Abstract
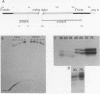
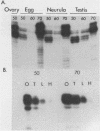
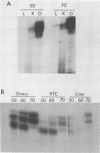
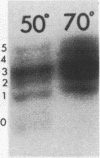
Primer extension analysis has been used to study the principal H4 mRNAs present at different developmental stages and in several adult tissues of Xenopus borealis and X. laevis. In X. borealis a single sequence class predominates in oocytes, tadpoles and cultured fibroblasts. There is also a polymorphic minor type which shows no developmental regulation. The primer extension bands obtained from adult liver and kidney RNA appear to be the same as ovary and therefore these tissues almost certainly contain the same major H4 mRNA species. This is confirmed by S1 mapping of the 3' end of the mRNA. Thus for H4 genes in X. borealis there is no evidence of the kind of switches in histone gene expression seen in sea urchins or certain protostomes. The situation in X. laevis is complicated by considerably higher gene variability both within and between individuals. Nevertheless, in this species, as in X. borealis, there seems to be no major developmental switch in the regulation of H4 gene expression, a conclusion that also holds for an H1C and an H3 gene.
Full text
PDF



















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adamson E. D., Woodland H. R. Histone synthesis in early amphibian development: histone and DNA syntheses are not co-ordinated. J Mol Biol. 1974 Sep 15;88(2):263–285. doi: 10.1016/0022-2836(74)90481-1. [DOI] [PubMed] [Google Scholar]
- Arceci R. J., Gross P. R. Histone gene expression: progeny of isolated early blastomeres in culture make the same change as in the embryo. Science. 1980 Aug 1;209(4456):607–609. doi: 10.1126/science.7394523. [DOI] [PubMed] [Google Scholar]
- Borun T. W., Scharff M. D., Robbins E. Rapidly labeled, polyribosome-associated RNA having the properties of histone messenger. Proc Natl Acad Sci U S A. 1967 Nov;58(5):1977–1983. doi: 10.1073/pnas.58.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler W. B., Mueller G. C. Control of histone synthesis in HeLa cells. Biochim Biophys Acta. 1973 Feb 4;294(1):481–496. doi: 10.1016/0005-2787(73)90104-4. [DOI] [PubMed] [Google Scholar]
- Casey J., Davidson N. Rates of formation and thermal stabilities of RNA:DNA and DNA:DNA duplexes at high concentrations of formamide. Nucleic Acids Res. 1977;4(5):1539–1552. doi: 10.1093/nar/4.5.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Cohen L. H., Newrock K. M., Zweidler A. Stage-specific switches in histone synthesis during embryogenesis of the sea urchin. Science. 1975 Dec 5;190(4218):994–997. doi: 10.1126/science.1237932. [DOI] [PubMed] [Google Scholar]
- DeLeon D. V., Cox K. H., Angerer L. M., Angerer R. C. Most early-variant histone mRNA is contained in the pronucleus of sea urchin eggs. Dev Biol. 1983 Nov;100(1):197–206. doi: 10.1016/0012-1606(83)90211-7. [DOI] [PubMed] [Google Scholar]
- DeLisle A. J., Graves R. A., Marzluff W. F., Johnson L. F. Regulation of histone mRNA production and stability in serum-stimulated mouse 3T6 fibroblasts. Mol Cell Biol. 1983 Nov;3(11):1920–1929. doi: 10.1128/mcb.3.11.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel J. D., Dodgson J. B. Histone genes are clustered but not tandemly repeated in the chicken genome. Proc Natl Acad Sci U S A. 1981 May;78(5):2856–2860. doi: 10.1073/pnas.78.5.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallwitz D. Kinetics of inactivation of histone mRNA in the cytoplasm after inhibition of DNA replication in synchronised HeLa cells. Nature. 1975 Sep 18;257(5523):247–248. doi: 10.1038/257247a0. [DOI] [PubMed] [Google Scholar]
- Gallwitz D., Mueller G. C. Histone synthesis in vitro on HeLa cell microsomes. The nature of the coupling to deoxyribonucleic acid synthesis. J Biol Chem. 1969 Nov 10;244(21):5947–5952. [PubMed] [Google Scholar]
- Heintz N., Sive H. L., Roeder R. G. Regulation of human histone gene expression: kinetics of accumulation and changes in the rate of synthesis and in the half-lives of individual histone mRNAs during the HeLa cell cycle. Mol Cell Biol. 1983 Apr;3(4):539–550. doi: 10.1128/mcb.3.4.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz N., Zernik M., Roeder R. G. The structure of the human histone genes: clustered but not tandemly repeated. Cell. 1981 Jun;24(3):661–668. doi: 10.1016/0092-8674(81)90092-1. [DOI] [PubMed] [Google Scholar]
- Hentschel C. C., Birnstiel M. L. The organization and expression of histone gene families. Cell. 1981 Aug;25(2):301–313. doi: 10.1016/0092-8674(81)90048-9. [DOI] [PubMed] [Google Scholar]
- Hereford L. M., Osley M. A., Ludwig T. R., 2nd, McLaughlin C. S. Cell-cycle regulation of yeast histone mRNA. Cell. 1981 May;24(2):367–375. doi: 10.1016/0092-8674(81)90326-3. [DOI] [PubMed] [Google Scholar]
- Jacob E. Histone-gene reiteration in the genome of mouse. Eur J Biochem. 1976 May 17;65(1):275–284. doi: 10.1111/j.1432-1033.1976.tb10415.x. [DOI] [PubMed] [Google Scholar]
- Kedes L. H., Gross P. R. Identification in cleaving embryos of three RNA species serving as templates for the synthesis of nuclear proteins. Nature. 1969 Sep 27;223(5213):1335–1339. doi: 10.1038/2231335a0. [DOI] [PubMed] [Google Scholar]
- Kedes L. H. Histone genes and histone messengers. Annu Rev Biochem. 1979;48:837–870. doi: 10.1146/annurev.bi.48.070179.004201. [DOI] [PubMed] [Google Scholar]
- Mackay S., Newrock K. M. Histone subtypes and switches in synthesis of histone subtypes during Ilyanassa development. Dev Biol. 1982 Oct;93(2):430–437. doi: 10.1016/0012-1606(82)90130-0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Maxson R., Mohun T., Gormezano G., Childs G., Kedes L. Distinct organizations and patterns of expression of early and late histone gene sets in the sea urchin. Nature. 1983 Jan 13;301(5896):120–125. doi: 10.1038/301120a0. [DOI] [PubMed] [Google Scholar]
- Moorman A. F., de Laaf R. T., Destrée O. H., Telford J., Birnstiel M. L. Histone genes from Xenopus laevis: molecular cloning and initial characterization. Gene. 1980 Aug;10(3):185–193. doi: 10.1016/0378-1119(80)90048-7. [DOI] [PubMed] [Google Scholar]
- Old R. W., Woodland H. R., Ballantine J. E., Aldridge T. C., Newton C. A., Bains W. A., Turner P. C. Organization and expression of cloned histone gene clusters from Xenopus laevis and X. borealis. Nucleic Acids Res. 1982 Dec 11;10(23):7561–7580. doi: 10.1093/nar/10.23.7561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry R. P., Kelley D. E. Messenger RNA turnover in mouse L cells. J Mol Biol. 1973 Oct 5;79(4):681–696. doi: 10.1016/0022-2836(73)90071-5. [DOI] [PubMed] [Google Scholar]
- Plumb M., Stein J., Stein G. Coordinate regulation of multiple histone mRNAs during the cell cycle in HeLa cells. Nucleic Acids Res. 1983 Apr 25;11(8):2391–2410. doi: 10.1093/nar/11.8.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst E., Kressmann A., Birnstiel M. L. Expression of sea urchin histone genes in the oocyte of Xenopus laevis. J Mol Biol. 1979 Dec 15;135(3):709–732. doi: 10.1016/0022-2836(79)90173-6. [DOI] [PubMed] [Google Scholar]
- Rickles R., Marashi F., Sierra F., Clark S., Wells J., Stein J., Stein G. Analysis of histone gene expression during the cell cycle in HeLa cells by using cloned human histone genes. Proc Natl Acad Sci U S A. 1982 Feb;79(3):749–753. doi: 10.1073/pnas.79.3.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins E., Borun T. W. The cytoplasmic synthesis of histones in hela cells and its temporal relationship to DNA replication. Proc Natl Acad Sci U S A. 1967 Feb;57(2):409–416. doi: 10.1073/pnas.57.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruberti I., Fragapane P., Pierandrei-Amaldi P., Beccari E., Amaldi F., Bozzoni I. Characterization of histone genes isolated from Xenopus laevis and Xenopus tropicalis genomic libraries. Nucleic Acids Res. 1982 Dec 11;10(23):7543–7559. doi: 10.1093/nar/10.23.7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruderman J. V., Pardue M. L. A portion of all major classes of histone messenger RNA in amphibian oocytes is polyadenylated. J Biol Chem. 1978 Mar 25;253(6):2018–2025. [PubMed] [Google Scholar]
- Ruderman J. V., Woodland H. R., Sturgess E. A. Modulations of histone messenger RNA during the early development of Xenopus laevis. Dev Biol. 1979 Jul;71(1):71–82. doi: 10.1016/0012-1606(79)90083-6. [DOI] [PubMed] [Google Scholar]
- Ruiz-Carrillo A., Affolter M., Renaud J. Genomic organization of the genes coding for the six main histones of the chicken: complete sequence of the H5 gene. J Mol Biol. 1983 Nov 15;170(4):843–859. doi: 10.1016/s0022-2836(83)80191-0. [DOI] [PubMed] [Google Scholar]
- Seiler-Tuyns A., Birnstiel M. L. Structure and expression in L-cells of a cloned H4 histone gene of the mouse. J Mol Biol. 1981 Oct 5;151(4):607–625. doi: 10.1016/0022-2836(81)90426-5. [DOI] [PubMed] [Google Scholar]
- Sierra F., Lichtler A., Marashi F., Rickles R., Van Dyke T., Clark S., Wells J., Stein G., Stein J. Organization of human histone genes. Proc Natl Acad Sci U S A. 1982 Mar;79(6):1795–1799. doi: 10.1073/pnas.79.6.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittman D. B., Chiu I. M., Pan C. J., Cohn R. H., Kedes L. H., Marzluff W. F. Isolation of two clusters of mouse histone genes. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4078–4082. doi: 10.1073/pnas.78.7.4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittman D. B., Graves R. A., Marzluff W. F. Histone mRNA concentrations are regulated at the level of transcription and mRNA degradation. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1849–1853. doi: 10.1073/pnas.80.7.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittman D. B., Graves R. A., Marzluff W. F. Structure of a cluster of mouse histone genes. Nucleic Acids Res. 1983 Oct 11;11(19):6679–6697. doi: 10.1093/nar/11.19.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein J. L., Thrall C. L., Park W. D., Mans R. J., Stein G. S. Hybridization analysis of histone messenger RNA: association with polyribosomes during the cell cycle. Science. 1975 Aug 15;189(4202):557–558. doi: 10.1126/science.1145212. [DOI] [PubMed] [Google Scholar]
- Stephenson E. C., Erba H. P., Gall J. G. Characterization of a cloned histone gene cluster of the newt Notophthalamus viridescens. Nucleic Acids Res. 1981 May 25;9(10):2281–2295. doi: 10.1093/nar/9.10.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson E. C., Erba H. P., Gall J. G. Histone gene clusters of the newt notophthalmus are separated by long tracts of satellite DNA. Cell. 1981 Jun;24(3):639–647. doi: 10.1016/0092-8674(81)90090-8. [DOI] [PubMed] [Google Scholar]
- Strausbaugh L. D., Weinberg E. S. Polymorphism and stability in the histone gene cluster of Drosophila melanogaster. Chromosoma. 1982;85(4):489–505. doi: 10.1007/BF00327345. [DOI] [PubMed] [Google Scholar]
- Tsang A. S., Mahbubani H., Williams J. G. Cell-type-specific actin mRNA populations in dictyostelium discoideum. Cell. 1982 Dec;31(2 Pt 1):375–382. doi: 10.1016/0092-8674(82)90131-3. [DOI] [PubMed] [Google Scholar]
- Turner P. C., Woodland H. R. H3 and H4 histone cDNA sequences from Xenopus: a sequence comparison of H4 genes. Nucleic Acids Res. 1982 Jun 25;10(12):3769–3780. doi: 10.1093/nar/10.12.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner P. C., Woodland H. R. Histone gene number and organisation in Xenopus: Xenopus borealis has a homogeneous major cluster. Nucleic Acids Res. 1983 Feb 25;11(4):971–986. doi: 10.1093/nar/11.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodland H. R. Histone synthesis during the development of Xenopus. FEBS Lett. 1980 Nov 17;121(1):1–10. doi: 10.1016/0014-5793(80)81252-x. [DOI] [PubMed] [Google Scholar]
- Wu R. S., Bonner W. M. Separation of basal histone synthesis from S-phase histone synthesis in dividing cells. Cell. 1981 Dec;27(2 Pt 1):321–330. doi: 10.1016/0092-8674(81)90415-3. [DOI] [PubMed] [Google Scholar]
- van Dongen W., de Laaf L., Zaal R., Moorman A., Destrée O. The organization of the histone genes in the genome of Xenopus laevis. Nucleic Acids Res. 1981 May 25;9(10):2297–2311. doi: 10.1093/nar/9.10.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]