Summary
PhoP is considered a virulence regulator despite being conserved in both pathogenic and non-pathogenic Enterobacteriaceae. While Escherichia coli strains represent non-pathogenic commensal isolates and numerous virulent pathotypes, the PhoP virulence regulator has only been studied in commensal E. coli. To better understand how conserved transcription factors contribute to virulence, we characterized PhoP in pathogenic E. coli. Deletion of phoP significantly attenuated E. coli during extraintestinal infection. This was not surprising since we demonstrated that PhoP differentially regulated the transcription of >600 genes. In addition to survival at acidic pH and resistance to polymyxin, PhoP was required for repression of motility and oxygen-independent changes in the expression of primary dehydrogenase and terminal reductase respiratory chain components. All phenotypes have in common a reliance on an energized membrane. Thus, we hypothesized that PhoP mediates these effects by regulating genes encoding proteins that generate proton motive force. Indeed, bacteria lacking PhoP exhibited a hyper-polarized membrane and dissipation of the transmembrane electrochemical gradient increased susceptibility of the phoP mutant to acidic pH, while inhibiting respiratory generation of the proton gradient restored resistance to antimicrobial peptides independent of lipopolysaccharide modification. These findings demonstrate a connection between PhoP, virulence, and the energized state of the membrane.
Keywords: uropathogenic, proton motive force, prophage, UTI, antimicrobial peptide
Introduction
Signal transduction in bacteria mediates changes in gene expression in response to specific external cues. Bacterial pathogens must respond to cues within the host environment to survive immune defense mechanisms and produce virulence factors. The response regulator PhoP has been studied primarily in Salmonella enterica serovar Typhimurium (herein referred to as Salmonella) where it regulates virulence in response to signals encountered by the bacterium within the mammalian host (Soncini et al., 1996, Prost et al., 2007, Bader et al., 2005). In contrast, PhoP has been proposed to regulate genes that promote survival in non-host environments for commensal E. coli (Zwir et al., 2005). Because PhoP and its cognate sensor kinase PhoQ are broadly conserved among many pathogenic and non-pathogenic bacterial species, it has been generally accepted that this two-component regulatory system has evolved to control virulence in pathogens while retaining core functions in non-pathogens (Winfield & Groisman, 2004).
PhoP was first identified and suggested to perform a regulatory function in Salmonella because a strain with a mutation in phoP demonstrated aberrant non-specific acid phosphatase activity (Kier et al., 1979). The relationship between PhoP and virulence was later established by the identification of phoP in a genetic screen that selected for mutants in Salmonella that had impaired survival in macrophages (Fields et al., 1986) and the finding that phoP mutants are attenuated in vivo (Fields et al., 1989, Miller et al., 1989, Galan & Curtiss, 1989). Attenuation of virulence is related to the phoP mutant’s exquisite susceptibility to extracts prepared from neutrophil granules and purified defensins (Fields et al., 1989, Miller et al., 1990). The mechanism responsible for the connection between PhoP and Salmonella virulence is the ability to activate lipopolysaccharide (LPS) modification (Bader et al., 2005, Guo et al., 1997, Bader et al., 2003) and acid resistance genes in direct response to cationic antimicrobial peptides and acidic pH produced by neutrophils and macrophages (Alpuche Aranda et al., 1992, Prost et al., 2007, Bearson et al., 1998). PhoP can also be phosphorylated and control its large regulatory network in response to low concentrations of Ca2+ and Mg2+ at the periplasmic face of the bacterial cytoplasmic membrane (Soncini et al., 1996).
Synergism between antimicrobial peptides, acidic pH, and the oxidative burst is critical for bacterial killing by phagocytes (Babior, 1978, Vazquez-Torres et al., 2000). Unlike antimicrobial peptides that either permeabilize or traverse the Gram negative bacterial outer membrane and facilitate acidification of the periplasm to directly activate PhoPQ (Bader et al., 2003, Bader et al., 2005), reactive oxygen species produced by phagocytes can freely access the bacterial periplasm (Borregaard et al., 1984, Korshunov & Imlay, 2002). In Salmonella, PhoP controls production of the phage-encoded superoxide dismutase SodCI (Golubeva & Slauch, 2006) that is required for host colonization because it protects the cell from killing by the phagocyte oxidase (Figueroa-Bossi & Bossi, 1999, De Groote et al., 1997). Collectively, periplasmic SodCI, enzymes that modify the outer leaflet LPS on the outer membrane, and acid resistance functions are well known PhoP-dependent responses that prevent pathogen killing by host neutrophils and macrophages.
Pathogens, whether intracellular or extracellular, must possess mechanisms to avoid being cleared by host neutrophils or macrophages. Although PhoP has been characterized extensively in the intracellular pathogen Salmonella, and in non-pathogenic strains of E. coli (Minagawa et al., 2003, Monsieurs et al., 2005), its role in controlling virulence gene expression in pathogenic E. coli is largely unknown. We reasoned that phoP would be critical for pathogenic E. coli virulence because of the central role played by the PhoP regulatory system in pathogen survival in a mammalian host (Miller et al., 1989, Oyston et al., 2000). The integration of pathogenicity islands and lysogenic bacteriophages into the E. coli genome has created distinct E. coli intestinal and extraintestinal pathotypes that cause a spectrum of diseases in humans (Welch et al., 2002). We chose to study PhoP in extraintestinal pathogenic E. coli because this pathotype is the only pathogenic E. coli exclusively capable of causing disseminated infections in humans.
Our findings demonstrate that loss of phoP attenuates uropathogenic E. coli virulence during extraintestinal infection. Indeed, among the hundreds of genes that were differentially expressed between wild-type bacteria and the phoP mutant are those required for motility, acid survival, and lipopolysaccharide modification. These findings paralleled phoP mutant phenotypes of hyper-motility, decreased survival at pH 2.5, and increased susceptibility to antimicrobial peptides, respectively. Alterations in the expression of genes encoding respiratory chain components also occurred in the phoP mutant, suggesting that the cytoplasmic membrane is hyper-polarized in the absence of PhoP. All of the described phenotypes have in common a reliance on an energized membrane. This led us to hypothesize that PhoP control of the energized bacterial membrane represents a conserved function that connects pathogen virulence and normal physiology. In this study, we provide evidence for this assertion.
Results
PhoP is required for E. coli virulence during acute urinary tract infection
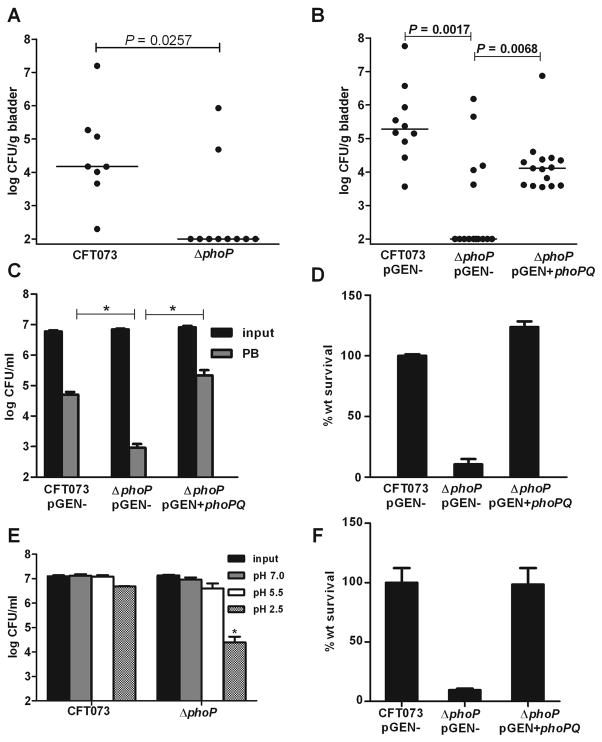
Although PhoP has not been studied in pathogenic E. coli, results from studies in Salmonella led to the assumption that PhoP is likely a regulator of virulence in E. coli. To address this, we generated a phoP deletion mutant in E. coli CFT073, a strain isolated from the blood and urine of a hospitalized patient with acute pyelonephritis (Welch et al., 2002, Mobley et al., 1990). Using a well-established murine model of ascending infection, we found that deletion of phoP resulted in nearly complete attenuation of strain CFT073 during acute cystitis, where 80% of mice lacked detectable bacteria in the bladder 48 h after transurethral independent challenge (P=0.0257) (Fig. 1A). The phoP mutant, complemented with phoPQ on a plasmid, had >100-fold increase in median CFU/g of bladder when compared to ΔphoP containing plasmid vector alone (P=0.0068). As for the wild-type strain, the complemented mutant had detectable bacteria in 100% of mice at 48 h post-inoculation (Fig. 1B).
Fig. 1.
PhoP is required for virulence, resistance to antimicrobial peptides, and survival at acidic pH in E. coli. A. Bladder colonization levels at 48 h post-transurethral inoculation following independent challenge with wild-type uropathogenic E. coli CFT073 and ΔphoP strains. B. In vivo complementation of ΔphoP with phoPQ (pGEN+phoPQ) restores colonization of female CBA/J mice at 48 h following independent challenge. C. CFU ml−1 and D. percent wild-type (wt) survival for CFT073, ΔphoP, and ΔphoP+phoPQ following 45 min incubation of 107 CFU logarithmic phase cells in fresh LB medium containing 10 μM FeCl3 and 2 μg/ml polymyxin B (PB). E. CFU ml−1 and F. percent wild-type (wt) survival for CFT073, ΔphoP, and ΔphoP+phoPQ at 1 h post-inoculation of buffered LB medium pH 2.5 with 107 CFU of bacteria. In A. and B. black dots represent the log CFU g−1 from individual mice and horizontal bars represent the median CFU g−1. P-values were determined using the non-parametric Mann Whitney significance test. In C.-F. Bars represent mean values and significant differences (P < 0.01) determined by Welch’s t test are indicated with an asterisk.
Identification of the pathogenic E. coli PhoP regulon
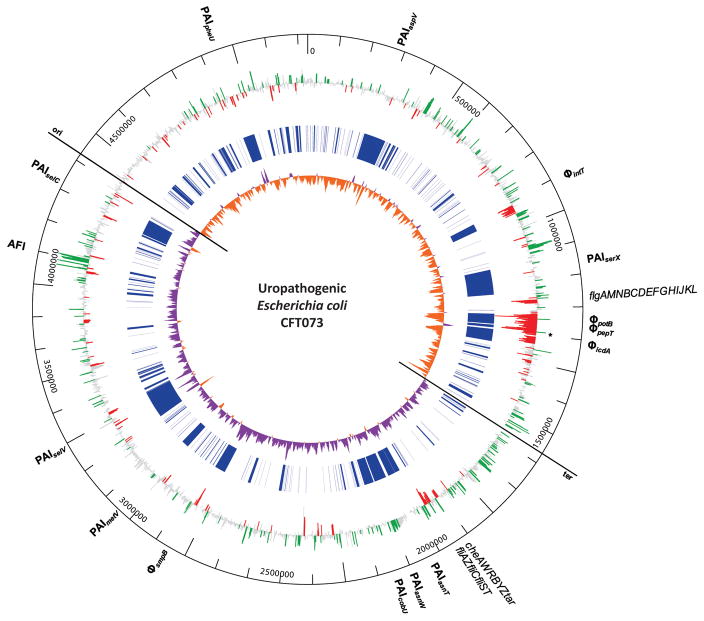
To seek an explanation for attenuation of the phoP mutant and to define the pathogenic E. coli PhoP regulon, we identified genes with significant changes in expression between the parental CFT073 strain and the phoP mutant using microarrays that contained probes representing all 5,379 CFT073 ORFs. The transcription profiles between the parental CFT073 strain and the phoP mutant were compared from aerated, mid-exponential phase cultures in LB medium (OD600=0.6). We reasoned this would identify genes regulated directly or indirectly by PhoP because (i) LB medium is sufficiently Mg2+-limited to activate phoPQ-dependent gene expression (Bishop et al., 2004, Jia et al., 2004, Garcia Vescovi et al., 1996), and (ii) each phoP-dependent phenotype was tested using bacteria cultured under these growth conditions. The microarray analysis showed that deletion of phoP resulted in the differential expression of 620 genes (P<0.05). Of these, 306 and 314 were directly or indirectly activated or repressed by PhoP, respectively (Fig. 2).
Fig. 2.
Whole genome view of PhoP-dependent gene expression and identification of the large PhoP regulon in pathogenic E. coli. Log2-fold changes in gene expression between exponential phase uropathogenic E. coli CFT073 wild-type and ΔphoP were determined using microarrays and plotted as rays from the outer ring. Individual genes that have significantly decreased transcription in the absence of phoP (*) are considered directly or indirectly activated by PhoP (green) while significant increases in gene expression represent directly or indirectly PhoP-repressed genes (red). Gene expression levels that were not considered significant are colored grey. Lines in the center ring (blue) mark uropathogenic E. coli CFT073 genes absent in commensal E. coli K12 strain MG1655 that were identified using Mauve whole genome sequence alignments. The inner-most ring represents positive (purple) and negative (orange) values for GC skew (C−G)/(C+G) calculated using a 10 kb sliding-window. Local changes in base composition bias indicate recent integration sites of foreign DNA or recombination events. The values switch polarity at the origin (ori) and terminus (ter). Coordinates from the CFT073 chromosome, the Acid Fitness Island (AFI), flagellar gene operons, pathogenecity (PAI), and prophage (Φ) islands are labeled on the periphery.
The transcription profile of the phoP mutant (Fig. 2, outer ring) demonstrated activation (green bars) of the colibactin polyketide genotoxin locus (Nougayrede et al., 2006) encoded on PAI-CFT073-asnW, activation of acid fitness island (AFI) genes, repression (red bars) of 36 flagellar and chemotaxis genes (Table S1), and repression of 139 genes encoded within prophage islands are PhoP-dependent. The CFT073 genome contains 14 pathogenicity islands including 5 prophage regions that are absent in the commensal E. coli K-12 strain MG1655 (Fig 2, central blue ring). The gene expression data are consistent with previous studies that described the activation and repression of a similarly large number of genes controlled by PhoP in commensal E. coli (Minagawa et al., 2003, Monsieurs et al., 2005) and Salmonella (Bader et al., 2003, Zwir et al., 2005).
Assessment of established PhoP phenotypes
As predicted by the established role of PhoP in regulating polymyxin and acid resistance genes, deletion of phoP in pathogenic E. coli resulted in significantly decreased transcription of arnBCADTEF, which encode enzymes that catalyze LPS modification by the addition of UDP-4-amino-4-deoxy-L-arabinose to lipid A (Gunn et al., 1998), and genes that encode structural components of the glutamate-, arginine-, and lysine-dependent acid resistance (AR) systems (Foster, 2004) (Table 1). Indeed, the LPS and AR phenotypes were clearly affected by mutation of phoP. E. coli strains produce all of the enzymes necessary for the aminoarabinose modification of LPS; this promotes resistance to host antimicrobial peptides (Trent et al., 2001). This modification to LPS is induced by PhoP in Salmonella, however, a role for PhoP in L-Ara4N modification of lipid A has been presumed to be absent in E. coli because low [Mg2+] failed to promote resistance to an antimicrobial peptide in a commensal strain, and E. coli lacks a functional PmrD that links PhoP to the PmrA regulon (Winfield & Groisman, 2004). Although low [Mg2+] can induce palmitoylation of lipid A (Guo et al., 1998) and antimicrobial peptides can induce expression of the PhoP regulon independent of [Mg2+] (Bader et al., 2005), a phoP mutant in E. coli has not been directly tested for resistance to antimicrobial peptides. To determine if PhoP affects this virulence property, we assessed the survival of the phoP mutant when exposed to bactericidal concentrations of the antimicrobial peptide polymyxin B. Following exposure to polymyxin B, we found that deletion of phoP in pathogenic E. coli resulted in a 100-fold reduction in CFU (Fig. 1C), where the phoP mutant displayed only 1% the survival rate of the wild-type strain or complemented mutant (Fig. 1D). The increased polymyxin sensitivity of the phoP mutant relative to wild-type was independent of [Fe3+] (Fig. S1), a signal known to induce expression of LPS modification genes through activation of the iron-responsive basR (pmrA) transcription factor (Winfield & Groisman, 2004).
Table 1.
Microarray result for lipopolysaccharide modification and acid resistance gene transcription in the absence of phoP.
| Gene | Function | log 2 fold-change | P-value |
|---|---|---|---|
| gadA | glutamate decarboxylase isozyme | −4.9026 | 0.0206 |
| gadY | ncRNA | −4.3170 | 0.0065 |
| gadE | DNA-binding transcriptional activator | −4.1352 | 0.0209 |
| gadB | glutamate decarboxylase isozyme | −4.1239 | 0.0240 |
| hdeD | acid-resistance membrane protein | −4.1196 | 0.0170 |
| gadC | acid sensitivity protein | −4.0879 | 0.0158 |
| hdeB | acid-resistance protein | −3.3836 | 0.0344 |
| hdeA | acid-resistance protein | −3.2784 | 0.0240 |
| gadW | putative transcriptional regulator GadW | −3.0689 | 0.0214 |
| gadX | DNA-binding transcriptional regulator GadX | −2.5727 | 0.0226 |
| arnB | UDP-4-amino-4-deoxy-L-arabinose-- aminotransferase | −1.5925 | 0.0375 |
| arnE | UDP-4-amino-4-deoxy-L-arabinose--flippase | −1.5471 | 0.0090 |
| arnD | UDP-4-amino-4-deoxy-L-arabinose--deformylase | −1.3614 | 0.0307 |
| arnA | UDP-4-amino-4-deoxy-L-arabinose--formyltransferase | −1.2314 | 0.0455 |
| arnC | UDP-4-amino-4-deoxy-L-arabinose--transferase | −1.3350 | 0.0786 |
| arnT | 4-amino-4-deoxy-L-arabinose transferase | −0.9219 | 0.0786 |
A key role for PhoP in Salmonella is to activate LPS modification and other virulence genes during exposure to acidic pH generated by neutrophils and macrophages in host tissues (Alpuche Aranda et al., 1992). A common link between PhoP in pathogens and non-pathogens may feature the response to acidic pH because commensal E. coli lacking phoP has impaired survival at pH 2.5 (Zwir et al., 2005). Consistent with PhoP regulating acid resistance genes, the pathogenic E. coli phoP mutant displayed decreased viability when shifted from neutral conditions to pH 2.5 (Fig. 1E); the increased susceptibility to extreme acid could be reversed by complementing the phoP mutation with phoPQ (Fig. 1F).
Deletion of phoP results in a hyper-motile phenotype
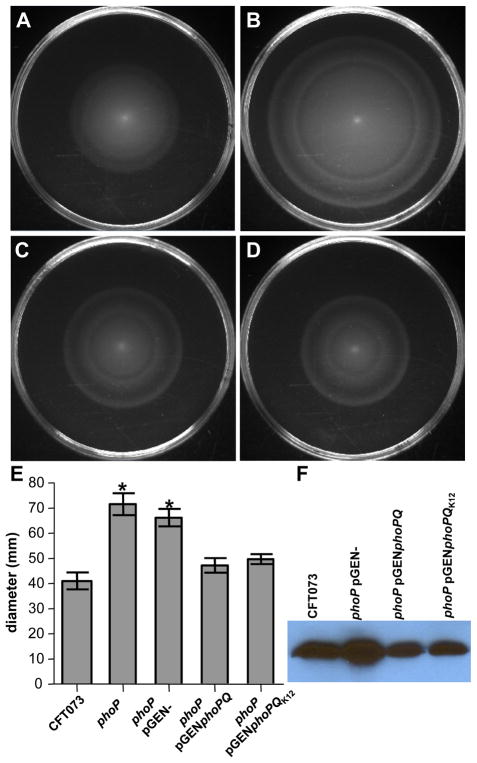
It was also possible to phenotypically validate the gene-chip data by examining motility in soft agar and assessing flagellin production by western blot. In these studies, as expected by the increased flagellar gene transcription in the absence of PhoP, the phoP mutant displayed a hyper-motile phenotype and increased production of FliC (Fig. 3). The increased expression of class II (fliA) and class III (fliC) flagellar genes was confirmed by qPCR and could be independent of class I transcription because expression of flhDC appeared unaffected in the phoP mutant (Fig. S2). Despite significant increases in the expression of 36/50 flagellar genes in the absence of phoP (Table S1), the precise PhoP-mediated control of motility also specifically excluded 5 of 6 type III protein export apparatus components (flhA, flhB, fliP, fliQ, fliR, but not fliO) (Fig. S3). Hyper-motility and increased FliC production in the phoP mutant, was restored to normal levels observed in the wild-type strain by complementation with the wild-type phoPQ allele from either the parental CFT073 or commensal K12 strain MG1655 (Fig. 3).
Fig. 3.
PhoP represses motility in pathogenic E. coli. Microarray analysis showed increased expression of 37/44 flagellar genes in the absence of phoP. A.-D. Soft agar motility plates of A. CFT073, B. ΔphoP, C. ΔphoP complemented with phoPQ or with D. phoPQ from E. coli K12 MG1655. E. Diameter of motility from triplicate experiments (* P < 0.05). F. Western blot with anti-H1 FliC antisera.
Loss of PhoP creates a hyper-polarized membrane
Decreased survival at acidic pH, greater susceptibility to polymyxin B, and increased motility, all directly resulted from the loss of PhoP. Each phenotype potentially shares a common dependence on the energized membrane or would be affected by perturbations in membrane potential. For example, hyper-motility, as observed in the absence of PhoP, could be due to an increase in membrane potential that would generate a large proton motive force and lead to increased proton translocation through the stator complex that drives rotation of the flagellum to propel bacteria. Consistent with the hypothesis that the PhoP regulon controls membrane potential, genes encoding proteins involved in electron transfer, respiration, and ion transport were also shown to be regulated by PhoP in the microarray experiments. Specifically, the altered expression of genes encoding alternative respiratory chain components in phoP mutant bacteria indicated a connection between PhoP and membrane energetics. For example, loss of PhoP resulted in significantly decreased expression of cytochrome bd-II oxidase subunits appB and appC, the electron transfer NADH:quinone oxidoreductase wrbA, and the NADPH:quinone oxidoreductase qor (P < 0.05) (Table 2). In bacteria with a functional PhoP response, PhoP-dependent activation of genes encoding terminal oxidase cytochrome bd-II App and quinone oxidoreductases WrbA and Qor would greatly reduce the extrusion of protons during respiration because these non-electrogenic respiratory components (0H+/e) do not pump protons (Bekker et al., 2009, Natalello et al., 2007). Despite aerobic logarithmic growth conditions in the absence of exogenous nitrate, microarray data also indicated that the respiratory nitrate reductase (NRZ) transcripts narU, narW, and narV were decreased 6.0- to 15.0-fold in the phoP mutant (P < 0.01), while the expression of periplasmic nitrate reductase cytochrome c (Nap) was increased 2.0- to 4.0-fold (P < 0.03) (Table 2). The anaerobic hydrogenase 1 b-type cytochrome hyaABCD that is normally induced under anaerobiosis and repressed by nitrate demonstrated a 6.0- to 12.0-fold decrease (P < 0.01) in transcription in the absence of PhoP (Table 2).
Table 2.
Microarray result for primary dehydrogenase, terminal reductase, and oxidation-related transcription in the absence of phoP.
| Gene | Function | Log 2 fold-change | P-value |
|---|---|---|---|
| yhiD | Mg(2+) transport ATPase inner membrane protein | −3.9398 | 0.0002 |
| narU | Nitrite extrusion protein 2 | −3.8395 | 0.0093 |
| hyaB | hydrogenase 1, large subunit | −3.6945 | 0.0076 |
| hyaA | hydrogenase 1, small subunit | −3.5637 | 0.0049 |
| narW | Respiratory nitrate reductase 2 delta chain | −3.4979 | 0.0042 |
| hyaD | Hydrogenase 1 maturation protease | −2.9709 | 0.0049 |
| sufB | component of SufBCD complex | −2.4839 | 0.0069 |
| narV | Respiratory nitrate reductase 2 gamma chain | −2.4373 | 0.0060 |
| hyaC | Ni/Fe-hydrogenase 1 B-type cytochrome subunit | −2.4161 | 0.0029 |
| sufA | iron-sulfur cluster assembly scaffold protein | −2.4119 | 0.0174 |
| sufE | cysteine desufuration protein SufE | −2.3753 | 0.0076 |
| sufD | component of SufBCD complex | −2.3616 | 0.0149 |
| appC | Cytochrome BD-II oxidase subunit I | −2.3207 | 0.0215 |
| katE | hydroperoxidase HPII(III) (catalase) | −2.3165 | 0.0396 |
| appB | Cytochrome BD-II oxidase subunit II | −2.2607 | 0.0515 |
| sodC | Superoxide dismutase [Cu-Zn] precursor | −2.0652 | 0.0132 |
| hyaF | protein involved in nickel incorporation into hydrogenase-1 proteins | −1.9418 | 0.0130 |
| sufC | cysteine desulfurase ATPase component | −1.7518 | 0.0051 |
| appA | phosphoanhydride phosphorylase | −1.6539 | 0.0429 |
| wrbA | electron transport component WrbA | −1.5563 | 0.0300 |
| ybaR | Copper-transporting P-type ATPase | −1.3787 | 0.0349 |
| yohF | oxidoreductase with NAD(P)-binding Rossmann-fold domain | −1.3696 | 0.0368 |
| fhlA | Formate hydrogenlyase transcriptional activator | −1.3173 | 0.0184 |
| hyaE | protein involved in processing of HyaA and HyaB proteins | −1.2780 | 0.0126 |
| nhaA | Na(+)/H(+) antiporter 1 | −1.2602 | 0.0213 |
| gcd | Glucose dehydrogenase | −1.1960 | 0.0375 |
| qor | quinone oxidoreductase, NADPH-dependent | −1.0977 | 0.0320 |
| ugd | UDP-glucose 6-dehydrogenase | −0.9449 | 0.0277 |
| yadQ | chloride channel protein | −0.9051 | 0.0396 |
| hemH | ferrochelatase | −0.8957 | 0.0269 |
| trkA | potassium transporter peripheral membrane component | −0.7195 | 0.0220 |
| frdC | fumarate reductase subunit C | 0.6357 | 0.0383 |
| frdB | fumarate reductase (anaerobic), Fe-S subunit | 0.6576 | 0.0442 |
| ccmA | Heme exporter protein A | 0.7094 | 0.0308 |
| napG | quinol dehydrogenase periplasmic component | 0.8784 | 0.0174 |
| napH | quinol dehydrogenase membrane component | 0.9033 | 0.0113 |
| pdhR | transcriptional regulator of pyruvate dehydrogenase complex | 1.0407 | 0.0348 |
| napC | nitrate reductase, cytochrome c-type, periplasmic | 1.1595 | 0.0259 |
| ykgF | electron transport amino acid dehydrogenase | 1.6450 | 0.0054 |
| ykgE | oxidoreductase | 1.6883 | 0.0033 |
| napB | nitrate reductase, small, cytochrome C550 subunit, periplasmic | 1.9532 | 0.0143 |
PhoP-dependent modulation of primary dehydrogenase and terminal reductase respiratory component gene expression would configure electron transfer systems that limit generation of the proton motive force (μH+). μH+ is composed of two components, ΔpH and Δψ, and reflects the electrochemical gradient of protons and charge, respectively, across the cytoplasmic membrane. Consistent with our assertion that PhoP controls the expression of genes that encode proteins that affect inner transmembrane potential, a number of ion transport genes [yhiD (mgtC) Mg2+; ybaR Cu2+; yadQ Cl−; and trkA K+] displayed significantly decreased transcription (P < 0.05) in the phoP mutant (Table 2).
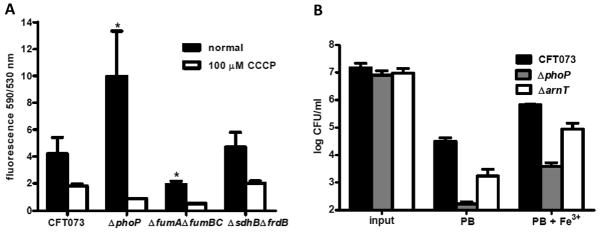
To determine if PhoP controls membrane potential in E. coli, we used the carbocyanine dye 3′3′-diethyloxacarbocyanine iodide (DiOC2) to measure relative changes in membrane potential between wild-type CFT073 and the phoP mutant. DiOC2 exhibits green fluorescence in all bacterial cells and its emission shifts to red fluorescence when the dye self-associates at high intracellular concentrations (Sims et al., 1974). Accumulation of intracellular DiOC2 is directly dependent on membrane potential (positive outside, negative inside). The red:green ratio of DiOC2 was over 2-fold greater for logarithmic phase ΔphoP cells than the ratio observed for wild-type CFT073 (P<0.05) (Fig. 4A). This difference between wild-type and phoP mutant bacteria was notable because CFT073 lacking fumarase activity (ΔfumAΔfumBΔfumC), which is defective in generating μH+ due to the disruption of aerobic and anaerobic fumarate conversions, exhibited a 2-fold reduction in red:green emission as compared to wild-type bacteria (Fig. 4A). Thus, the increased DiOC2 uptake in phoP mutant bacteria results from a hyper-polarized membrane caused by the absence of PhoP. CFT073 mutant bacteria lacking non-electrogenic respiratory components of the TCA cycle (ΔsdhBΔfrdB), which do not extrude protons during electron transfer, do not exhibit any difference in the ratio of red:green fluorescence (Fig. 4A). Depolarizing the electrochemical gradient across the inner membrane using the protonophore m-chlorophenyl carbonyl cyanide hydrozone (CCCP) collapsed the proton gradient and resulted in the expected loss of red fluorescence for all strains (Fig. 4A). These data demonstrated that controlling membrane potential or limiting generation of μH+ is a PhoP-dependent function that could also contribute to PhoP-mediated activities such as survival at acidic pH or resistance to cationic antimicrobial peptides.
Fig. 4.
Prevention of membrane hyper-polarization and lipid A modification with L-Ara-4N are necessary for PhoP-dependent antimicrobial peptide resistance phenotype. A. Ratio of red:green fluorescence for 107 CFU of logarithmic phase CFT073, ΔphoP, ΔfumAΔfumBΔfumC, and ΔsdhBΔfrdA incubated in 30 μM 3′3′-diethyloxacarbocyanine iodide (DiOC2) (black bars). Depolarized control cells were incubated in 30 μM DiOC2 and 100 μM carbonyl cyanide 3-chlorophenylhydrazone (CCCP) (white bars). The green fluorescent carbocyanine dye (DiOC2) shifts to red fluorescence when the dye self-associates at high intracellular concentrations. Bars represent mean values and significant differences determined by Welch’s t test are indicated with an asterisk. B. CFU/ml for CFT073, ΔphoP, and ΔarnT following 45 min incubation of 107 CFU logarithmic phase cells in fresh LB medium containing 10 μM FeCl3 and 2 μg ml−1 polymyxin B (PB) or LB medium containing only 2 μg ml−1 PB. Viable counts were determined from triplicate experiments by plating serial dilutions on LB agar.
PhoP-dependent LPS modification is necessary but not sufficient for resistance to polymyxin B
The most widely accepted strategy to resist killing by cationic bactericidal compounds is to reduce the negative charge of the outer membrane and decrease the affinity of the charged peptides for the outer membrane by modification of the outer leaflet LPS. Aminoarabinose modification of the LPS structure requires arnBCADTEF and has been shown previously to be induced by low pH in E. coli (Trent et al., 2001). Because the operon that encodes the enzymes required to synthesize L-Ara4N was induced by PhoP (Table 1), we generated an arnT deletion mutant in CFT073 to determine if bacteria lacking lipid A modification with L-Ara4N would have a polymyxin B susceptibility phenotype similar to bacteria lacking PhoP. We chose to inactivate arnT because 4-amino-4-deoxy-L-arabinose transferase activity is required for the attachment of L-Ara4N to lipid A and is critical for resistance to polymyxin B (Breazeale et al., 2005). Experiments were performed in the presence of 10 μM Fe3+ to induce expression of pmrA (basR) because PmrA-dependent lipid A modification can occur independently from PhoP due to a non-functional pmrD in E. coli (Wosten et al., 2000, Winfield & Groisman, 2004). We found that deletion of arnT in CFT073 resulted in an intermediate level of polymyxin B sensitivity between wild-type and the phoP mutant (Fig. 4B). We reasoned that an additional modification of lipid A or the loss of the PhoP-dependent alteration of potential across the inner membrane could be responsible for the difference in antimicrobial peptide sensitivity between ΔphoP and ΔarnT bacteria.
Respiratory generation of proton motive force is required for bactericidal activity of polymyxin B
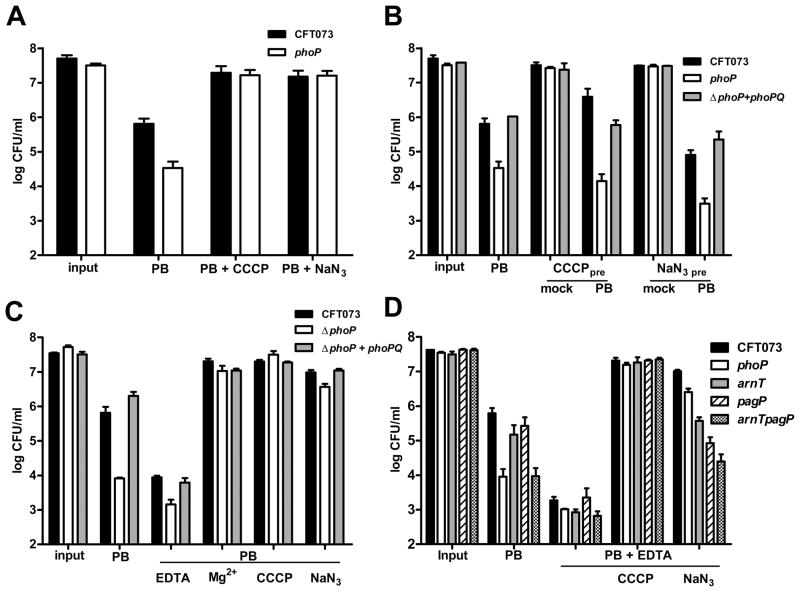
We tested the relationship between μH+ and PhoP by disrupting the electrochemical gradient and determining resistance to polymyxin B. By uncoupling oxidative phosphorylation from μH+ generation using sodium azide (NaN3) or depolarizing the membrane using CCCP, we observed an increase in the phoP mutant strain’s resistance to antimicrobial peptides to wild-type levels (Fig 5A). In addition, finding that NaN3 or CCCP also increased wild-type resistance to polymyxin B supported the notion that, in general, reducing the proton gradient across the bacterial inner membrane is an effective means to abrogate the bactericidal effects of cationic antimicrobial compounds (Matsuzaki et al., 1995). Dissipating the electrochemical gradient with CCCP or NaN3 and washing the cells to remove uncoupler prior to exposing either CFT073 or phoP mutant bacteria to polymyxin B was not sufficient to protect the cells (Fig 5B). This suggested that the protective effect of reducing μH+ is not a pre-set state of the membrane.
Fig. 5.
Influence of the transmembrane proton motive force on E. coli susceptibility to antimicrobial peptides. A. Viable counts of CFT073 (black bars) and ΔphoP (white bars) following 45 min incubation in 2 μg ml−1 PB with and without 50 μM m-chlorophenyl carbonyl cyanide hydrozone (CCCP) or 0.1% sodium azide (NaN3). B. CFT073, ΔphoP, and ΔphoP+phoPQ cells grown to OD600 = 0.8 were diluted 1:10 (input) and pre-treated with CCCP or NaN3 for 1 h. Following pre-treatment, cells were washed with LB medium and incubated in fresh LB medium (mock) or LB containing 2 μg/ml PB for 45 min. C. CFT073, ΔphoP, and ΔphoP+phoPQ were grown and incubated in PB alone or PB containing CCCP or NaN3 as described in (A) or in PB containing 0.5 mM EDTA or 10 mM MgCl2. D. CFT073, ΔphoP, ΔarnT, ΔpagP, and ΔarnTΔpagP were incubated in LB medium containing 10 μM FeCl3 and 2 μg ml−1 PB or LB medium containing 0.5 mM EDTA, 2 μg ml−1 PB and CCCP or NaN3. Bacteria treated with EDTA were pre-cultured in LB medium with 0.5 mM EDTA. Viable counts in A.–D. were determined from triplicate experiments by plating serial dilutions on LB agar.
Excess Mg2+ most likely protects cells from antimicrobial peptides through competitive inhibition at negatively charged sites on the LPS. We found that excess Mg2+, which would increase the net surface charge of outer membrane LPS (Vaara, 1992) and could contribute to reversal of inner transmembrane Δψ, protected both wild-type and ΔphoP bacteria from killing by polymyxin B, similar to the protection afforded by the addition of CCCP and NaN3 (Fig. 5C). This Mg2+- protection against polymyxin B would be predicted to be independent of phoP, which would be repressed under this condition. Because PhoP is involved in the maintenance of a robust permeability barrier (Murata et al., 2007), we also tested polymyxin resistance in combination with EDTA, a compound that chelates divalent cations and renders the bacterial LPS and outer membrane more permeable for antimicrobial peptides. We found that EDTA increased sensitivity to polymyxin B for both wild-type CFT073 and the phoP mutant and that CFT073 maintained greater resistance to polymyxin than the phoP mutant strain (Fig. 5C).
Treatment of E. coli or Salmonella with millimolar concentrations of EDTA is known to induce lipid A palmitoylation mediated by PagP (Bishop et al., 2000). We constructed CFT073 strains lacking pagP (crcA in E. coli) and a double mutant lacking pagP and arnT to further differentiate the contribution of LPS modifications or permeability defects at the outer membrane from changes in the transmembrane potential across the inner membrane. Susceptibility to polymyxin B was determined in the presence of 10μM Fe3+ to activate PmrA and to avoid potential cross-regulatory effects between PhoP and PmrA. In the absence of EDTA, the arnT mutant bacteria displayed a 5-fold decrease in polymyxin B resistance relative to wild-type, while the arnTpagP double mutant demonstrated susceptibility similar to the phoP mutant despite the pagP mutant being nearly indistinguishable from wild-type (Fig. 5D). All the bacteria tested, when pre-cultured in 0.5 mM EDTA and exposed to polymyxin B and EDTA demonstrated a marked increase in susceptibility to polymyxin B with 0.5 mM EDTA, (Fig. 5D). Interestingly, while CCCP abolished killing by polymyxin B for all strains and constructs tested, only the LPS modification mutants lacked the same magnitude increase in resistance to killing by polymyxin B as the phoP mutant bacteria when respiratory cytochromes were uniformly inhibited by NaN3 (Fig. 5D). These changes in both outer membrane LPS charge and inner membrane potential appear to reflect an active response because pre-treatment or culturing CFT073 and ΔphoP with EDTA, NaN3, CCCP, or excess [Mg2+] alone did not significantly affect bacterial viability, nor did pre-treatment alter the polymyxin resistance phenotype (Fig. S4). Together, these findings provide evidence that controlling μH+ represents a mechanism for PhoP-mediated antimicrobial peptide resistance, independent from LPS modification and maintaining a robust permeability barrier in E. coli.
Acid resistance requires PhoP-dependent changes in μH+
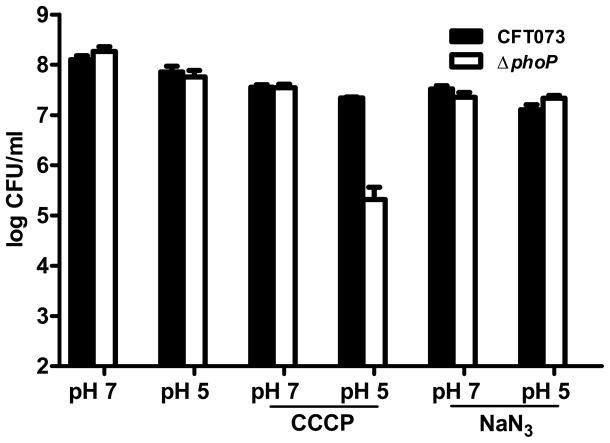
Because resistance to acidic pH has been shown to involve enzymatic consumption of cytoplasmic protons and a reversal in membrane potential (Foster, 2004, Richard & Foster, 2004), we determined if the survival defect of the phoP mutant at acidic pH is due to the inability to alter respiratory components, reverse membrane potential, or consume intracellular protons in response to acid induction of PhoP. CCCP specifically decreased the ability of the phoP mutant but not that of wild-type bacteria to survive when shifted from neutral pH to pH 5.0 (Fig. 6). These findings demonstrated that the role for PhoP-activated genes encoding glutamate-, lysine-, and arginine-decarboxylases that consume cytoplasmic protons is distinct from PhoP regulation of genes encoding proteins that control membrane potential because wild-type CFT073 was unaffected by CCCP in these experiments (Fig. 6). The increased sensitivity to acid for phoP mutant bacteria was independent of respiratory generation of μH+ because NaN3 did not alter susceptibility to acid for either wild-type or the mutant bacteria (Fig. 6). The lack of resistance to moderate acid for the phoP mutant during membrane depolarization is notable because phoP mutant bacteria did not have a survival defect at pH 5.0 in the absence of CCCP nor did CCCP treatment at neutral pH cause a reduction in viability (Fig. 6).
Fig. 6.
PhoP-mediated control of membrane potential is required for E. coli resistance to acidic pH. CFT073 (black bars) and ΔphoP (white bars) were cultured to OD600 = 0.8 were diluted and incubated in buffered LB medium at pH 7 and pH 5 with and without CCCP and NaN3 for 1 h. Viable counts were determined from triplicate experiments by plating serial dilutions on LB agar.
PhoP represses the induction of a fully functional lysogenic bacteriophage
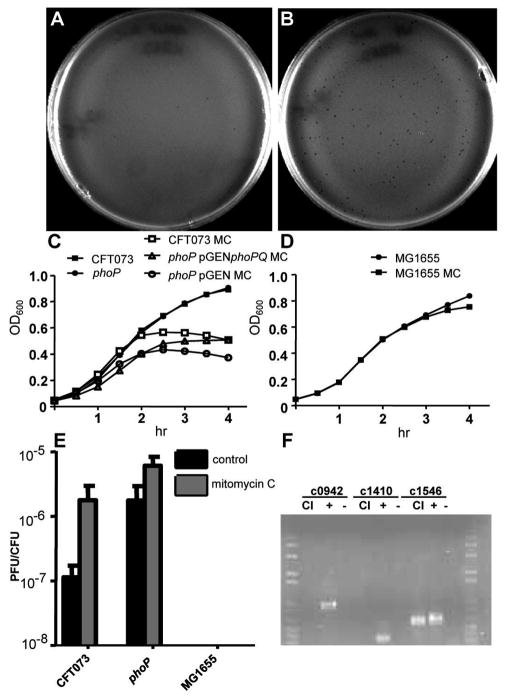
Synergism between acidification, antimicrobial peptides, and reactive oxygen species allows neutrophils and macrophages to eradicate bacterial pathogens from host tissues (Babior, 1978, Vazquez-Torres et al., 2000). Consistent with a role for PhoP regulating genes that encode proteins for bacterial defense against oxidative attack, expression of superoxide dismutase (sodC) and catalase (katG) genes was significantly decreased >4.0-fold (P < 0.05) in the absence of PhoP (Table 2). PhoP was also required for induction of genes that code for the Suf system that is involved in Fe-S cluster repair during oxidative damage (Table 2). When reactive oxygen species breach the inner membrane, superoxides damage Fe-S-containing proteins (Jang & Imlay, 2007) and promote the release of free iron, the latter rapidly induces DNA damage by driving Fenton reactions (Park & Imlay, 2003). Oxidative DNA damage can cause prophage induction (Little, 2005), which could occur during exposure to reactive oxygen species generated by the phagocyte NADPH oxidase. In keeping with the hypothesis that PhoP controls a large number of genes that encode a diverse set of proteins involved in comprehensive protection against bactericidal activities of phagocytes (acidification, antimicrobial peptides, and the oxidative burst), the microarray results also indicated that more than 100 genes in prophage islands are repressed by PhoP (Fig. 2). To test phage induction in the absence of PhoP-mediated repression of prophage genes, we assayed culture supernatants from both the phoP mutant and parental CFT073 for the ability to form plaques on a restriction/modification (r−/m−) -deficient E. coli strain (Bertani & Weigle, 1953). Supernatant prepared from logarithmic phase CFT073 produced few (<10) plaques (Fig. 7A), while supernatant from phoP mutant bacteria consistently produced significantly more plaques (>100) (Fig. 7B). That CFT073 carries an active phage was not previously appreciated. By quantifying burst size (no. plaques/CFU), we found that mitomycin C (MMC), a potent inducer of DNA damage and phage lambda in E. coli (Little, 2005), caused maximal prophage induction for both CFT073 and phoP mutant bacteria. However, the burst size in the absence of MMC induction was over 10-fold greater for the phoP mutant than for wild-type bacteria (Fig. 7E). Finding that bacteria lacking PhoP have increased phage induction suggests that PhoP-mediated repression of prophage genes would increase bacterial survival by preventing lytic phage replication during oxidative damage caused by the phagocyte NADPH oxidase. To exclude the possibility that the plaques originated from the indicator strain, PCR amplification of phage lysate produced in the (r−/m−) strain using probes specific for each predicted cI repressor gene from the CFT073 genome (Fig. 7F) indicated that the active prophage (ΦicdA) originated from the CFT073 chromosome.
Fig. 7.
PhoP represses prophage induction. A. CFT073 and B. ΔphoP supernatants prepared from cells cultured in LB medium to OD600 = 1.0 were mixed in molten top agar containing >109 CFU E. coli strain C and poured onto TCMG agar plates to visualize plaques. C. Growth of CFT073 (□), ΔphoP (○), and complemented mutant (Δ) in LB medium containing 5 μg ml−1 mitomycin C (MMC). CFT073 (■) and ΔphoP (●) growth in LB medium alone is shown for comparison. D. Growth of MG1655, which lacks a functional prophage, in the presence (■) and absence of MMC (●). E. Burst size (no. plaques CFU−1) for CFT073, ΔphoP, and MG1655 cultured in LB or LB medium containing 5 μg ml−1 MMC. F. CFT073 genes homologous to lambda cI were amplified by PCR from E. coli C lysate (CI) following incubation with plaques isolated from MC induced CFT073 supernatant. Genomic DNA purified from CFT073 (+) and E. coli C (−) were included as controls.
Discussion
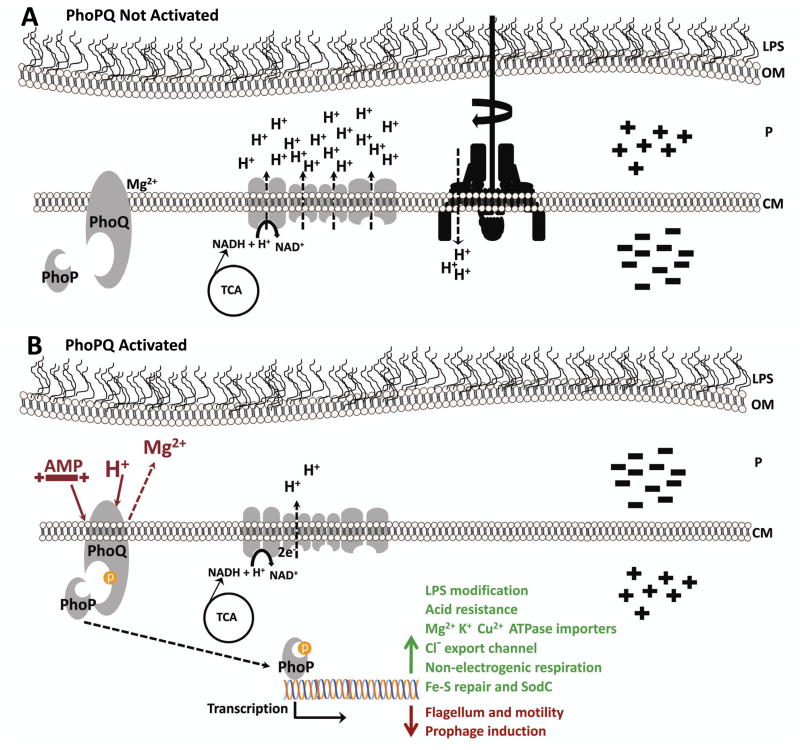
Our results suggest that PhoP targets, represented by seemingly unrelated outputs of PhoP activity (sensitivity to acid, antimicrobial peptide resistance, and motility) act through a common mechanism that is dependent on the energized state of the membrane (Glagolev & Skulachev, 1978, Foster, 2004, Grundling et al., 2001). Coordinating gene expression with changes in membrane potential or regulating genes that encode proteins that control the state of the energized membrane is relevant for both pathogens avoiding host defenses and for non-pathogens surviving in the environment. PhoP regulation of genes encoding products that control membrane potential is supported by PhoP-dependent activation of transcription of genes encoding non-electrogenic respiratory chain components that do not pump protons, ion importers that actively transport cations against an electrochemical gradient, and cytoplasmic enzymes that mediate acid resistance (Fig. 8). The loss of PhoP-mediated control of the energized membrane is manifested by decreased survival at acidic pH, increased susceptibility to polymyxin B, and hyper-motility in phoP mutant bacteria.
Fig. 8.
The PhoP regulon controls energy transformations and membrane potential as one protective counter-measure against mammalian host defenses. A. PhoPQ is not activated. Proton motive force (μH+), consisting of a gradient of protons (ΔpH) and charge (Δψ), across the cytoplasmic membrane (CM), is generated by respiration using electron transfer systems. Protons are pumped into the periplasm (P) by electrogenic respiratory chain components concomitant with the re-oxidation of NAD+ from NADH produced by the tricarboxylic acid (TCA) cycle. The transmembrane potential (inside-negative) allows protons to diffuse into the cytoplasm, with the [H+] gradient, to drive processes including ATP synthesis and rotation of the flagellum and motility. B. In response to antimicrobial activities of host phagocytes, PhoPQ is activated by acidification and cationic antimicrobial peptides (AMP) that have traversed the bacterial outer membrane (OM). PhoP-mediated induction (green arrow and text) of acid resistance responses, non-electrogenic respiratory chain components that do not pump protons, active import of cations, and efflux of Cl− create a reversed membrane potential (inside positive) and reduce the bactericidal effects of AMPs and inorganic acid by reducing their ability to disrupt the CM or acidify the cytoplasm. Activated PhoP represses (red arrow and text) the transcription of flagellar and prophage genes, expression of the latter can be induced by oxidative DNA damage caused by Fenton chemistry during assault by the phagocyte NADPH oxidase.
By decreasing flagellar gene transcription, reducing motility, and activating acid resistance responses, proteins encoded by genes regulated by PhoP would affect ΔpH by limiting an increase in internal [H+] and preventing acidification of the cytoplasm. The results of experiments where maintenance of cytoplasmic pH was abolished by membrane depolarization with CCCP (Matin et al., 1982, Krulwich et al., 1978) suggest that the loss of PhoP-activation of genes encoding the cytoplasmic enzymes glutamate-, lysine-, and arginine-decarboxylases and antiporters that consume protons and extrude decarboxylated products, respectively, could account for the specificity of the protonophore’s effect on acid survival for bacteria lacking phoP. In addition to consuming intracellular protons, AR responses in E. coli and other bacteria have been previously shown to promote survival in extreme acid by reversing membrane potential Δψ to reduce proton influx (Richard & Foster, 2004, Konings et al., 2002, Michels & Bakker, 1985). Because the weak acid CCCP freely diffuses through phospholipid bilayers, discharges ΔpH by release of the ionizable H+ inside the cytoplasm, and leaves the cell as an anion, destroying Δψ, the specific mechanism controlled by PhoP to promote survival during exposure to acidic pH, appears dependent on both ΔpH and Δψ. The ΔpH-dependence of the PhoP acid response is demonstrated by reduced survival of the ΔphoP mutant at pH 5.0 only when the ΔpH is discharged with CCCP. This defect is caused by the inability to activate genes that encode cytoplasmic enzymes that consume protons, while the inability to regulate genes that encode functions required for Δψ reversal results in reduced survival at pH 2.5, in the absence of PhoP. Consistent with this hypothesis, using the membrane potential-dependent uptake of DiOC2 we demonstrated that loss of PhoP creates a hyper-polarized membrane (more negative-inside); PhoP-dependent activation of the Acid Fitness Island (AFI) genes, and genes encoding P-type ATPases that import cations: YhiD (MgtC), YbaR, TrkA, and the Cl−anion efflux channel YadQ would prevent hyperpolarization during acid stress (positive-inside) and would contribute to Δψ reversal (Iyer et al., 2002). It is possible that bacteria lacking phoP could have increased DiOC2 uptake due to an altered permeability barrier (Murata et al., 2007). However, hyper-motility, acid susceptibility, decreased expression of genes encoding non-electrogenic respiratory components, regulation of genes encoding ion transporters, and PhoP-dependent manipulation of polymyxin resistance by poisoning respiration, taken together, add up to compelling evidence that PhoP control of membrane potential represents the unifying mechanism to resist host antibacterial defenses.
PhoP-mediated control of membrane potential is further supported by the well-known PhoP activation of mgtA and genes encoding additional magnesium transporters during growth in the absence of divalent cations (Soncini et al., 1996). These P-type ATPases mediate active import of ions against an electrochemical gradient. Upon first consideration, this may seem like an unusual property since bacterial cells are normally inside-negative allowing polyvalent cations like magnesium to freely diffuse into the cell through the constitutively active CorA channel (Maguire, 2006). However, a PhoP-activated cell, in our model, would be positive-inside and thus would require active transport to import these positively charged ions (Fig. 8). Indeed, active transport of ions against an electrochemical gradient is particularly intriguing given that activated phagocytes, which generate acidification and antimicrobial peptides to kill microbes, require counter-ion conductance and voltage-gated channels that prevent host cell membrane depolarization to sustain NADPH oxidase function and effectively control bacterial infections (DeCoursey et al., 2003, Di et al., 2006, Forbes & Gros, 2001). Specifically, phagocyte killing of bacteria is known to require the cation/H+ transporter Nramp1 (Gruenheid et al., 1997, Forbes & Gros, 2003) and the CFTR Cl− channel (Di et al., 2006). Bacteria, in turn, would benefit by reversing membrane potential or reducing their inside-negative potential in order to resist killing by these host defenses. Furthermore, ion transporters in both Salmonella and E. coli have been shown to be involved in the response to reactive oxygen (Kehres et al., 2000). The importance of energy transformations by the bacterial membrane having a key role during survival against host defenses is also supported by a recent study by Karlinsey et al. that demonstrated maintenance of proton motive force, μH+, is required for Salmonella virulence in Nramp1-expressing mice (Karlinsey et al., 2010).
Self-promoted uptake of host-derived antimicrobial peptides by bacteria has been proposed to be μH+-dependent (Falla et al., 1996, Matsuzaki et al., 1995). Consistent with this, we found that dissipating μH+ with CCCP abrogated the bactericidal effects of polymyxin B. The contribution of charge modifications to the LPS in the outer membrane to resistance to antimicrobial peptides is well documented; in this study, in contrast to phoP mutant bacteria, the susceptibilities of the ΔarnT and ΔpagP mutant bacteria or the ΔarnTΔpagP double mutant to polymyxin B were unaffected by uniformly inhibiting respiratory cytochromes with NaN3. To distinguish PhoP-dependent sensitivity to polymyxin from PmrA-dependent effects, we maintained high [Fe3+] throughout our polymyxin experiments to activate PmrA. Because E. coli lacks a functional PmrD (Winfield & Groisman, 2004), PmrA-dependent L-Ara4N modification of lipid A can occur independent of PhoP in both the wild-type and phoP mutant bacteria that were both protected by poisoning respiration with NaN3. Interestingly, the increase in free iron following phagocyte-induced oxidative damage to bacterial iron-sulfur clusters would also activate the iron-responsive PmrA and PmrA-dependent polymyxin resistance genes independent from PhoP. Similarly, PagP-mediated palmitate addition to lipid A, which confers resistance to antimicrobial peptides other than polymyxin B (Murray et al., 2007), can be induced by NaCl, EDTA, or ammonium metavanadate independent of PhoP (Jia et al., 2004, Zhou et al., 1999). Collectively, these findings suggest that the mechanism responsible for susceptibility of the phoP mutant to polymyxin B is distinct from a defect in LPS modification alone. Thus, resistance to polymyxin B involves both charge modifications that occur at the outer leaflet LPS of the outer membrane and modulation of the respiratory generation of the transmembrane electrochemical gradient across the inner membrane.
Synergism between acidification, antimicrobial peptides, and reactive oxygen species allows neutrophils and macrophages to eradicate bacterial pathogens from host tissues (Babior, 1978, Vazquez-Torres et al., 2000). This assault on bacteria leads to the penetration of superoxide into the bacterial periplasm and cytoplasm. The PhoP-activated sodC gene product can detoxify superoxide in the periplasm (Kim et al., 2010). When reactive oxygen species breach the inner membrane, superoxide and hydrogen peroxide damage iron-sulfur clusters (Jang & Imlay, 2007) and promote the release of free iron, which rapidly induces DNA damage and bacterial cell death by driving Fenton reactions (Park & Imlay, 2003). Consistent with a defense against superoxide, in the absence of PhoP, we found decreased expression of sodC and the suf genes; the latter is required for repair and activation of Fe-S-containing enzymes (Imlay, 2008). In addition, PhoP was required for uniform repression of prophage genes in uropathogenic E. coli; in the absence of PhoP, we observed an increase in the frequency of spontaneous prophage induction and following induction with the potent DNA-damaging agent MMC. It is reasonable to speculate that a complete PhoP-mediated defense against killing by phagocytes would involve repression of prophage in addition to resistance to antimicrobial peptides and survival at acidic pH because oxidative DNA damage is known to cause prophage induction (Little, 2005).
These results provide compelling evidence linking PhoP, generation of μH+, and resistance to host defenses such as acidification, antimicrobial peptides, and reactive oxygen species. In our view, this is clearly reflected by the fact that phoP was originally identified in genetic screens that selected for Salmonella mutants with increased sensitivity to phagocyte granule extracts and reduced intracellular survival in macrophages (Fields et al., 1989, Fields et al., 1986). For E. coli, it has been demonstrated that the antimicrobial peptide LL-37 cathelicidin contributes to protection of the host during invasive urinary tract infection (Chromek et al., 2006) and extraintestinal E. coli infections in the urinary tract elicit a massive inflammatory response characterized by neutrophil influx (Godaly et al., 2001, Bergsten et al., 2004). Additionally, bacterial survival during bacteremia is required for extraintestinal virulence, because these E. coli strains are confronted with antimicrobial effectors during meningitis or urosepsis (Barondess & Beckwith, 1990, Smith et al., 2010, Wooster et al., 2006). We speculate that the conserved processes controlled by PhoP for E. coli to evade host defenses within the bladder are responsible for the attenuation of phoP mutant bacteria during extraintestinal infection because PhoP controls a plethora of fitness traits centered on surviving bactericidal activities of phagocytic cells in a large number of enteric pathogens.
Our findings demonstrate that a basic physiological process, modulating the electrochemical gradient across the inner membrane, can be an integral component of bacterial pathogenesis. Specifically, the PhoP regulon activates layers of protective counter-measures against mammalian host defenses including i) modification of lipid A at the outer membrane, ii) superoxide dismutase and ion transport in the periplasm, iii) energy transformations by the cytoplasmic membrane, and iv) cytoplasmic responses to oxidative damage. Basic principles of physiology, shared by nearly all living cells, are beginning to be appreciated as playing a key role in processes that are essential for pathogenesis. For example, bacterial respiration coordinates the timing of type three secretion system (TTSS) assembly with movement of Shigella from the intestinal lumen to the mucosa (Marteyn et al., 2010). Salmonella pathogenicity island (SPI-2) TTSS stability and effector translocation are determined by changes in the external pH inside Salmonella-containing host cell vacuoles (Yu et al., 2010). These recent reports in conjunction with our findings reveal a relationship between fundamental bioenergetics and pathogenesis. The connection between PhoP and μH+ matches the large PhoPQ regulatory circuit with physiological changes that are most likely to occur through stress encountered during host-pathogen interactions.
Experimental Procedures
Bacterial strains and culture conditions
E. coli strain CFT073 was isolated from the blood and urine of a patient with acute pyelonephritis (Mobley et al., 1990). Unless otherwise noted, bacteria were cultured in Lysogeny broth (LB) medium containing appropriate antibiotics (100 μg ml−1 ampicillin and/or 25 μg ml−1 kanamycin) at 37°C with aeration.
Construction of mutants and complementation
Deletion mutants were generated using lambda red recombinase (Datsenko & Wanner, 2000). Primers homologous to sequences within the 5′ and 3′ end of phoP, fumC, crcA (pagP), or arnT, respectively, were designed and used to replace the target genes with a nonpolar kanamycin resistance cassette (Datsenko & Wanner, 2000). To determine whether the kanamycin resistance cassette recombinedwithin the target gene site, primers that flank the target gene sequence were designed and used for PCR. After amplification, each PCR product was compared to wild-type PCR product when visualized on a 0.8% agarose gel stained with ethidium bromide. For ΔsdhBΔfrdB, homologous sequences flanking the frdA gene were used to remove the target gene and replace it with a nonpolar chloramphenicol resistance cassette in a kanamycin resistant ΔsdhB strain of CFT073 (Alteri et al., 2009). For the fumarase triple mutant, the kanamycin cassette that replaced fumC was unmarked using the FRT sites within the cassette (Datsenko & Wanner, 2000), and fumB and fumA were replaced using chloramphenicol and kanamycin cassettes, respectively. For phoP complementation, the phoPQ genes were amplified from CFT073 genomic DNA using Easy-A high-fidelity polymerase (Stratagene) and cloned into pGEN-MCS (Lane et al., 2007) using appropriate restriction enzymes as previously described (Alteri et al., 2009). The sequence of pGEN-phoPQ was verified by DNA sequence analysis prior to electroporation into the CFT073 ΔphoP mutant strain.
Murine infection model
Six- to 8-week old female CBA/J mice (20 to 22 g; Jackson Laboratories) were transurethrally inoculated as previously described (Hagberg et al., 1983). Mice were anesthetized with 100 mg ketamine, 10 mg xylazine kg−1 body weight. Prior to inoculation, overnight cultures of strain CFT073 were pelleted and resuspended in PBS. Animals were inoculated transurethrally over a 30 sec period with a 50 μl bacterial suspension per mouse using a sterile polyethylene catheter (I.D. 0.28 mm x O.D. 0.61 mm) connected to an infusion pump (Harvard Apparatus), with total inoculum equaling 1×108 CFU/mouse. For determination of CFUs, bladders and kidneys were harvested from euthanized animals at 48 h post-inoculation, weighed, and homogenized in PBS with a GLH homogenizer (Omni International). Tissue homogenates were analyzed for colonization by plating on LB agar using an Autoplate 4000 spiral plater (Spiral Biotech). Colonies were enumerated using a QCount automated plate counter (Spiral Biotech) and P-values were determined using the non-parametric Mann Whitney significance test. All procedures were conducted according to protocols approved by University Committee on the Care and Use of Animals at the University of Michigan.
Polymyxin B resistance and acid survival assays
CFU ml−1 and percent wild-type survival were determined for CFT073, ΔphoP, and ΔphoP+phoPQ by incubating 107 CFU logarithmic phase cells in LB medium containing 10 μM FeCl3 and 2 μg ml−1 polymyxin B (PB) for 45 min at 37°C or in LB medium buffered to pH 7.0, 5.0, or 2.5 with 2-(N-morpholino) ethanesulfonic acid for 1 h at 37°C. For some experiments, 10 mM MgCl2, 0.5 mM EDTA, 50 μM m-chlorophenyl carbonyl cyanide hydrozone (CCCP) or 0.1% sodium azide (NaN3) were added to cells simultaneously with the addition of polymyxin B or a shift in pH. Pre-treatment of cells prior to exposure to polymyxin B or acidic pH was performed by incubating a 1:10 dilution of cells cultured to OD600 = 0.8 in LB medium containing 10 mM MgCl2, 0.5 mM EDTA, 50 μM CCCP or 0.1% NaN3 for 1 h. Following pre-treatment, cells were washed with LB medium and incubated in fresh LB containing 2 μg ml−1 PB, buffered LB pH 7.0, or pH 5.0 as described above. Viable counts were determined from triplicate experiments by plating serial dilutions on LB agar and significance was assessed using Welch’s t test.
Microarrays and quantitative real-time PCR
For the extraction of total RNA, 1 ml of bacteria culture was added to 0.125 ml of ice cold phenol-ethanol stop solution (5% phenol in ethanol), and the bacteria were collected by centrifugation. Cells were lysed and RNA was extracted using the RNeasy kit (Qiagen) following the manufacturer’s recommended protocol. Following elution, nucleic acid concentrations were determined by spectrophotometry (NanoDrop) and residual DNA contamination was removed by incubating the samples with 4U of TURBO DNase (Ambion). After DNase inactivation, the RNA was recovered, quantified, and used as a template for PCR to confirm inactivation of contaminating DNA. For microarrays, RNA purity and integrity were verified by using a BioAnalyzer 2100 (Agilent). RNA was labeled, hybridized, and scanned by using Affymetrix E. coli Genome 2.0 GeneChips according to the manufacturer's recommendations. Expression values for each gene were assessed using a robust multi-array average (RMA) (Irizarry et al., 2003). A principal components analysis (PCA) was fit onto the expression values to determine that the replicated samples grouped together and a linear model designed specifically for microarray analysis was fit to the data to compute the contrast of interest (Smyth, 2004) and filter out probesets with a variance < 0.05. Probesets were adjusted for multiple comparisons using a false discovery rate (FDR) of 0.05 (Klipper-Aurbach et al., 1995). Expression levels with a fold-change >2.0 and an adjusted P-value <0.05 from three independent replicates of each strain were compared by using the Affymetrix and Limma packages of Bioconductor implemented in the R statistical environment. For qPCR, transcripts were quantified on an MX3000P real-time PCR machine (Stratagene) using Brilliant SYBR green QPCR mix (Stratagene). Transcript levels were normalized to the level of gapA (glyceraldehyde 3-phosphate dehydrogenase A) and changes were determined using an experiment-specific calibrator using MXPro v 3.00 software package (Stratagene).
Bacteriophage induction
Supernatants prepared from cells cultured in LB medium or LB medium containing 5 μg ml−1 mitomycin C (MMC) were mixed in molten top agar containing >109 CFU E. coli strain C (Bertani & Weigle, 1953) and poured onto TCMG (1% tryptone, 0.5% NaCl, 0.1M MgSO4, 0.85% Difco Agar) to visualize plaques following overnight incubation at 37°C. Burst size was calculated as the ratio of plaque forming units (PFU) to CFU. Phage lysate was prepared by culturing E. coli C in LB medium with a re-isolated plaque selected on a lawn of E. coli C.
Soft agar motility and detection of FliC
For the assessment of swimming motility, 50 μl of overnight cultures of wild-type CFT073, ΔphoP, ΔphoP containing pGEN+phoPQCFT073, pGEN+phoPQK12, or pGEN were reinoculated into 2 ml of sterile LB medium and incubated at 37°C with aeration (200 rpm) to an OD600 of 1.0. Cultures were stabbed into 0.25% tryptone agar plates (1% tryptone, 0.5% NaCl, 0.25% Difco Agar) using a sterile inoculating needle and incubated at 30°C for 18 hr. For preparation of flagella, 1 ml aliquots of bacteria cultured to OD600 = 1.0 were vortexed for 5 min, centrifuged to remove bacterial cells, and supernatants were quantified and standardized using the BCA total protein kit (Thermo Scientific). Detection of FliC by Western blotting was performed using anti-H1 antisera.
Analysis of membrane potential
To determine relative membrane potential between CFT073 wild-type, ΔphoP, ΔfumAΔfumBΔfumC, and ΔsdhBΔfrdA, 107 CFU of logarithmic phase bacteria were diluted into 1 ml sterile PBS containing 10 μl of 3 mM 3,3′-diethyloxacarbocyanine iodide (DiOC2) (Sims et al., 1974) (Sigma) in DMSO. 10 μl of 10 mM carbonyl cyanide 3-chlorophenylhydrazone (CCCP) (Sigma) was added to depolarized control samples. Samples were incubated at 37°C for 15 min, dispensed into opaque black microtiter plates, and fluorescence was measured using 488 nm excitation with fluorescein and Texas Red emission filters on a Synergy HT plate-reader operating KC4 software (Bio-Tek). Measurements were normalized using the emission from DiOC2 blank wells and the ratio of red:green fluorescence for each sample was determined using arbitrary units calculated by dividing the mean fluorescence by CFU for each channel.
Supplementary Material
Acknowledgments
We are grateful to David Friedman for many helpful discussions and guidance in performing the phage experiments. We also thank Guy Plunkett III for generously sharing genomics expertise, and Erika Flannery for thoughtful review of the manuscript. This work was supported by public health service grants AI43363 and AI59722 from the National Institutes of Health to H.L.T.M
References
- Alpuche Aranda CM, Swanson JA, Loomis WP, Miller SI. Salmonella typhimurium activates virulence gene transcription within acidified macrophage phagosomes. Proc Natl Acad Sci U S A. 1992;89:10079–10083. doi: 10.1073/pnas.89.21.10079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alteri CJ, Smith SN, Mobley HL. Fitness of Escherichia coli during urinary tract infection requires gluconeogenesis and the TCA cycle. PLoS Pathog. 2009;5:e1000448. doi: 10.1371/journal.ppat.1000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babior BM. Oxygen-dependent microbial killing by phagocytes (first of two parts) N Engl J Med. 1978;298:659–668. doi: 10.1056/NEJM197803232981205. [DOI] [PubMed] [Google Scholar]
- Bader MW, Navarre WW, Shiau W, Nikaido H, Frye JG, McClelland M, Fang FC, Miller SI. Regulation of Salmonella typhimurium virulence gene expression by cationic antimicrobial peptides. Mol Microbiol. 2003;50:219–230. doi: 10.1046/j.1365-2958.2003.03675.x. [DOI] [PubMed] [Google Scholar]
- Bader MW, Sanowar S, Daley ME, Schneider AR, Cho U, Xu W, Klevit RE, Le Moual H, Miller SI. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell. 2005;122:461–472. doi: 10.1016/j.cell.2005.05.030. [DOI] [PubMed] [Google Scholar]
- Barondess JJ, Beckwith J. A bacterial virulence determinant encoded by lysogenic coliphage lambda. Nature. 1990;346:871–874. doi: 10.1038/346871a0. [DOI] [PubMed] [Google Scholar]
- Bearson BL, Wilson L, Foster JW. A low pH-inducible, PhoPQ-dependent acid tolerance response protects Salmonella typhimurium against inorganic acid stress. J Bacteriol. 1998;180:2409–2417. doi: 10.1128/jb.180.9.2409-2417.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker M, de Vries S, Ter Beek A, Hellingwerf KJ, de Mattos MJ. Respiration of Escherichia coli can be fully uncoupled via the nonelectrogenic terminal cytochrome bd-II oxidase. J Bacteriol. 2009;191:5510–5517. doi: 10.1128/JB.00562-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsten G, Samuelsson M, Wullt B, Leijonhufvud I, Fischer H, Svanborg C. PapG-dependent adherence breaks mucosal inertia and triggers the innate host response. J Infect Dis. 2004;189:1734–1742. doi: 10.1086/383278. [DOI] [PubMed] [Google Scholar]
- Bertani G, Weigle JJ. Host controlled variation in bacterial viruses. J Bacteriol. 1953;65:113–121. doi: 10.1128/jb.65.2.113-121.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop RE, Gibbons HS, Guina T, Trent MS, Miller SI, Raetz CR. Transfer of palmitate from phospholipids to lipid A in outer membranes of gram-negative bacteria. EMBO J. 2000;19:5071–5080. doi: 10.1093/emboj/cdd507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop RE, Lo EI, Khan MA, El Zoeiby A, Jia W. Enzymology of lipid A palmitoylation in bacterial outer membranes. J Endotoxin Res. 2004;10:107–112. doi: 10.1179/096805104225004004. [DOI] [PubMed] [Google Scholar]
- Borregaard N, Schwartz JH, Tauber AI. Proton secretion by stimulated neutrophils. Significance of hexose monophosphate shunt activity as source of electrons and protons for the respiratory burst. J Clin Invest. 1984;74:455–459. doi: 10.1172/JCI111442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breazeale SD, Ribeiro AA, McClerren AL, Raetz CR. A formyltransferase required for polymyxin resistance in Escherichia coli and the modification of lipid A with 4-Amino-4-deoxy-L-arabinose. Identification and function of UDP-4-deoxy-4-formamido-L-arabinose. J Biol Chem. 2005;280:14154–14167. doi: 10.1074/jbc.M414265200. [DOI] [PubMed] [Google Scholar]
- Chromek M, Slamova Z, Bergman P, Kovacs L, Podracka L, Ehren I, Hokfelt T, Gudmundsson GH, Gallo RL, Agerberth B, Brauner A. The antimicrobial peptide cathelicidin protects the urinary tract against invasive bacterial infection. Nat Med. 2006;12:636–641. doi: 10.1681/01.asn.0000926856.92699.53. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Groote MA, Ochsner UA, Shiloh MU, Nathan C, McCord JM, Dinauer MC, Libby SJ, Vazquez-Torres A, Xu Y, Fang FC. Periplasmic superoxide dismutase protects Salmonella from products of phagocyte NADPH-oxidase and nitric oxide synthase. Proc Natl Acad Sci U S A. 1997;94:13997–14001. doi: 10.1073/pnas.94.25.13997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCoursey TE, Morgan D, Cherny VV. The voltage dependence of NADPH oxidase reveals why phagocytes need proton channels. Nature. 2003;422:531–534. doi: 10.1038/nature01523. [DOI] [PubMed] [Google Scholar]
- Di A, Brown ME, Deriy LV, Li C, Szeto FL, Chen Y, Huang P, Tong J, Naren AP, Bindokas V, Palfrey HC, Nelson DJ. CFTR regulates phagosome acidification in macrophages and alters bactericidal activity. Nat Cell Biol. 2006;8:933–944. doi: 10.1038/ncb1456. [DOI] [PubMed] [Google Scholar]
- Falla TJ, Karunaratne DN, Hancock RE. Mode of action of the antimicrobial peptide indolicidin. J Biol Chem. 1996;271:19298–19303. doi: 10.1074/jbc.271.32.19298. [DOI] [PubMed] [Google Scholar]
- Fields PI, Groisman EA, Heffron F. A Salmonella locus that controls resistance to microbicidal proteins from phagocytic cells. Science. 1989;243:1059–1062. doi: 10.1126/science.2646710. [DOI] [PubMed] [Google Scholar]
- Fields PI, Swanson RV, Haidaris CG, Heffron F. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc Natl Acad Sci U S A. 1986;83:5189–5193. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa-Bossi N, Bossi L. Inducible prophages contribute to Salmonella virulence in mice. Mol Microbiol. 1999;33:167–176. doi: 10.1046/j.1365-2958.1999.01461.x. [DOI] [PubMed] [Google Scholar]
- Forbes JR, Gros P. Divalent-metal transport by NRAMP proteins at the interface of host-pathogen interactions. Trends Microbiol. 2001;9:397–403. doi: 10.1016/s0966-842x(01)02098-4. [DOI] [PubMed] [Google Scholar]
- Forbes JR, Gros P. Iron, manganese, and cobalt transport by Nramp1 (Slc11a1) and Nramp2 (Slc11a2) expressed at the plasma membrane. Blood. 2003;102:1884–1892. doi: 10.1182/blood-2003-02-0425. [DOI] [PubMed] [Google Scholar]
- Foster JW. Escherichia coli acid resistance: tales of an amateur acidophile. Nat Rev Microbiol. 2004;2:898–907. doi: 10.1038/nrmicro1021. [DOI] [PubMed] [Google Scholar]
- Galan JE, Curtiss R., 3rd Virulence and vaccine potential of phoP mutants of Salmonella typhimurium. Microb Pathog. 1989;6:433–443. doi: 10.1016/0882-4010(89)90085-5. [DOI] [PubMed] [Google Scholar]
- Garcia Vescovi E, Soncini FC, Groisman EA. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell. 1996;84:165–174. doi: 10.1016/s0092-8674(00)81003-x. [DOI] [PubMed] [Google Scholar]
- Glagolev AN, Skulachev VP. The proton pump is a molecular engine of motile bacteria. Nature. 1978;272:280–282. doi: 10.1038/272280a0. [DOI] [PubMed] [Google Scholar]
- Godaly G, Bergsten G, Hang L, Fischer H, Frendeus B, Lundstedt AC, Samuelsson M, Samuelsson P, Svanborg C. Neutrophil recruitment, chemokine receptors, and resistance to mucosal infection. J Leukoc Biol. 2001;69:899–906. [PubMed] [Google Scholar]
- Golubeva YA, Slauch JM. Salmonella enterica serovar Typhimurium periplasmic superoxide dismutase SodCI is a member of the PhoPQ regulon and is induced in macrophages. J Bacteriol. 2006;188:7853–7861. doi: 10.1128/JB.00706-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenheid S, Pinner E, Desjardins M, Gros P. Natural resistance to infection with intracellular pathogens: the Nramp1 protein is recruited to the membrane of the phagosome. J Exp Med. 1997;185:717–730. doi: 10.1084/jem.185.4.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundling A, Manson MD, Young R. Holins kill without warning. Proc Natl Acad Sci U S A. 2001;98:9348–9352. doi: 10.1073/pnas.151247598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunn JS, Lim KB, Krueger J, Kim K, Guo L, Hackett M, Miller SI. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol Microbiol. 1998;27:1171–1182. doi: 10.1046/j.1365-2958.1998.00757.x. [DOI] [PubMed] [Google Scholar]
- Guo L, Lim KB, Gunn JS, Bainbridge B, Darveau RP, Hackett M, Miller SI. Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science. 1997;276:250–253. doi: 10.1126/science.276.5310.250. [DOI] [PubMed] [Google Scholar]
- Guo L, Lim KB, Poduje CM, Daniel M, Gunn JS, Hackett M, Miller SI. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell. 1998;95:189–198. doi: 10.1016/s0092-8674(00)81750-x. [DOI] [PubMed] [Google Scholar]
- Hagberg L, Engberg I, Freter R, Lam J, Olling S, Svanborg Eden C. Ascending, unobstructed urinary tract infection in mice caused by pyelonephritogenic Escherichia coli of human origin. Infect Immun. 1983;40:273–283. doi: 10.1128/iai.40.1.273-283.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay JA. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem. 2008;77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Iyer R, Iverson TM, Accardi A, Miller C. A biological role for prokaryotic ClC chloride channels. Nature. 2002;419:715–718. doi: 10.1038/nature01000. [DOI] [PubMed] [Google Scholar]
- Jang S, Imlay JA. Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron-sulfur enzymes. J Biol Chem. 2007;282:929–937. doi: 10.1074/jbc.M607646200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia W, El Zoeiby A, Petruzziello TN, Jayabalasingham B, Seyedirashti S, Bishop RE. Lipid trafficking controls endotoxin acylation in outer membranes of Escherichia coli. J Biol Chem. 2004;279:44966–44975. doi: 10.1074/jbc.M404963200. [DOI] [PubMed] [Google Scholar]
- Karlinsey JE, Maguire ME, Becker LA, Crouch ML, Fang FC. The phage shock protein PspA facilitates divalent metal transport and is required for virulence of Salmonella enterica sv. Typhimurium. Mol Microbiol. 2010;78:669–685. doi: 10.1111/j.1365-2958.2010.07357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehres DG, Zaharik ML, Finlay BB, Maguire ME. The NRAMP proteins of Salmonella typhimurium and Escherichia coli are selective manganese transporters involved in the response to reactive oxygen. Mol Microbiol. 2000;36:1085–1100. doi: 10.1046/j.1365-2958.2000.01922.x. [DOI] [PubMed] [Google Scholar]
- Kier LD, Weppelman RM, Ames BN. Regulation of nonspecific acid phosphatase in Salmonella: phoN and phoP genes. J Bacteriol. 1979;138:155–161. doi: 10.1128/jb.138.1.155-161.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B, Richards SM, Gunn JS, Slauch JM. Protecting against antimicrobial effectors in the phagosome allows SodCII to contribute to virulence in Salmonella enterica serovar Typhimurium. J Bacteriol. 2010;192:2140–2149. doi: 10.1128/JB.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klipper-Aurbach Y, Wasserman M, Braunspiegel-Weintrob N, Borstein D, Peleg S, Assa S, Karp M, Benjamini Y, Hochberg Y, Laron Z. Mathematical formulae for the prediction of the residual beta cell function during the first two years of disease in children and adolescents with insulin-dependent diabetes mellitus. Med Hypotheses. 1995;45:486–490. doi: 10.1016/0306-9877(95)90228-7. [DOI] [PubMed] [Google Scholar]
- Konings WN, Albers SV, Koning S, Driessen AJ. The cell membrane plays a crucial role in survival of bacteria and archaea in extreme environments. Antonie Van Leeuwenhoek. 2002;81:61–72. doi: 10.1023/a:1020573408652. [DOI] [PubMed] [Google Scholar]
- Korshunov SS, Imlay JA. A potential role for periplasmic superoxide dismutase in blocking the penetration of external superoxide into the cytosol of Gram-negative bacteria. Mol Microbiol. 2002;43:95–106. doi: 10.1046/j.1365-2958.2002.02719.x. [DOI] [PubMed] [Google Scholar]
- Krulwich TA, Davidson LF, Filip SJ, Jr, Zuckerman RS, Guffanti AA. The protonmotive force and beta-galactoside transport in Bacillus acidocaldarius. J Biol Chem. 1978;253:4599–4603. [PubMed] [Google Scholar]
- Lane MC, Alteri CJ, Smith SN, Mobley HL. Expression of flagella is coincident with uropathogenic Escherichia coli ascension to the upper urinary tract. Proc Natl Acad Sci U S A. 2007;104:16669–16674. doi: 10.1073/pnas.0607898104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little JW. Lysogeny, prophage induction, and lysogenic conversion. In: Waldor DIFMK, Adhya SL, editors. Phages, Their Role in Pathogenesis and Biotechnology. Washington, D.C.: ASM Press; 2005. [Google Scholar]
- Maguire ME. Magnesium transporters: properties, regulation and structure. Front Biosci. 2006;11:3149–3163. doi: 10.2741/2039. [DOI] [PubMed] [Google Scholar]
- Marteyn B, West NP, Browning DF, Cole JA, Shaw JG, Palm F, Mounier J, Prevost MC, Sansonetti P, Tang CM. Modulation of Shigella virulence in response to available oxygen in vivo. Nature. 2010;465:355–358. doi: 10.1038/nature08970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matin A, Wilson B, Zychlinsky E, Matin M. Proton motive force and the physiological basis of delta pH maintenance in thiobacillus acidophilus. J Bacteriol. 1982;150:582–591. doi: 10.1128/jb.150.2.582-591.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki K, Sugishita K, Fujii N, Miyajima K. Molecular basis for membrane selectivity of an antimicrobial peptide, magainin 2. Biochemistry. 1995;34:3423–3429. doi: 10.1021/bi00010a034. [DOI] [PubMed] [Google Scholar]
- Michels M, Bakker EP. Generation of a large, protonophore-sensitive proton motive force and pH difference in the acidophilic bacteria Thermoplasma acidophilum and Bacillus acidocaldarius. J Bacteriol. 1985;161:231–237. doi: 10.1128/jb.161.1.231-237.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SI, Kukral AM, Mekalanos JJ. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc Natl Acad Sci U S A. 1989;86:5054–5058. doi: 10.1073/pnas.86.13.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SI, Pulkkinen WS, Selsted ME, Mekalanos JJ. Characterization of defensin resistance phenotypes associated with mutations in the phoP virulence regulon of Salmonella typhimurium. Infect Immun. 1990;58:3706–3710. doi: 10.1128/iai.58.11.3706-3710.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minagawa S, Ogasawara H, Kato A, Yamamoto K, Eguchi Y, Oshima T, Mori H, Ishihama A, Utsumi R. Identification and molecular characterization of the Mg2+ stimulon of Escherichia coli. J Bacteriol. 2003;185:3696–3702. doi: 10.1128/JB.185.13.3696-3702.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobley HL, Green DM, Trifillis AL, Johnson DE, Chippendale GR, Lockatell CV, Jones BD, Warren JW. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect Immun. 1990;58:1281–1289. doi: 10.1128/iai.58.5.1281-1289.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsieurs P, De Keersmaecker S, Navarre WW, Bader MW, De Smet F, McClelland M, Fang FC, De Moor B, Vanderleyden J, Marchal K. Comparison of the PhoPQ regulon in Escherichia coli and Salmonella typhimurium. J Mol Evol. 2005;60:462–474. doi: 10.1007/s00239-004-0212-7. [DOI] [PubMed] [Google Scholar]
- Murata T, Tseng W, Guina T, Miller SI, Nikaido H. PhoPQ-mediated regulation produces a more robust permeability barrier in the outer membrane of Salmonella enterica serovar typhimurium. J Bacteriol. 2007;189:7213–7222. doi: 10.1128/JB.00973-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray SR, Ernst RK, Bermudes D, Miller SI, Low KB. pmrA(Con) confers pmrHFIJKL-dependent EGTA and polymyxin resistance on msbB Salmonella by decorating lipid A with phosphoethanolamine. J Bacteriol. 2007;189:5161–5169. doi: 10.1128/JB.01969-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natalello A, Doglia SM, Carey J, Grandori R. Role of flavin mononucleotide in the thermostability and oligomerization of Escherichia coli stress-defense protein WrbA. Biochemistry. 2007;46:543–553. doi: 10.1021/bi061769c. [DOI] [PubMed] [Google Scholar]
- Nougayrede JP, Homburg S, Taieb F, Boury M, Brzuszkiewicz E, Gottschalk G, Buchrieser C, Hacker J, Dobrindt U, Oswald E. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science. 2006;313:848–851. doi: 10.1126/science.1127059. [DOI] [PubMed] [Google Scholar]
- Oyston PC, Dorrell N, Williams K, Li SR, Green M, Titball RW, Wren BW. The response regulator PhoP is important for survival under conditions of macrophage-induced stress and virulence in Yersinia pestis. Infect Immun. 2000;68:3419–3425. doi: 10.1128/iai.68.6.3419-3425.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Imlay JA. High levels of intracellular cysteine promote oxidative DNA damage by driving the fenton reaction. J Bacteriol. 2003;185:1942–1950. doi: 10.1128/JB.185.6.1942-1950.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prost LR, Daley ME, Le Sage V, Bader MW, Le Moual H, Klevit RE, Miller SI. Activation of the bacterial sensor kinase PhoQ by acidic pH. Mol Cell. 2007;26:165–174. doi: 10.1016/j.molcel.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Richard H, Foster JW. Escherichia coli glutamate- and arginine-dependent acid resistance systems increase internal pH and reverse transmembrane potential. J Bacteriol. 2004;186:6032–6041. doi: 10.1128/JB.186.18.6032-6041.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims PJ, Waggoner AS, Wang CH, Hoffman JF. Studies on the mechanism by which cyanine dyes measure membrane potential in red blood cells and phosphatidylcholine vesicles. Biochemistry. 1974;13:3315–3330. doi: 10.1021/bi00713a022. [DOI] [PubMed] [Google Scholar]
- Smith SN, Hagan EC, Lane MC, Mobley HL. Dissemination and Systemic Colonization of Uropathogenic Escherichia coli in a Murine Model of Bacteremia. MBio. 2010;1 doi: 10.1128/mBio.00262-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Soncini FC, Garcia Vescovi E, Solomon F, Groisman EA. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J Bacteriol. 1996;178:5092–5099. doi: 10.1128/jb.178.17.5092-5099.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trent MS, Ribeiro AA, Lin S, Cotter RJ, Raetz CR. An inner membrane enzyme in Salmonella and Escherichia coli that transfers 4-amino-4-deoxy-L-arabinose to lipid A: induction on polymyxin-resistant mutants and role of a novel lipid-linked donor. J Biol Chem. 2001;276:43122–43131. doi: 10.1074/jbc.M106961200. [DOI] [PubMed] [Google Scholar]
- Vaara M. Agents that increase the permeability of the outer membrane. Microbiol Rev. 1992;56:395–411. doi: 10.1128/mr.56.3.395-411.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Torres A, Jones-Carson J, Mastroeni P, Ischiropoulos H, Fang FC. Antimicrobial actions of the NADPH phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. I. Effects on microbial killing by activated peritoneal macrophages in vitro. J Exp Med. 2000;192:227–236. doi: 10.1084/jem.192.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch RA, Burland V, Plunkett G, 3rd, Redford P, Roesch P, Rasko D, Buckles EL, Liou SR, Boutin A, Hackett J, Stroud D, Mayhew GF, Rose DJ, Zhou S, Schwartz DC, Perna NT, Mobley HL, Donnenberg MS, Blattner FR. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci USA. 2002;99:17020–17024. doi: 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winfield MD, Groisman EA. Phenotypic differences between Salmonella and Escherichia coli resulting from the disparate regulation of homologous genes. Proc Natl Acad Sci U S A. 2004;101:17162–17167. doi: 10.1073/pnas.0406038101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooster DG, Maruvada R, Blom AM, Prasadarao NV. Logarithmic phase Escherichia coli K1 efficiently avoids serum killing by promoting C4bp-mediated C3b and C4b degradation. Immunology. 2006;117:482–493. doi: 10.1111/j.1365-2567.2006.02323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wosten MM, Kox LF, Chamnongpol S, Soncini FC, Groisman EA. A signal transduction system that responds to extracellular iron. Cell. 2000;103:113–125. doi: 10.1016/s0092-8674(00)00092-1. [DOI] [PubMed] [Google Scholar]
- Yu XJ, McGourty K, Liu M, Unsworth KE, Holden DW. pH sensing by intracellular Salmonella induces effector translocation. Science. 2010;328:1040–1043. doi: 10.1126/science.1189000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Lin S, Cotter RJ, Raetz CR. Lipid A modifications characteristic of Salmonella typhimurium are induced by NH4VO3 in Escherichia coli K12. Detection of 4-amino-4-deoxy-L-arabinose, phosphoethanolamine and palmitate. J Biol Chem. 1999;274:18503–18514. doi: 10.1074/jbc.274.26.18503. [DOI] [PubMed] [Google Scholar]
- Zwir I, Shin D, Kato A, Nishino K, Latifi T, Solomon F, Hare JM, Huang H, Groisman EA. Dissecting the PhoP regulatory network of Escherichia coli and Salmonella enterica. Proc Natl Acad Sci U S A. 2005;102:2862–2867. doi: 10.1073/pnas.0408238102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.