.1467-789x.2002.00052.x. [DOI] [PubMed] [Google Scholar]
- 37.Willer C.J., Speliotes E.K., Loos R.J., Li S., Lindgren C.M., Heid I.M., Berndt S.I., Elliott A.L., Jackson A.U., Lamina C., Wellcome Trust Case Control Consortium. Genetic Investigation of ANthropometric Traits Consortium Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat. Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raychaudhuri S., Stuart J.M., Altman R.B. Principal components analysis to summarize microarray experiments: application to sporulation time series. Pac. Symp. Biocomput. 2000;2000:455–466. doi: 10.1142/9789814447331_0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gateva V., Sandling J.K., Hom G., Taylor K.E., Chung S.A., Sun X., Ortmann W., Kosoy R., Ferreira R.C., Nordmark G. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat. Genet. 2009;41:1228–1233. doi: 10.1038/ng.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han J.W., Zheng H.F., Cui Y., Sun L.D., Ye D.Q., Hu Z., Xu J.H., Cai Z.M., Huang W., Zhao G.P. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat. Genet. 2009;41:1234–1237. doi: 10.1038/ng.472. [DOI] [PubMed] [Google Scholar]
- 41.Harley J.B., Alarcón-Riquelme M.E., Criswell L.A., Jacob C.O., Kimberly R.P., Moser K.L., Tsao B.P., Vyse T.J., Langefeld C.D., Nath S.K., International Consortium for Systemic Lupus Erythematosus Genetics (SLEGEN) Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat. Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hom G., Graham R.R., Modrek B., Taylor K.E., Ortmann W., Garnier S., Lee A.T., Chung S.A., Ferreira R.C., Pant P.V. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N. Engl. J. Med. 2008;358:900–909. doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- 43.Kozyrev S.V., Abelson A.K., Wojcik J., Zaghlool A., Linga Reddy M.V., Sanchez E., Gunnarsson I., Svenungsson E., Sturfelt G., Jönsen A. Functional variants in the B-cell gene BANK1 are associated with systemic lupus erythematosus. Nat. Genet. 2008;40:211–216. doi: 10.1038/ng.79. [DOI] [PubMed] [Google Scholar]
- 44.Stahl E.A., Raychaudhuri S., Remmers E.F., Xie G., Eyre S., Thomson B.P., Li Y., Kurreeman F.A., Zhernakova A., Hinks A., BIRAC Consortium. YEAR Consortium Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat. Genet. 2010;42:508–514. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhernakova A., Stahl E.A., Trynka G., Raychaudhuri S., Festen E.A., Franke L., Westra H.J., Fehrmann R.S., Kurreeman F.A., Thomson B. Meta-analysis of genome-wide association studies in celiac disease and rheumatoid arthritis identifies fourteen non-HLA shared loci. PLoS Genet. 2011;7:e1002004. doi: 10.1371/journal.pgen.1002004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chan O.T., Madaio M.P., Shlomchik M.J. The central and multiple roles of B cells in lupus pathogenesis. Immunol. Rev. 1999;169:107–121. doi: 10.1111/j.1600-065x.1999.tb01310.x. [DOI] [PubMed] [Google Scholar]
- 47.Navarra S.V., Guzmán R.M., Gallacher A.E., Hall S., Levy R.A., Jimenez R.E., Li E.K., Thomas M., Kim H.Y., León M.G., BLISS-52 Study Group Efficacy and safety of belimumab in patients with active systemic lupus erythematosus: a randomised, placebo-controlled, phase 3 trial. Lancet. 2011;377:721–731. doi: 10.1016/S0140-6736(10)61354-2. [DOI] [PubMed] [Google Scholar]
- 48.Zheng Y., Liu H., Coughlin J., Zheng J., Li L., Stone J.C. Phosphorylation of RasGRP3 on threonine 133 provides a mechanistic link between PKC and Ras signaling systems in B cells. Blood. 2005;105:3648–3654. doi: 10.1182/blood-2004-10-3916. [DOI] [PubMed] [Google Scholar]
- 49.Cho J.H. The genetics and immunopathogenesis of inflammatory bowel disease. Nat. Rev. Immunol. 2008;8:458–466. doi: 10.1038/nri2340. [DOI] [PubMed] [Google Scholar]
- 50.Baumgart D.C., Carding S.R. Inflammatory bowel disease: cause and immunobiology. Lancet. 2007;369:1627–1640. doi: 10.1016/S0140-6736(07)60750-8. [DOI] [PubMed] [Google Scholar]
- 51.Cooney R., Baker J., Brain O., Danis B., Pichulik T., Allan P., Ferguson D.J., Campbell B.J., Jewell D., Simmons A. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat. Med. 2010;16:90–97. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- 52.Travis M.A., Reizis B., Melton A.C., Masteller E., Tang Q., Proctor J.M., Wang Y., Bernstein X., Huang X., Reichardt L.F. Loss of integrin alpha(v)beta8 on dendritic cells causes autoimmunity and colitis in mice. Nature. 2007;449:361–365. doi: 10.1038/nature06110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Franke L., van Bakel H., Fokkens L., de Jong E.D., Egmont-Petersen M., Wijmenga C. Reconstruction of a functional human gene network, with an application for prioritizing positional candidate genes. Am. J. Hum. Genet. 2006;78:1011–1025. doi: 10.1086/504300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen R., Morgan A.A., Dudley J., Deshpande T., Li L., Kodama K., Chiang A.P., Butte A.J. FitSNPs: highly differentially expressed genes are more likely to have variants associated with disease. Genome Biol. 2008;9:R170. doi: 10.1186/gb-2008-9-12-r170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


