Abstract
As transcriptional regulators, the genes responsible for maintaining circadian rhythm exert influence in a variety of biological processes. Recently, it has been suggested that the core circadian genes may play a role in breast tumorigenesis, possibly by influencing hormone regulation or other cancer-relevant pathways. Here, we examine the role of the central circadian regulator CLOCK in breast cancer by conducting a genetic and epigenetic association study, as well as transcriptional profiling arrays and a pathway-based network analysis. Significant associations were detected between CLOCK tagging SNPs and breast cancer risk, with apparent effect modification by ER/PR status. Furthermore, hypermethylation in the CLOCK promoter was found to reduce breast cancer risk, and these findings were corroborated by publicly available tissue array data, which showed lower levels of CLOCK expression in healthy controls relative to normal or tumor tissue from breast cancer patients. Finally, we silenced CLOCK in vitro and performed a whole genome expression microarray and pathway analysis, which identified a cancer-relevant network of transcripts with altered expression following CLOCK gene knockdown. These findings support the hypothesis that circadian genes may be relevant for tumorigenesis, and suggest that circadian gene variants may represent a novel panel of breast cancer susceptibility biomarkers.
Keywords: CLOCK, Circadian Genetics, Breast Cancer, Genetic variants, Epigenetic variants
Introduction
Nearly all of the planet’s organisms have adapted to the alternating day/night pattern which accompanies the ≈24-hour rotation of the earth. Adaptations to this naturally occurring circadian cycle influence nearly every biological pathway, and disruption of the circadian cycle may negatively affect cellular function, potentially leading to increased susceptibility to certain malignancies, including breast cancer (1, 2). Epidemiological studies have demonstrated that women who work the night shift are at increased risk of developing breast cancer (3-6), leading to the hypothesis that variants in the genes responsible for maintaining circadian rhythm may also influence cancer susceptibility.
The human molecular clockwork is regulated by transcription-translation feedback loops among a small set of core circadian genes (reviewed in (7)). These genes may have an extensive regulatory role, as studies in mice have demonstrated that up to 10% of all genes in the mammalian genome are under some form of circadian control (8). Emerging evidence from animal models suggests that circadian genes may function as oncogenes or tumor suppressors at the systemic, cellular and molecular levels due to their involvement in cell proliferation, apoptosis, cell cycle control, and DNA damage response (reviewed in (9)). The first evidence linking a circadian gene to breast cancer in humans came from an epidemiologic study which demonstrated an association between a structural genetic variant (54bp InDel in exon 18) in PER3 and risk of breast cancer (10). A non-synonymous polymorphism (Ala394Thr) in the circadian gene NPAS2 has also been associated with breast cancer risk (11).
The current study investigates the role of the core circadian gene CLOCK in breast tumorigenesis. CLOCK belongs to the bHLH-PAS family of transcription factors, which, when dimerized with ARNTL, binds to E-box regulatory elements in target promoter regions and enhances target gene expression (12). As the primary stimulus behind the positive component of the circadian feedback system, CLOCK and ARNTL are considered the heart of the circadian molecular autoregulatory loop (13). Here, we report epidemiologic findings from genetic and epigenetic analyses of CLOCK and breast cancer risk. Moreover, we performed a whole genome expression microarray to determine the effect of CLOCK silencing on the expression of cancer-related genes, and to determine whether CLOCK influences biological pathways which may be relevant for breast tumorigenesis. We also searched the public database for gene expression arrays involving normal breast tissue drawn from individuals with breast cancer and healthy controls, in order to investigate whether CLOCK gene alterations were observed in clinical samples.
Materials and Methods
Study population
The study subjects consisted of participants previously enrolled in a Connecticut breast cancer case-control study. The study was approved by the Institutional Review Boards (IRB) at Yale University, the Connecticut Department of Public Health, and the National Cancer Institute. Participation was voluntary, and written informed consent was obtained. The details of the study population, including recruitment details and participant characteristics, have been described previously (14). Briefly, breast cancer patients were identified from computerized patient records at Yale-New Haven Hospital (YNHH) in New Haven County, Connecticut, and from hospital records in Tolland County Connecticut, by the Rapid Case Ascertainment Shared Resource at the Yale Cancer Center. All cases were incident and histologically confirmed (International Classification of Diseases for Oncology, 174.0 –174.9). Patients had no previous history of cancer apart from non-melanoma skin cancer, were between the ages of 30 and 80, and were alive at the time of the interview. Controls at YNHH were identified through computerized files as patients who underwent breast-related surgery at YNHH, but who had histologically confirmed benign breast disease. Tolland County controls younger than 65 were identified through random digit dialing, and those over 65 were identified through Health Care Finance Administration files. Permission to contact the subject was obtained from the hospital, as well as the personal physician for all cases. Potential participants were then contacted first by letter, and then by telephone, if necessary. Subjects who agreed to participate were interviewed by a trained interviewer, who administered a standardized questionnaire and collected blood samples into sodium-heparinized tubes for immediate DNA isolation and subsequent analyses. Participation rates were: 77% for YNHH cases, 71% for YNHH controls, 74% for Tolland County cases, and 61% for Tolland County Controls. Estrogen receptor (ER) and progesterone receptor (PR) status were determined immunohistochemically at YNHH, as previously described (15), with an H-score greater than 75 considered receptor positive. A total of 441 cases and 479 controls had DNA samples available for the current study. Supplementary Table 1 presents the distribution of selected baseline characteristics for all participants.
SNP selection and genotyping
CLOCK gene SNPs were identified using the Haploview interface (16) of HapMap’s genome browser, Release 22 (http://www.hapmap.org/cgi-perl/gbrowse/hapmap22_B36/). Tag SNPs were chosen using the Tagger algorithm (17) employing the pairwise tagging method with the CEU population, an r2 cutoff of 0.8, and a minimum minor allele frequency (MAF) of 0.2. In addition, all SNPs from the CLOCK 3’UTR with allele frequency data available in the dbSNP database and MAF>0.2 in European populations were also included. Genomic DNA was extracted using standard methods and genotyping was performed using the Sequenom MassARRAY multiplex genotyping platform (Sequenom, Inc., San Diego, CA) at Yale University’s W.M. Keck Foundation Biotechnology Research Laboratory. Duplicate samples from 17 study subjects were interspersed throughout the genotyping assays, and the concordance rate for these QC samples was 98%. All genotyping calls, including quality control data, were re-checked by different laboratory personnel and genotyping scores were reproduced with 100% accuracy.
Promoter CpG island identification and methylation analysis
A CpG island in the promoter region of CLOCK was identified using the CpG Island Searcher web tool (http://www.cpgislands.com/). Methylation-specific PCR primers for this region were then designed using the MethPrimer program (www.urogene.org/methprimer), with one pair designed to amplify methylated DNA, and one pair designed to amplify unmethylated DNA. The two methylated primer sequences were: L: 5′-TCGTTTTTTCGGTTTTTTAGTAATC-3′ and R: 5′-CTTACCCCGTTAAACAACACG-3′; and the two unmethylated primer sequences were: L: 5′-TTGTTTTTTTGGTTTTTTAGTAATT-3′ and R: 5′-CTTACCCCATTAAACAACACA-3′. Since radio- and chemotherapy can affect DNA methylation, only patients who had not undergone these treatments were included in this portion of the analysis (n=80), along with an equal number of age-matched controls. Genomic DNA extracted from blood of these subjects was bisulfite treated using the EZ DNA Methylation Kit (Zymo Research, Orange, CA) according to the manufacturer’s protocol, which converts unmethylated cytosines into uracil, and leaves methylated cytosines unchanged. Following the treatment, quantitative PCR was performed using the primers described above and the Power SYBR Green Kit (Applied Biosystems, Foster City, CA) according to the manufacturer’s protocol, in order to distinguish methylated from unmethylated DNA sequences. In order to assign a quantitative measure to the level of methylation, a methylation index (MI) was calculated for each sample using the formula: MI = [1 / (1+2−(CTu-CTme)] × 100%, as previously described (18), where CTu = the average cycle threshold (CT) obtained from duplicate qPCRs using the unmethylated primer pair, and CTme = the average CT obtained using the methylated primer pair. Untreated cases were compared to treated cases on a number of patient characteristics using a chi-square test in order to determine whether the untreated cases examined in the methylation analysis were representative of all cases in the sample.
Expression analysis of CLOCK in breast tumor tissues
We used the Atlas of Gene Expression function implemented in the Array Express database (www.ebi.ac.uk/arrayexpress; accessed on February 21, 2009) to search for expression array comparisons involving breast tissue drawn from breast cancer patients and healthy controls. The keywords used were: Gene: “Clock”; Conditions: “Breast Cancer”; and the results were filtered by species to include only Homo Sapiens. This search returned only one experiment. Further details regarding tissue collection and the experimental protocol of this array are available at the Array Express database under accession number E-TABM-276, or from the primary publication (19).
Cell culture and treatments
Human breast adenocarcinoma cells (MCF-7) (American Type Culture Collection, Manassas, VA) were maintained in Dulbecco’s modified Eagle medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (Invitrogen), 0.01 mg/ml bovine insulin, and 1% penicillin/streptomycin (Sigma-Aldrich, St. Louis, MO). siRNA oligos targeting CLOCK (Catalog #18394) and a scrambled sequence negative control siRNA were designed and manufactured by Ambion, Inc. (Ambion/Applied Biosystems). Each oligo was reverse transfected in 12-well plates with approximately 10,000 cells at a final concentration of 10nM using the Lipofectamine RNAiMax transfection reagent (Invitrogen).
RNA isolation and quantitation
RNA was isolated using the RNA Mini Kit (Qiagen, Valencia, CA), with on-column digestion, according to the manufacturers’ protocol for mammalian cells. RNA was quantified using a NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE), and first-strand cDNA was synthesized using the AffinityScript cDNA kit (Stratagene, La Jolla, CA) with random ninemer primers. Quantitative real-time PCR was performed in duplicate using the Power SYBR Green PCR master mix (Applied Biosystems) with gene-specific primers, and a standard thermal cycling procedure on an ABI 7500 instrument (Applied Biosystems). The primers used for CLOCK amplification were: L: 5′-CAAGGAAATGTGCACTGTTGA-3′, R: 5′-TATTATGGGTGGTGCCCTGT-3′. RNA quantity was normalized using HPRT1 content, and CLOCK silencing was quantified according to the 2−ΔΔCt method.
Gene expression microarray and pathway analysis
Gene expression differences in normal MCF7 cells, and those with reduced CLOCK levels were examined by whole genome microarray (Agilent, Inc. 44k chip, performed by MoGene, LC, St Louis, MO). RNA was isolated from separate cell populations from biological replicates of each treatment condition (CLOCK-targeting or scrambled negative siRNA). Each knockdown experiment and microarray was repeated twice, and the log ratio and gene expression fold change in CLOCK knockdown cells relative to the mock siRNA-treated negative control population were determined for each replicate. Differentially expressed transcripts were interrogated for network and functional inter-relatedness using the Ingenuity Pathway Analysis software tool (Ingenuity Systems, www.ingenuity.com). This software employs an extensive database of functional interactions which are drawn from peer-reviewed publications and are manually maintained (20). P-values for individual networks were obtained by comparing the likelihood of obtaining the same number of transcripts or greater in a random gene set as are actually present in the input set (i.e. the set of genes differentially expressed following CLOCK knockdown) using a Fisher’s exact test, based on the hypergeometric distribution. All microarray data were uploaded to the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/projects/geo/); accession #GSE17766.
Statistical analyses
All statistical analyses were performed using the SAS statistical software, version 9.1 (SAS Institute, Cary, NC), unless otherwise noted. CLOCK knockdown was assessed using the 2−ΔΔCt method with RNA content normalized to the housekeeping gene HPRT1. For the case-control analyses, allelic distributions for all SNPs were tested by goodness-of-fit chi-square for compliance with Hardy-Weinberg equilibrium (HWE) among the controls. Odds ratios and 95% confidence intervals were determined for each SNP-disease association by unconditional multivariate logistic regression including the following covariates: age (continuous), race, family history of cancer in a first-degree relative, study site, menopausal status, parity, and age at menarche. Other covariates, such as alcohol use and smoking, did not alter the parameter estimates and were therefore not included in the final model.
To further explore the relationship between SNPs in the CLOCK gene, all nine CLOCK gene variants were used to construct haplotypes using the PHASE program (21). Each of these was individually analyzed for their association with breast cancer risk, both in the full population and in a stratified analysis by joint ER/PR status. Each haplotype was compared to all other haplotypes in order to determine odds ratios for each SNP combination. Odds ratios and 95% confidence intervals for each haplotype were determined by unconditional multivariate logistic regression, using the same covariates as the main effects model, and with all other haplotypes as the referent category. Odds ratios and 95% confidence intervals for the methylation analysis, in which participants were matched on age, were determined by conditional logistic regression with the same set of covariates.
Due to the multiple comparisons inherent in the microarray analysis, adjustments were made to control for false discoveries. A multiple comparisons correction was applied to each observation using the Benjamini-Hochberg method, as previously described (22), to obtain a false discovery rate-adjusted p-value (referred to as the Q-value). Alpha was set at 0.05, and in order to be considered statistically significant, each transcript had to meet the criteria of Q-value less than 0.05 in both biological replicates. In addition, in order to further reduce false positives, and to enrich for biologically relevant expression changes, only transcripts with a mean fold change greater than ∣2∣ were considered to be “significantly differentially expressed”. Samples with inadequate signal intensity (intensity<50 in both the Cy3 and Cy5 channels) were discarded.
Results
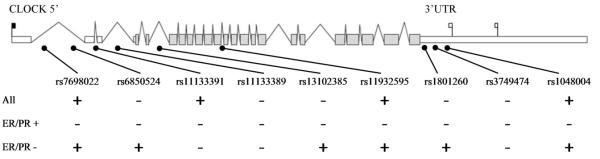
Compared to controls, breast cancer cases were more likely to be post-menopausal (P<0.001), but no other demographic characteristic differed significantly between cases and controls (Supplementary Table 1). Six intronic tag SNPs (rs7698022, rs6850524, rs11133391, rs11133389, rs13102385, and rs11932595) were identified as representative of all variation found within the exonic and intronic regions of the CLOCK gene with r2 ≥ 0.8. Three additional SNPs (rs1801260, rs3749474, and rs1048004) with MAF>0.2 were identified in the CLOCK 3’UTR, and these were also included, as variants in this region may impact phenotype by interrupting microRNA binding sites. All nine SNPs were successfully genotyped, and no departures from Hardy-Weinberg equilibrium were observed among the controls (P>0.05).
CLOCK variants influence breast cancer risk with effect modification by ER/PR status
Of the nine SNPs investigated, four were found to be significantly associated with breast cancer risk assuming a dominant model, including three tagging SNPs (rs7698022, OR=1.34, (95% CI: 1.02-1.76); rs11133391, OR=0.75 (0.56-0.99); and rs11932595, OR=1.43 (1.07-1.91)) and one 3’UTR SNP (rs1048004, OR=1.34 (1.02-1.76). The positions of the genotyped variants and a summary of these results are illustrated in Figure 1. In addition to the main effects model, controls were simultaneously compared to breast cancer cases stratified into two groups corresponding to tumors which were ER and PR positive (H-score>75; n=84) or ER and PR negative (H-score≤75; n=86). Interestingly, six of the nine SNPs were significantly associated with ER/PR negative cases, including three tag SNPs assuming a dominant model (rs6850524, OR=0.45 (0.27-0.76); rs13102385, OR=0.46 (0.27-0.76); and rs11932595, OR=1.88 (1.06-3.31)), as well as the homozygous variant genotypes of one tag SNP (rs7698022, OR=2.87 (1.25-6.59)), and two 3’UTR SNPs (rs1801260, OR=2.57 (1.14-5.82) and rs1048004, OR=2.69 (1.18-6.13). However, none of the nine SNPs were significantly associated with ER/PR positive cancers, despite having approximately equal number of cases in both groups. In addition, the effect of the combined variant genotypes varied significantly according to receptor status for two tag SNPs: rs6850524 (P for interaction=0.021) and rs13102385 (P for interaction=0.027). Of note, these contrasts were less pronounced when examining ER alone, indicating a potentially distinct phenotype in the double negative tumors. Similar results were obtained when restricting the population to Caucasians only. Full genotyping results can be found in Supplementary Table 2.
Figure 1.
CLOCK SNP locations and association with breast cancer risk. SNPs which were significantly associated with breast cancer risk, assuming either a dominant or co-dominant model in each subtype are depicted with a plus sign, while non-significant associations are shown with a minus.
CLOCK haplotypes and breast cancer risk
A total of 27 haplotypes were identified in the full population including all nine CLOCK gene variants. Of these, six appeared at greater than 2% frequency in the population, and greater than 96% of all subjects carried one of these common haplotypes. Since the variant allele in one SNP (rs11133391) was significantly protective in the population, all haplotypes containing this variant were combined, as were any haplotypes containing one or more, two or more, or all three risk alleles (rs7698022, rs11932595, and rs1048004). The association with breast cancer risk of each of these individual haplotypes, as well as the haplotype combinations, is presented in Table 1. In the full population, one haplotype was significantly associated with decreased risk (OR=0.78 (0.61-1.00)), and one haplotype was significantly associated with increased risk (OR=1.87 (1.10-3.18). As expected, the protective haplotype contained the protective allele of rs11133391 and none of the risk alleles, while the haplotype associated with increased risk contained all three risk alleles and did not contain the protective allele. Similarly, having one or more, two or more, or all three risk alleles was significantly associated with increased risk among all women (OR1+=1.23 (1.01-1.48), OR2+=1.31 (1.06-1.61), OR3=1.28 (1.04-1.58)); however, there was no apparent interaction or dose-response relationship. It should be noted that a portion of our control population consists of women who underwent surgery for benign breast conditions. It is possible that some of these women may be at increased risk of developing breast cancer, and assuming that the observed genetic associations are true, inclusion of these women in the control population may bias our estimates toward the null (i.e. the true effect sizes are stronger than those observed).
Table 1.
Haplotypes in CLOCK gene and breast cancer risk
| ER/PR + | ER/PR − | |||||||
|---|---|---|---|---|---|---|---|---|
| Common or Variant Allele* |
Frequency % |
Controls N |
Cases N |
OR† (95% CI) |
Cases N |
OR† (95% CI) |
Cases N |
OR†(95% CI) |
| CVCCVCCCC | 33.48% | 321 | 286 | 0.95 (0.78-1.16) | 54 | 0.94 (0.66-1.36) | 44 | 0.68 (0.46-0.99) |
| VCCCCVVCV | 23.65% | 216 | 215 | 1.18 (0.95-1.48) | 38 | 1.13 (0.75-1.70) | 48 | 1.54 (1.04-2.27) |
| CCVVCCCVC | 18.47% | 191 | 143 | 0.78 (061-1.00) | 29 | 0.79 (0.51-1.24) | 31 | 0.88 (0.57-1.36) |
| CCVVCVCVC | 12.74% | 120 | 110 | 0.97 (0.73-1.28) | 22 | 1.02 (0.62-1.70) | 25 | 1.17 (0.72-1.91) |
| CCVCCCCVC | 4.27% | 40 | 35 | 0.98 (0.61-1.57) | 5 | 0.78 (0.29-2.07) | 5 | 0.87 (0.32-2.32) |
| VVCCCVVCV | 3.56% | 24 | 38 | 1.87 (1.10-3.18) | 11 | 2.25 (1.04-4.87) | 6 | 1.33 (0.52-3.43) |
|
Protective
Allele |
36.49% | 363 | 293 | 0.81 (0.66-0.98) | 58 | 0.82 (0.57-1.17) | 61 | 0.91 (0.63-1.29) |
| 1+ Risk Alleles | 42.81% | 386 | 389 | 1.23 (1.01-1.48) | 76 | 1.27 (0.90-1.79) | 85 | 1.51 (1.07-2.14) |
| 2+ Risk Alleles | 28.43% | 265 | 250 | 1.31 (1.06-1.61) | 51 | 1.32 (0.90-1.92) | 57 | 1.59 (1.10-2.30) |
| 3 Risk Alleles | 28.11% | 249 | 260 | 1.28 (1.04-1.58) | 49 | 1.26 (0.86-1.84) | 57 | 1.60 (1.11-2.32) |
SNP order: rs7698022, rs6850524, rs11133391, rs11133389, rs13102385, rs11932595, rs1801260, rs3749474, rs1048004
Adjusted for age, race, family history of breast cancer, study site, menopausal status, age at menarche, and parity.
Interestingly, the two most common haplotypes, which were present on more than 57% of the total chromosomes, were each significantly associated with breast cancer risk among women with ER/PR negative tumors. The most common haplotype (33.5% frequency) was significantly associated with reduced risk in the ER/PR negative group (OR=0.68 (0.46-0.99)), while the second most common haplotype (23.7% frequency) was significantly associated with increased risk (OR=1.54 (1.04-2.27)). Haplotypes containing one or more, two or more, or all three risk alleles were all significantly associated with ER/PR negative cancer risk (OR=1.51 (1.07-2.14), OR=1.59 (1.10-2.30), and OR=1.60 (1.11-2.32), respectively). Only one relatively rare haplotype (3.6% frequency) was significantly associated with increased risk in the ER/PR positive group (OR=2.25 (1.04-4.87)).
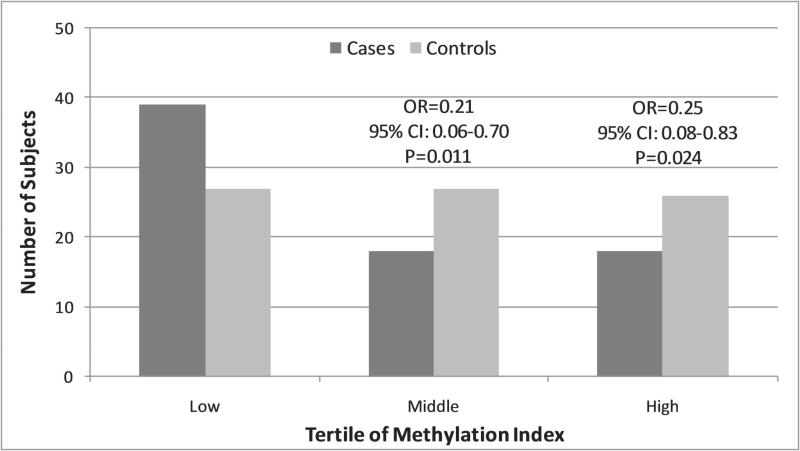
CLOCK promoter hypermethylation reduces breast cancer risk
Although only untreated cases could be used for methylation analysis, no differences were detected among several characteristics between untreated and treated cases (Supplementary Table 3). Subjects were arranged into tertiles of low, mid, and high CLOCK promoter methylation, based on the distribution of the methylation indices among the controls. Interestingly, the methylation index among controls fit very closely to a normal distribution (P=0.990 for Shapiro-Wilk test for normality), while among cases, the distribution was highly right-skewed, with the mass of the distribution concentrated at the lower methylation indices (Shapiro-Wilk P=0.001). Both mid and high methylation were significantly associated with reduced breast cancer risk, (OR=0.21 (0.06-0.70) and OR=0.25 (0.08-0.83), respectively; Figure 2).
Figure 2.
Distribution of CLOCK promoter methylation among breast cancer cases and controls. Overall, cases were more likely to have lower methylation indices at the CLOCK promoter relative to controls. Odds ratios and 95% confidence intervals are for breast cancer risk in the middle and highest tertile, relative to the lowest tertile of methylation index.
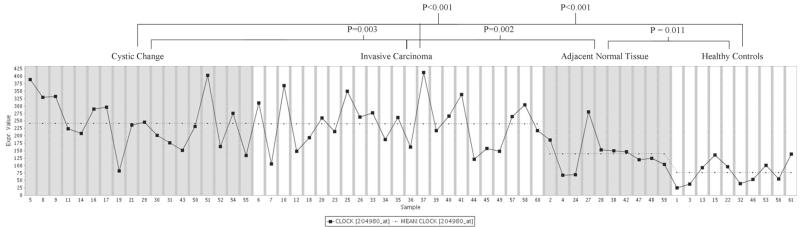
CLOCK is over-expressed in breast tumor tissues using data from a public resource
Since RNA was not available from subjects in our case-control population, we used the Atlas of Gene Expression tool, implemented in the Array Express microarray database (23) to determine whether CLOCK expression was altered in breast cancer patients relative to controls. Searching for “CLOCK” in “breast cancer” returned one array (accession # E-TABM-276), which examined gene expression in: 1) breast tumor tissue (invasive carcinoma), 2) adjacent tissue with cystic changes, mild ductal hyperplasia, or other nonproliferative changes (labeled “cystic change”), 3) adjacent normal breast tissue, and 4) tissue drawn from healthy controls undergoing breast reduction mammoplasty. Normalized mean CLOCK gene expression values for tumor tissue, cystic change, adjacent normal tissue and healthy controls were: 241.2, 242.9, 140.2, and 77.3, respectively (Figure 3). Breast tissue from healthy controls had significantly lower CLOCK gene expression than all breast tissue from breast cancer patients, including adjacent normal tissue (P=0.011; Wilcoxon two-sample test), as well as invasive carcinoma and cystic change (P<0.001 for both comparisons). In addition, CLOCK expression was significantly lower in adjacent normal tissue relative to invasive carcinoma and tissue with cystic change (P=0.003 and P=0.002; respectively). These data suggest that aberrant overexpression of CLOCK may be an early event in cancer development, as there was a clear trend of increasing CLOCK levels in tissue from healthy controls to normal breast tissue from breast cancer patients to nonproliferative tissue and tumor tissue. No difference was observed between the latter two categories. Further comparison by hormone receptor status showed that patients with ER/PR negative tumors had even higher levels of CLOCK gene expression than patients with ER/PR positive tumors (mean expression values of 261.8 and 206.0, respectively; P=0.049), which is consistent with our previous findings suggesting that CLOCK is particularly relevant for ER/PR negative tumorigenesis.
Figure 3.
CLOCK gene expression in breast tissue from breast cancer patients and healthy controls. An array experiment was identified in the publicly accessible Array Express database (accession #E-TABM-276) which compared CLOCK gene expression in breast tissue from patients with breast cancer to CLOCK expression in breast tissue from healthy controls. Mean normalized expression was 241.2 for invasive carcionomas, 242.9 in tissues with a nonproliferative change (cystic change), 140.2 in normal tissue adjacent to the tumor, and 77.3 for healthy controls undergoing breast reduction.
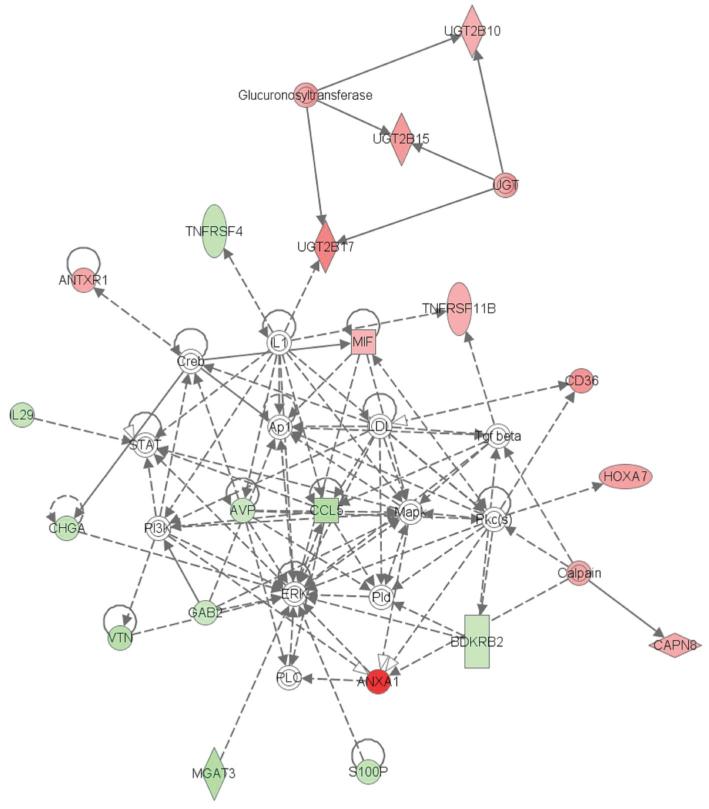
Cancer-related network formed by CLOCK influenced genes
Since CLOCK is a transcriptional regulator, we performed a loss-of-function analysis using CLOCK-targeting siRNA oligos, followed by a whole genome expression microarray to determine which genes and biological pathways may be regulated, directly or indirectly, by CLOCK. Prior to each microarray, CLOCK knockdown was confirmed by qPCR, and in each replicate CLOCK was reduced by 82% (ΔΔCt=2.5). In the array, 154 transcripts fit our significance criteria fordifferential expression following CLOCK knockdown (Q<0.05 and mean fold change ≥ ∣2∣), and this gene set was examined for functional inter-relatedness using the Ingenuity Pathway Analysis (IPA) software tool. The highest confidence functional network (P=1.0E−41) associated with the CLOCK-affected genes was defined as “Cellular Growth and Proliferation, Cell Signaling and Interaction”, and contained several transcripts which may be relevant for breast carcinogenesis and tumor progression (Figure 4). Notable breast cancer-related genes that were up-regulated following CLOCK-knockdown included ANXA1 (5.6-fold increase, Q=1.67E−11), and CD36 (2.9-fold increase, Q=1.25E−5) both of which have protective roles in breast carcinogenesis and are differentially expressed according to estrogen and progesterone receptor status (24, 25). Notable down-regulated genes included CCL5 (2.9-fold decrease, Q=3.17E−6), BDKRB2 (2.1-fold decrease, Q=1.63E−3), and SP100 (2.3-fold decrease, Q=5.00E−5); all of which are positively associated with cell proliferation, and encourage breast tumor promotion or progression (26-28). A summary of breast cancer-relevant genes in this network, along with a brief description of relevant functions and fold-changes following CLOCK knockdown, is presented in Table 2.
Figure 4.
Highest confidence network of genes influenced by CLOCK knockdown. The network is relevant to “Cellular Growth and Proliferation, Cell Signaling and Interaction”. Transcripts which were up-regulated following CLOCK-knockdown are depicted in red, and down-regulated molecules are shown in green. Each interaction is supported by at least one literature reference identified in the Ingenuity Pathway Knowledge Base, with solid lines representing direct interactions, and dashed lines representing indirect interactions.
Table 2.
Molecules in the highest confidence (P=1.0E−41) network of genes differentially expressed following CLOCK knockdown.
| Symbol | Accession | Relevant Functions | Fold change |
Q-Value |
|---|---|---|---|---|
| ANTXR1 | NM_032208 | Angiogenesis, Cell Migration - Elevated in Tumors | 2.39 | 4.97E-05 |
| ANXA1 | NM_000700 | Malignant Transformation - Lost in Most Breast Carcionomas | 5.62 | 1.67E-11 |
| Ap1 | NM_002228 | Transcription Factor, Tumor Promotion | 1.30 | 3.18E-01 |
| AVP | NM_000490 | Growth Factor, Cell Cycle Progression | −2.00 | 1.18E-03 |
| BDKRB2 | NM_000623 | Breast Cell Proliferation | −2.06 | 1.63E-03 |
| CAPN8 | XM_938885 | Membrane Trafficking, Estrogen Responsive | 2.36 | 8.29E-04 |
| CCL5 | NM_002985 | Cell Cycle Progression, Elevated in Breast Carcinomas | −2.94 | 3.17E-06 |
| CD36 | NM_001001547 | Inhibitor of Angiogenesis, Down-Regulated by Estradiol | 2.94 | 1.25E-05 |
| CHGA | NM_001275 | Estrogen Receptor Responsive, Associated with ERa positivity | −2.27 | 8.12E-04 |
| GAB2 | NM_012296 | Cell Proliferation and Breast Carcinogenesis | −2.02 | 1.76E-03 |
| HOXA7 | NM_006896 | Transcription Factor, Inhibits Differentiation | 2.54 | 1.21E-03 |
| IL29 | NM_172140 | Cytokine with Antiviral and Antiproliferative Activity | −2.20 | 3.99E-03 |
| MGAT3 | AK125361 | Glycosyltransferase, Inhibits Tumor Metastisis | −2.91 | 1.02E-03 |
| MIF | NM_002415 | Cytokine, Enhances Tumor Growth and Angiogenesis | 2.04 | 7.63E-05 |
| S100P | NM_005980 | Induction of Metastisis and Breast Tumor Progression | −2.29 | 5.99E-05 |
| TNFRSF11B | NM_002546 | Cancer Cell Migration and Differentiation | 2.18 | 1.65E-08 |
| TNFRSF4 | NM_003327 | Member of the TNFR Superfamily, Facilitates Tumor Rejection | −2.32 | 1.57E-02 |
| UGT2B10 | NM_001075 | Phase II Metabolic Enzyme | 2.20 | 2.78E-04 |
| UGT2B15 | NM_001076 | Phase II Metabolic Enzyme, Regulated by Androgen and Estrogen | 2.74 | 2.44E-06 |
| UGT2B17 | NM_001077 | Phase II Metabolic Enzyme, Regulated by Androgen | 3.36 | 6.37E-05 |
| VTN | NM_000638 | Cellular Adhesion and Migration | −2.69 | 3.78E-04 |
Note: The following transcripts were included in the network, but refer to a gene family or other molecule with no unique mRNA transcript for assignment of fold change or P-value, and were thus excluded from the table: Glucuronosyltransferase, Calpain, Creb, ERK, MAPK, IL1, LDL, PI3K Pkc(s), PLC, PLD, STAT, Tgf beta, and UGT.
Discussion
Our understanding of the complex mechanics behind circadian rhythmicity, including the interplay between environmental cues and endogenous molecular timekeepers, is continually evolving. However, it is clear that CLOCK is at the heart of the molecular autoregulatory feedback loop, and that, in addition to maintaining the circadian cycle, CLOCK is responsible, directly or indirectly, for regulating a number of clock-controlled genes, with a wide variety of biological functions, including those with relevance for carcinogenesis (12, 29). In its capacity as a transcriptional enhancer, CLOCK has been shown to directly mediate genes important for cell cycle control (29), and fibroblasts derived from CLOCK-deficient mice had significantly inhibited cell growth and proliferation relative to wild type (30). A similar study also found that lymphoid tissues from mice with a functional deficiency in CLOCK have diminished proliferation and increased apoptotic activity (31). The rhythmic expression of several cyclins, as well as other transcripts involved in cell cycle control, are regulated by the circadian clock in humans (32), and a recent study also shows that CLOCK is a histone acetyltransferase, adding another potential avenue by which CLOCK may regulate transcriptional activation (33).
These previous findings, which consistently suggest that CLOCK plays an important role in encouraging cell cycle progression, are consistent with the results from our methylation analysis, which demonstrate that increased methylation in the promoter region of CLOCK is associated with decreased breast cancer risk. Of note, due to the potential for radio- or chemotherapy to influence global methylation patterns, only women who had not undergone these treatments at the time of blood collection were eligible for the methylation analysis. As such, our results may not be readily generalizeable to all breast cancer cases, and should be interpreted accordingly; although no significant demographic differences were apparent between untreated and treated cases. Another potential concern is whether the observed epigenetic changes in surrogate tissue (peripheral blood lymphocytes; PBLs) accurately reflect changes in the target tissue. A previous study showed good agreement between methylation of IGF2 in PBLs and colon tissue (kappa statistic = 86.5%, p < 0.0001) (34), and a recent large scale case-control study of breast cancer also detected a significant association between the methylation of several ER-α target (ERT) genes measured in PBLs and human breast cancer risk (35). While these studies demonstrate, in principle, that methylation in PBLs may be reasonable surrogates for use in association analyses, we do not have RNA available for patients in our sample, and it is therefore difficult to determine the phenotypic impact of hypermethylation in this region. Our preliminary hypothesis is that increased methylation would lead to decreased gene expression, thereby diminishing the proliferative effect of CLOCK. This is consistent with the results obtained from a publicly available tissue expression array, which showed that breast tissue samples taken from healthy controls had significantly lower CLOCK expression than tissue from breast cancer patients, and that tumor tissue had higher CLOCK levels than adjacent normal tissue.
The implication that CLOCK may have oncogenic properties is further supported by the findings from our whole genome expression microarray experiment, which showed that expression of several cancer-related transcripts is significantly altered following CLOCK gene knockdown. Some of the genes most relevant for breast carcinogenesis which were down-regulated following CLOCK gene silencing included CCL5 (2.9-fold decrease, Q=3.17E−6), which is associated with cell cycle regulation and breast cancer progression (26), BDKRB2 (2.1-fold decrease, Q=1.63E−3), which induces proliferation in human epithelial breast cells (27), and SP100 (2.3-fold decrease, Q=5.00E−5), which is associated with induction of metastasis, breast tumor progression, and poor survival (28). Genes which were up-regulated following CLOCK knockdown included ANXA1 (5.6-fold increase, Q=1.67E−11), which is often lost in breast carcinomas, but is maintained in many ER and PR negative tumors (24) and CD36 (2.9-fold increase, Q=1.25E−5) which has anti-angiogenic activity and may also be differentially expressed in ER/PR negative tumors (25). The direction of each of these alterations is consistent with CLOCK operating to encourage cell proliferation, as well as breast tumor promotion and progression.
An interesting and unexpected finding was that the effect of CLOCK SNPs on breast cancer risk appeared to be mediated by estrogen and progesterone receptor status, with the strongest associations observed among cases with ER/PR negative tumors. While the direct effect of CLOCKon estrogen-response pathways remains unclear, data from the transcriptional profiling element of our analysis showed that CLOCK gene expression was significantly higher in tissue extracted from patients with ER/PR negative tumors relative to those with ER/PR positive cancers. Moreover, two common haplotypes were significantly associated with ER/PR negative breast cancer risk, indicating that these markers may have broad public health impact. It should be noted, however, that the cells used for our microarray analysis (MCF-7) do express ER and PR. It is therefore unclear whether the regulatory influence of CLOCK is altered in the absence of these receptors, and future investigations may wish to focus on these relationships in order to further characterize the interactions between the circadian system and hormone signaling pathways. Mechanistic data in this area may provide new insights in the development of effective therapeutic strategies for tumors which do not respond to currently available therapies such as tamoxifen, which operates by interfering with the estrogen receptor, and is therefore only effective in ER positive tumors. Furthermore, since RNA was not available for the participants in our study, we are not able to characterize any relationship between genotype or epitype and gene expression. As such, in addition to a direct examination of the effect of promoter methylation on CLOCK expression, future investigations may focus on determining whether variants in the 3′ UTR can influence miRNA binding capacity, thereby affecting translation; particularly since two significant associations were detected for variants in this region.
In conclusion, these findings provide further evidence in support of a role for circadian genes in breast cancer development, and suggest that CLOCK may play a particularly prominent role in regulating breast cancer-related biological pathways. The finding that CLOCK gene variants were of particular significance for ER/PR negative tumors is especially notable, as women with these tumors have the poorest prognosis, and do not benefit from treatment with selective estrogen receptor modulators. As such, further mechanistic investigation into the impact of CLOCK is warranted in order to advance our understanding of the role of circadian rhythm in breast tumorigenesis, and to aid in the development of novel and targeted therapeutic strategies.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (grants CA122676, and CA110937). We would also like to thank Irina Tikhonova at Yale University’s W.M. Keck Foundation Biotechnology Research Laboratory for Sequenom genotyping analysis.
References
- 1.Stevens RG. Electric power use and breast cancer: a hypothesis. Am J Epidemiol. 1987;125:556–61. doi: 10.1093/oxfordjournals.aje.a114569. [DOI] [PubMed] [Google Scholar]
- 2.Stevens RG. Circadian disruption and breast cancer: from melatonin to clock genes. Epidemiology. 2005;16:254–8. doi: 10.1097/01.ede.0000152525.21924.54. [DOI] [PubMed] [Google Scholar]
- 3.Stevens RG, Rea MS. Light in the built environment: potential role of circadian disruption in endocrine disruption and breast cancer. Cancer Causes Control. 2001;12:279–87. doi: 10.1023/a:1011237000609. [DOI] [PubMed] [Google Scholar]
- 4.Hansen J. Increased breast cancer risk among women who work predominantly at night. Epidemiology. 2001;12:74–7. doi: 10.1097/00001648-200101000-00013. [DOI] [PubMed] [Google Scholar]
- 5.Davis S, Mirick DK, Stevens RG. Night shift work, light at night, and risk of breast cancer. J Natl Cancer Inst. 2001;93:1557–62. doi: 10.1093/jnci/93.20.1557. [DOI] [PubMed] [Google Scholar]
- 6.Schernhammer ES, Laden F, Speizer FE, et al. Rotating night shifts and risk of breast cancer in women participating in the nurses’ health study. J Natl Cancer Inst. 2001;93:1563–8. doi: 10.1093/jnci/93.20.1563. [DOI] [PubMed] [Google Scholar]
- 7.Young MW, Kay SA. Time zones: a comparative genetics of circadian clocks. Nat Rev Genet. 2001;2:702–15. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- 8.Storch KF, Lipan O, Leykin I, et al. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 9.Fu L, Lee CC. The circadian clock: pacemaker and tumour suppressor. Nat Rev Cancer. 2003;3:350–61. doi: 10.1038/nrc1072. [DOI] [PubMed] [Google Scholar]
- 10.Zhu Y, Brown HN, Zhang Y, Stevens RG, Zheng T. Period3 structural variation: a circadian biomarker associated with breast cancer in young women. Cancer Epidemiol Biomarkers Prev. 2005;14:268–70. [PubMed] [Google Scholar]
- 11.Zhu Y, Stevens RG, Leaderer D, et al. Non-synonymous polymorphisms in the circadian gene NPAS2 and breast cancer risk. Breast Cancer Res Treat. 2008;107:421–5. doi: 10.1007/s10549-007-9565-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gekakis N, Staknis D, Nguyen HB, et al. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–9. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 13.Kondratov RV, Antoch MP. The clock proteins, aging, and tumorigenesis. Cold Spring Harb Symp Quant Biol. 2007;72:477–82. doi: 10.1101/sqb.2007.72.050. [DOI] [PubMed] [Google Scholar]
- 14.Zheng T, Holford TR, Mayne ST, et al. Risk of female breast cancer associated with serum polychlorinated biphenyls and 1,1-dichloro-2,2′-bis(p-chlorophenyl)ethylene. Cancer Epidemiol Biomarkers Prev. 2000;9:167–74. [PubMed] [Google Scholar]
- 15.McCarty KS, Jr., Miller LS, Cox EB, Konrath J, McCarty KS., Sr. Estrogen receptor analyses. Correlation of biochemical and immunohistochemical methods using monoclonal antireceptor antibodies. Arch Pathol Lab Med. 1985;109:716–21. [PubMed] [Google Scholar]
- 16.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 17.de Bakker PI, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nature Genetics. 2005;37:1217–23. doi: 10.1038/ng1669. [see comment] [DOI] [PubMed] [Google Scholar]
- 18.Lu L, Katsaros D, de la Longrais IA, Sochirca O, Yu H. Hypermethylation of let-7a-3 in epithelial ovarian cancer is associated with low insulin-like growth factor-II expression and favorable prognosis. Cancer Res. 2007;67:10117–22. doi: 10.1158/0008-5472.CAN-07-2544. [DOI] [PubMed] [Google Scholar]
- 19.Cheng AS, Culhane AC, Chan MW, et al. Epithelial progeny of estrogen-exposed breast progenitor cells display a cancer-like methylome. Cancer Res. 2008;68:1786–96. doi: 10.1158/0008-5472.CAN-07-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Calvano SE, Xiao W, Richards DR, et al. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–7. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 21.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–89. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 23.Parkinson H, Kapushesky M, Kolesnikov N, et al. ArrayExpress update--from an archive of functional genomics experiments to the atlas of gene expression. Nucleic Acids Res. 2009;37:D868–72. doi: 10.1093/nar/gkn889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao Y, Li Y, Edelweiss M, et al. Loss of annexin A1 expression in breast cancer progression. Appl Immunohistochem Mol Morphol. 2008;16:530–4. doi: 10.1097/PAI.0b013e31817432c3. [DOI] [PubMed] [Google Scholar]
- 25.Uray IP, Liang Y, Hyder SM. Estradiol down-regulates CD36 expression in human breast cancer cells. Cancer Lett. 2004;207:101–7. doi: 10.1016/j.canlet.2003.10.021. [DOI] [PubMed] [Google Scholar]
- 26.Azenshtein E, Luboshits G, Shina S, et al. The CC chemokine RANTES in breast carcinoma progression: regulation of expression and potential mechanisms of promalignant activity. Cancer Res. 2002;62:1093–102. [PubMed] [Google Scholar]
- 27.Greco S, Muscella A, Elia MG, Romano S, Storelli C, Marsigliante S. Mitogenic signalling by B2 bradykinin receptor in epithelial breast cells. J Cell Physiol. 2004;201:84–96. doi: 10.1002/jcp.20052. [DOI] [PubMed] [Google Scholar]
- 28.Wang G, Platt-Higgins A, Carroll J, et al. Induction of metastasis by S100P in a rat mammary model and its association with poor survival of breast cancer patients. Cancer Res. 2006;66:1199–207. doi: 10.1158/0008-5472.CAN-05-2605. [DOI] [PubMed] [Google Scholar]
- 29.Matsuo T, Yamaguchi S, Mitsui S, Emi A, Shimoda F, Okamura H. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302:255–9. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- 30.Miller BH, McDearmon EL, Panda S, et al. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc Natl Acad Sci U S A. 2007;104:3342–7. doi: 10.1073/pnas.0611724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Antoch MP, Gorbacheva VY, Vykhovanets O, et al. Disruption of the circadian clock due to the Clock mutation has discrete effects on aging and carcinogenesis. Cell Cycle. 2008;7:1197–204. doi: 10.4161/cc.7.9.5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bjarnason GA, Jordan RC, Sothern RB. Circadian variation in the expression of cell-cycle proteins in human oral epithelium. Am J Pathol. 1999;154:613–22. doi: 10.1016/S0002-9440(10)65306-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 34.Cui H, Cruz-Correa M, Giardiello FM, et al. Loss of IGF2 imprinting: a potential marker of colorectal cancer risk. Science. 2003;299:1753–5. doi: 10.1126/science.1080902. [DOI] [PubMed] [Google Scholar]
- 35.Widschwendter M, Apostolidou S, Raum E, et al. Epigenotyping in peripheral blood cell DNA and breast cancer risk: a proof of principle study. PLoS ONE. 2008;3:e2656. doi: 10.1371/journal.pone.0002656. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.