Completion of the Human Genome Project in 2003 ushered in a wave of optimism and anticipation that new therapies and even cures for many diseases would soon be forthcoming [1]. Aside from impressive progress in reducing the costs of genotyping [2], the promise offered by the Human Genome Project has been largely unrealized, particularly in relation to stroke [3]. More than 100 Genome-Wide Association studies [4] made possible with the new information provided by the Human Genome Project have yielded many interesting findings about the genetics of stroke-related brain injury, but all have generally fallen far short of identifying a genetic basis for vulnerability to cerebral ischemia [5]. With the notable exception of monogenic diseases, Genome-Wide Association studies have generally not been an efficient strategy to elucidate the genetic mechanisms of disease, particularly in complex pathologies such as ischemic cerebral injury [6]. These studies have taught us that most disease pathologies, including those associated with cerebral ischemia, are polygenic and involve highly variable contributions from the genes involved. Such findings raise the important question: why are the genetic components of complex diseases so variable?
The Three Categories of Epigenetic Mechanisms
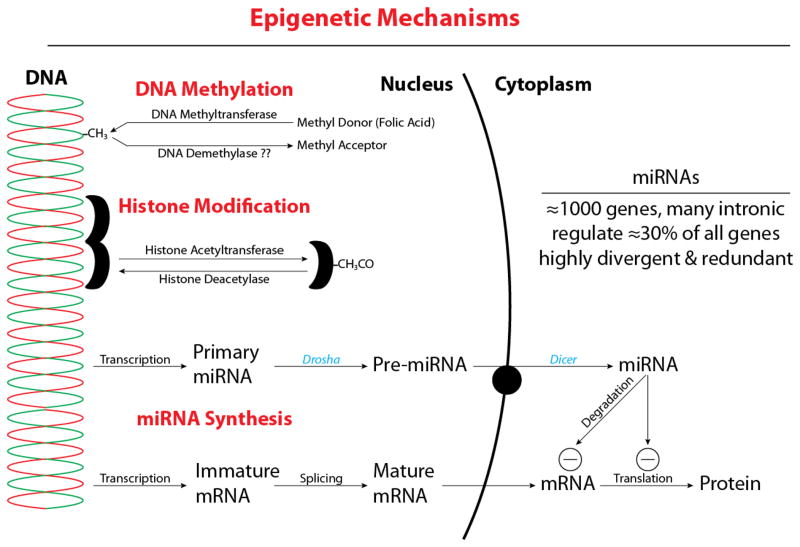
Long before the Human Genome Project was completed, it was recognized that genetic factors were not the only, or perhaps even not the most important, determinants of responses to some diseases. Indeed, in the same issue of Nature Genetics that was dedicated to completion of the Human Genome Project, it was already recognized that “epigenetic” factors were major players in the etiology and progression of many diseases [7]. Following completion of the Human Genome Project, understanding of epigenetic mechanisms has expanded rapidly and it is now recognized that epigenetic regulation involves three main categories of mechanisms [8, 9], as summarized in Figure 1.
Figure 1.
Epigenetic mechanisms can be divided into three main categories. The first includes the mechanisms mediating DNA methylation, typically at cytosine residues in gene promoter regions. These reactions attenuate gene expression and are catalyzed by multiple different isoforms of DNA Methyltransferases. An important requirement for these reactions is a methyl donor, typically folic acid supplied through the diet. DNA demethylation has been proposed, but has not yet been demonstrated in vivo. The second epigenetic category of mechanisms includes the enzymes that acetylate and deacetylate lysine residues on histone proteins. These enzymes regulate chromatin structure and include Histone Acetyltransferases and Histone Deacetylases. In general, histone acetylation promotes dissociation from DNA and facilitates gene expression, whereas deacetylation promotes reassociation and reduced gene expression. The third epigenetic category includes the pathways that transcribe, process, and transport miRNA, which arises from more than 1000 genes. In many cases, miRNA is coded by intronic regions of DNA, and regulates the gene coded by adjacent exons. In general, miRNAs are highly divergent, such that one miRNA can bind multiple different mRNAs. In addition, miRNAs are also highly redundant; many different miRNAs can regulate a single gene mRNA. Overall, approximately 30% of all genes are regulated by miRNAs.
The most widely studied category of epigenetic mechanisms includes the enzymes that mediate DNA methylation. These reactions are catalyzed by DNA Methyltransferases [10], which play a critical role in many cancers [11]. DNA methylation occurs most often on cytosine residues in promoter regions of regulated genes, and can greatly attenuate gene expression. Many DNA methylation reactions exhibit specific developmental timing and require methyl donors, such as folic acid, for their function. This is one reason why dietary folic acid is so important during pregnancy [12]. Work with DNA Methyltransferases has helped identify multiple inhibitors of DNA methylation, many of which have found use in clinical studies of cancer [13]. DNA demethylation reactions have been proposed, but clear evidence for enzymes reversing DNA methylation, in vivo, is not yet well established [14].
A second category of epigenetic mechanisms involves the enzymes that reversibly acetylate and methylate the histone proteins that tightly bind DNA. This large and diverse group of enzymes plays a key role chromatin structure and gene expression [15, 16], although the basis for their gene-specificity remains unclear [17]. As a general mechanism, acetylation of the lysine residues on histone proteins adds negative charge, which in turn promotes their dissociation from DNA; deacetylation reverses this process and promotes gene “silencing” [18]. Studies of histone acetyltransferases and deacetylases have also helped identify a broad variety of selective inhibitors, many of which are in clinical trials [19, 20].
The third and most recently recognized category of epigenetic mechanisms includes the pathways involved in the transcription, processing, and action of a class of short (≈20–25 nucleotides) RNA molecules identified as micro-RNAs (miRNAs) [21]. In contrast to DNA methylation and histone modification, the main function of miRNAs is involved much more with message translation than with gene transcription. The miRNA molecules directly bind mRNA and either retard or accelerate its degradation. In addition, miRNA binding to mRNA can block message translation [22]. The sequences coding for miRNAs often arise from intronic DNA, and regulate the gene products coded by adjacent exons. More than 1000 unique sequences of miRNA have been identified, and together these regulate approximately 30% of all mammalian genes [22]. A single miRNA can help regulate multiple different gene products, and a single gene product can be regulated by multiple different miRNAs. As such, miRNAs play key roles in many cellular functions and are particularly important in cardiovascular biology [23–28]. Expression and action of miRNAs change with development and in response to nutritional stress [29]. The actions of specific miRNA molecules can be inhibited by reverse-sense antagomirs, and these have proven useful in many studies of miRNA function [30, 31].
Epigenetic Mechanisms in Physiological Regulation
Apart from the molecular mechanisms responsible for epigenetic regulation, a broad variety of evidence has implicated epigenetic regulation in long-term environmental influences on gene regulation. One of the best-known such examples is the epidemiological work of Barker, who identified a cohort of Dutch individuals with a uniquely elevated risk of coronary artery disease [32, 33]. The common feature among this cohort was maternal food restriction during the Dutch famine in World War II. These early studies established that fetal nutritional stress could produce life-long changes in the vulnerability to cardiovascular disease, and subsequent work has further established the epigenetic basis of such “vascular programming” [34]. Similarly, other studies have implicated epigenetic mechanisms in long-term responses to hypoxia [35–38] and ischemia [39–41]. Of particular relevance to stroke are findings that miRNA is involved in ischemic preconditioning [42–48] and may even play a role in ischemic postconditioning [49]. Together, these results emphasize that environmental influences can produce long-term changes in physiological patterns of gene expression through epigenetic mechanisms.
Epigenetic Mechanisms in Stroke
Appreciation of the potential involvement of epigenetic mechanisms in the incidences and outcomes of stroke has begun to motivate studies of these mechanisms in relation to cerebral ischemia and stroke [50]. DNA methylation has been suggested to contribute to delayed ischemic brain injury in mice [51] and has been correlated with stroke risk in humans [52, 53]. Histone modifications have been implicated in LPS-induced cerebral inflammation [54] and oxidative neuronal injury [55], and may be neuroprotective following ischemia [56] in rodent brains. Correspondingly, inhibitors of histone modification have been suggested to be neuroprotective in animal models of cerebral ischemia [57–61] and intracranial hemorrhage [62]. In turn, miRNAs have been shown to play diverse roles in neuronal [63–66], glial [67] and endothelial [68, 69] responses to stroke. In addition, miRNAs have been suggested to regulate the effects of ischemia on aquaporin expression and function [70] and in some cases may be neuroprotective [71]. miRNAs may also help explain gender-based differences in responses to cerebral ischemia [72]. Although studies of epigenetic mechanisms in stroke are still in the early stages, the data accumulated to date strongly suggest that further studies of these mechanisms are well justified and that future publications resulting from these studies are worthy of careful attention.
Epigenetic Mechanisms and Cell-Based Therapies
Outside the brain, epigenetic mechanisms may contribute to the cerebral vulnerability to ischemia through many different pathways. Epigenetic mechanisms appear to play a major role in the development and progression of diabetes [73–75], and are intimately involved in regulation of angiogenesis [30, 76–83] and growth factor function [84]. Correspondingly, epigenetic mechanisms can play key roles in cell proliferation and differention [85–93]. These diverse findings suggest that the success of cell-based therapies, such as those discussed in this issue of Translational Stroke Research, may also depend, at least in part, on epigenetic mechanisms. Without doubt, studies of epigenetics are expanding rapidly and offer great promise for improved understanding of stroke pathology. Translational Stroke Research has already included two manuscripts on this topic [94–95], and another by Baltan appears in this issue. More are sure to come.
References
- 1.Botstein D, Risch N. Discovering genotypes underlying human phenotypes: Past successes for mendelian disease, future approaches for complex disease. Nat Genet. 2003;33 (Suppl):228–237. doi: 10.1038/ng1090. [DOI] [PubMed] [Google Scholar]
- 2.Ng PC, Kirkness EF. Whole genome sequencing. Methods Mol Biol. 2010;628:215–226. doi: 10.1007/978-1-60327-367-1_12. [DOI] [PubMed] [Google Scholar]
- 3.Lanktree MB, Dichgans M, Hegele RA. Advances in genomic analysis of stroke: What have we learned and where are we headed? Stroke. 2010;41:825–832. doi: 10.1161/STROKEAHA.109.570523. [DOI] [PubMed] [Google Scholar]
- 4.Marnellos G. High-throughput snp analysis for genetic association studies. Curr Opin Drug Discov Devel. 2003;6:317–321. [PubMed] [Google Scholar]
- 5.Meschia JF, Worrall BB, Rich SS. Genetic susceptibility to ischemic stroke. Nat Rev Neurol. 2011;7:369–378. doi: 10.1038/nrneurol.2011.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glaser B. Genetic analysis of complex disease--a roadmap to understanding or a colossal waste of money. Pediatr Endocrinol Rev. 2010;7:258–265. [PubMed] [Google Scholar]
- 7.Jaenisch R, Bird A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33 (Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 8.Hamilton JP. Epigenetics: Principles and practice. Dig Dis. 2011;29:130–135. doi: 10.1159/000323874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ballestar E. An introduction to epigenetics. Adv Exp Med Biol. 2011;711:1–11. doi: 10.1007/978-1-4419-8216-2_1. [DOI] [PubMed] [Google Scholar]
- 10.Jurkowska RZ, Jurkowski TP, Jeltsch A. Structure and function of mammalian DNA methyltransferases. Chembiochem. 2011;12:206–222. doi: 10.1002/cbic.201000195. [DOI] [PubMed] [Google Scholar]
- 11.Khandige S, Shanbhogue VV, Chakrabarty S, Kapettu S. Methylation markers: A potential force driving cancer diagnostics forward. Oncol Res. 2011;19:105–110. doi: 10.3727/096504011x12935427587641. [DOI] [PubMed] [Google Scholar]
- 12.McKay JA, Waltham KJ, Williams EA, Mathers JC. Folate depletion during pregnancy and lactation reduces genomic DNA methylation in murine adult offspring. Genes Nutr. 2011;6:189–196. doi: 10.1007/s12263-010-0199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mani S, Herceg Z. DNA demethylating agents and epigenetic therapy of cancer. Adv Genet. 2010;70:327–340. doi: 10.1016/B978-0-12-380866-0.60012-5. [DOI] [PubMed] [Google Scholar]
- 14.He XJ, Chen T, Zhu JK. Regulation and function of DNA methylation in plants and animals. Cell Res. 2011;21:442–465. doi: 10.1038/cr.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Selvi BR, Mohankrishna DV, Ostwal YB, Kundu TK. Small molecule modulators of histone acetylation and methylation: A disease perspective. Biochim Biophys Acta. 2010;1799:810–828. doi: 10.1016/j.bbagrm.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 16.Khan SN, Khan AU. Role of histone acetylation in cell physiology and diseases: An update. Clin Chim Acta. 2010;411:1401–1411. doi: 10.1016/j.cca.2010.06.020. [DOI] [PubMed] [Google Scholar]
- 17.Cho Y, Griswold A, Campbell C, Min KT. Individual histone deacetylases in drosophila modulate transcription of distinct genes. Genomics. 2005;86:606–617. doi: 10.1016/j.ygeno.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 18.Wanczyk M, Roszczenko K, Marcinkiewicz K, Bojarczuk K, Kowara M, Winiarska M. Hdaci--going through the mechanisms. Front Biosci. 2011;16:340–359. doi: 10.2741/3691. [DOI] [PubMed] [Google Scholar]
- 19.Chuang DM, Leng Y, Marinova Z, Kim HJ, Chiu CT. Multiple roles of hdac inhibition in neurodegenerative conditions. Trends Neurosci. 2009;32:591–601. doi: 10.1016/j.tins.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iwamoto FM, Lamborn KR, Kuhn JG, Wen PY, Yung WK, Gilbert MR, Chang SM, Lieberman FS, Prados MD, Fine HA. A phase i/ii trial of the histone deacetylase inhibitor romidepsin for adults with recurrent malignant glioma: North american brain tumor consortium study 03-03. Neuro Oncol. 2011;13:509–516. doi: 10.1093/neuonc/nor017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sato F, Tsuchiya S, Meltzer SJ, Shimizu K. Micrornas and epigenetics. FEBS J. 2011;278:1598–1609. doi: 10.1111/j.1742-4658.2011.08089.x. [DOI] [PubMed] [Google Scholar]
- 22.Felekkis K, Touvana E, Stefanou C, Deltas C. Micrornas: A newly described class of encoded molecules that play a role in health and disease. Hippokratia. 2010;14:236–240. [PMC free article] [PubMed] [Google Scholar]
- 23.Pan ZW, Lu YJ, Yang BF. Micrornas: A novel class of potential therapeutic targets for cardiovascular diseases. Acta Pharmacol Sin. 2010;31:1–9. doi: 10.1038/aps.2009.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu N, Olson EN. Microrna regulatory networks in cardiovascular development. Dev Cell. 2010;18:510–525. doi: 10.1016/j.devcel.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng Y, Zhang C. Microrna-21 in cardiovascular disease. J Cardiovasc Transl Res. 2010;3:251–255. doi: 10.1007/s12265-010-9169-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Small EM, Olson EN. Pervasive roles of micrornas in cardiovascular biology. Nature. 2011;469:336–342. doi: 10.1038/nature09783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen KC, Wang YS, Hu CY, Chang WC, Liao YC, Dai CY, Juo SH. Oxldl up-regulates microrna-29b, leading to epigenetic modifications of mmp-2/mmp-9 genes: A novel mechanism for cardiovascular diseases. FASEB J. 2011 doi: 10.1096/fj.10-174904. [DOI] [PubMed] [Google Scholar]
- 28.Han M, Toli J, Abdellatif M. Micrornas in the cardiovascular system. Curr Opin Cardiol. 2011;26:181–189. doi: 10.1097/HCO.0b013e328345983d. [DOI] [PubMed] [Google Scholar]
- 29.Khorram O, Han G, Bagherpour R, Magee TR, Desai M, Ross MG, Chaudhri AA, Toloubeydokhti T, Pearce WJ. The effect of maternal undernutrition on vascular expression of micro and messenger rna in newborn and aging offspring. Am J Physiol Regul Integr Comp Physiol. 2010;298:R1366–1374. doi: 10.1152/ajpregu.00704.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Solingen C, Seghers L, Bijkerk R, Duijs JM, Roeten MK, van Oeveren-Rietdijk AM, Baelde HJ, Monge M, Vos JB, de Boer HC, Quax PH, Rabelink TJ, van Zonneveld AJ. Antagomir-mediated silencing of endothelial cell specific microrna-126 impairs ischemia-induced angiogenesis. J Cell Mol Med. 2009;13:1577–1585. doi: 10.1111/j.1582-4934.2008.00613.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ye Y, Hu Z, Lin Y, Zhang C, Perez-Polo JR. Downregulation of microrna-29 by antisense inhibitors and a ppar-gamma agonist protects against myocardial ischaemia-reperfusion injury. Cardiovasc Res. 2010;87:535–544. doi: 10.1093/cvr/cvq053. [DOI] [PubMed] [Google Scholar]
- 32.Barker DJ, Osmond C, Golding J, Kuh D, Wadsworth ME. Growth in utero, blood pressure in childhood and adult life, and mortality from cardiovascular disease. BMJ. 1989;298:564–567. doi: 10.1136/bmj.298.6673.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McMillen IC, Robinson JS. Developmental origins of the metabolic syndrome: Prediction, plasticity, and programming. Physiol Rev. 2005;85:571–633. doi: 10.1152/physrev.00053.2003. [DOI] [PubMed] [Google Scholar]
- 34.Geelhoed JJ, Jaddoe VW. Early influences on cardiovascular and renal development. Eur J Epidemiol. 2010;25:677–692. doi: 10.1007/s10654-010-9510-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watson JA, Watson CJ, McCann A, Baugh J. Epigenetics, the epicenter of the hypoxic response. Epigenetics. 2010;5:293–296. doi: 10.4161/epi.5.4.11684. [DOI] [PubMed] [Google Scholar]
- 36.Chen S, Sang N. Histone deacetylase inhibitors: The epigenetic therapeutics that repress hypoxia-inducible factors. J Biomed Biotechnol. 2011;2011:197946. doi: 10.1155/2011/197946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Biswas S, Roy S, Banerjee J, Hussain SR, Khanna S, Meenakshisundaram G, Kuppusamy P, Friedman A, Sen CK. Hypoxia inducible microrna 210 attenuates keratinocyte proliferation and impairs closure in a murine model of ischemic wounds. Proc Natl Acad Sci U S A. 2010;107:6976–6981. doi: 10.1073/pnas.1001653107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan SY, Loscalzo J. Microrna-210: A unique and pleiotropic hypoxamir. Cell Cycle. 2010;9 doi: 10.4161/cc.9.6.11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stowell C, Wang L, Arbogast B, Lan JQ, Cioffi GA, Burgoyne CF, Zhou A. Retinal proteomic changes under different ischemic conditions - implication of an epigenetic regulatory mechanism. Int J Physiol Pathophysiol Pharmacol. 2010;2:148–160. [PMC free article] [PubMed] [Google Scholar]
- 40.Fasanaro P, Greco S, Ivan M, Capogrossi MC, Martelli F. Microrna: Emerging therapeutic targets in acute ischemic diseases. Pharmacol Ther. 2010;125:92–104. doi: 10.1016/j.pharmthera.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 41.Hsieh CH, Jeng JC, Jeng SF, Wu CJ, Lu TH, Liliang PC, Rau CS, Chen YC, Lin CJ. Microrna profiling in ischemic injury of the gracilis muscle in rats. BMC Musculoskelet Disord. 2010;11:123. doi: 10.1186/1471-2474-11-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yin C, Salloum FN, Kukreja RC. A novel role of microrna in late preconditioning: Upregulation of endothelial nitric oxide synthase and heat shock protein 70. Circ Res. 2009;104:572–575. doi: 10.1161/CIRCRESAHA.108.193250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim HW, Haider HK, Jiang S, Ashraf M. Ischemic preconditioning augments survival of stem cells via mir-210 expression by targeting caspase-8-associated protein 2. J Biol Chem. 2009;284:33161–33168. doi: 10.1074/jbc.M109.020925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lusardi TA, Farr CD, Faulkner CL, Pignataro G, Yang T, Lan J, Simon RP, Saugstad JA. Ischemic preconditioning regulates expression of micrornas and a predicted target, mecp2, in mouse cortex. J Cereb Blood Flow Metab. 2010;30:744–756. doi: 10.1038/jcbfm.2009.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng Y, Zhu P, Yang J, Liu X, Dong S, Wang X, Chun B, Zhuang J, Zhang C. Ischaemic preconditioning-regulated mir-21 protects heart against ischaemia/reperfusion injury via anti-apoptosis through its target pdcd4. Cardiovasc Res. 2010;87:431–439. doi: 10.1093/cvr/cvq082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee ST, Chu K, Jung KH, Yoon HJ, Jeon D, Kang KM, Park KH, Bae EK, Kim M, Lee SK, Roh JK. Micrornas induced during ischemic preconditioning. Stroke. 2010;41:1646–1651. doi: 10.1161/STROKEAHA.110.579649. [DOI] [PubMed] [Google Scholar]
- 47.Saugstad JA. Micrornas as effectors of brain function with roles in ischemia and injury, neuroprotection, and neurodegeneration. J Cereb Blood Flow Metab. 2010;30:1564–1576. doi: 10.1038/jcbfm.2010.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salloum FN, Yin C, Kukreja RC. Role of mirs in cardiac preconditioning. J Cardiovasc Pharmacol. 2010 doi: 10.1097/FJC.0b013e3181f581ba. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He B, Xiao J, Ren AJ, Zhang YF, Zhang H, Chen M, Xie B, Gao XG, Wang YW. Role of mir-1 and mir-133a in myocardial ischemic postconditioning. J Biomed Sci. 2011;18:22. doi: 10.1186/1423-0127-18-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qureshi IA, Mehler MF. Emerging role of epigenetics in stroke: Part 1: DNA methylation and chromatin modifications. Arch Neurol. 2010;67:1316. doi: 10.1001/archneurol.2010.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Endres M, Meisel A, Biniszkiewicz D, Namura S, Prass K, Ruscher K, Lipski A, Jaenisch R, Moskowitz MA, Dirnagl U. DNA methyltransferase contributes to delayed ischemic brain injury. J Neurosci. 2000;20:3175–3181. doi: 10.1523/JNEUROSCI.20-09-03175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim M, Long TI, Arakawa K, Wang R, Yu MC, Laird PW. DNA methylation as a biomarker for cardiovascular disease risk. PLoS One. 2010;5:e9692. doi: 10.1371/journal.pone.0009692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baccarelli A, Wright R, Bollati V, Litonjua A, Zanobetti A, Tarantini L, Sparrow D, Vokonas P, Schwartz J. Ischemic heart disease and stroke in relation to blood DNA methylation. Epidemiology. 2010;21:819–828. doi: 10.1097/EDE.0b013e3181f20457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suuronen T, Huuskonen J, Pihlaja R, Kyrylenko S, Salminen A. Regulation of microglial inflammatory response by histone deacetylase inhibitors. J Neurochem. 2003;87:407–416. doi: 10.1046/j.1471-4159.2003.02004.x. [DOI] [PubMed] [Google Scholar]
- 55.Soriano FX, Papadia S, Bell KF, Hardingham GE. Role of histone acetylation in the activity-dependent regulation of sulfiredoxin and sestrin 2. Epigenetics. 2009;4:152–158. doi: 10.4161/epi.4.3.8753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Langley B, Brochier C, Rivieccio MA. Targeting histone deacetylases as a multifaceted approach to treat the diverse outcomes of stroke. Stroke. 2009;40:2899–2905. doi: 10.1161/STROKEAHA.108.540229. [DOI] [PubMed] [Google Scholar]
- 57.Langley B, Gensert JM, Beal MF, Ratan RR. Remodeling chromatin and stress resistance in the central nervous system: Histone deacetylase inhibitors as novel and broadly effective neuroprotective agents. Curr Drug Targets CNS Neurol Disord. 2005;4:41–50. doi: 10.2174/1568007053005091. [DOI] [PubMed] [Google Scholar]
- 58.Faraco G, Pancani T, Formentini L, Mascagni P, Fossati G, Leoni F, Moroni F, Chiarugi A. Pharmacological inhibition of histone deacetylases by suberoylanilide hydroxamic acid specifically alters gene expression and reduces ischemic injury in the mouse brain. Mol Pharmacol. 2006;70:1876–1884. doi: 10.1124/mol.106.027912. [DOI] [PubMed] [Google Scholar]
- 59.Kim HJ, Rowe M, Ren M, Hong JS, Chen PS, Chuang DM. Histone deacetylase inhibitors exhibit anti-inflammatory and neuroprotective effects in a rat permanent ischemic model of stroke: Multiple mechanisms of action. J Pharmacol Exp Ther. 2007;321:892–901. doi: 10.1124/jpet.107.120188. [DOI] [PubMed] [Google Scholar]
- 60.Baltan S, Murphy SP, Danilov CA, Bachleda A, Morrison RS. Histone deacetylase inhibitors preserve white matter structure and function during ischemia by conserving atp and reducing excitotoxicity. J Neurosci. 2011;31:3990–3999. doi: 10.1523/JNEUROSCI.5379-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lv L, Tang YP, Han X, Wang X, Dong Q. Therapeutic application of histone deacetylase inhibitors for stroke. Cent Nerv Syst Agents Med Chem. 2011 doi: 10.2174/187152411796011330. [DOI] [PubMed] [Google Scholar]
- 62.Sinn DI, Kim SJ, Chu K, Jung KH, Lee ST, Song EC, Kim JM, Park DK, Kun Lee S, Kim M, Roh JK. Valproic acid-mediated neuroprotection in intracerebral hemorrhage via histone deacetylase inhibition and transcriptional activation. Neurobiol Dis. 2007;26:464–472. doi: 10.1016/j.nbd.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 63.Dharap A, Bowen K, Place R, Li LC, Vemuganti R. Transient focal ischemia induces extensive temporal changes in rat cerebral micrornaome. J Cereb Blood Flow Metab. 2009;29:675–687. doi: 10.1038/jcbfm.2008.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu DZ, Tian Y, Ander BP, Xu H, Stamova BS, Zhan X, Turner RJ, Jickling G, Sharp FR. Brain and blood microrna expression profiling of ischemic stroke, intracerebral hemorrhage, and kainate seizures. J Cereb Blood Flow Metab. 2010;30:92–101. doi: 10.1038/jcbfm.2009.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yin KJ, Deng Z, Huang H, Hamblin M, Xie C, Zhang J, Chen YE. Mir-497 regulates neuronal death in mouse brain after transient focal cerebral ischemia. Neurobiol Dis. 2010;38:17–26. doi: 10.1016/j.nbd.2009.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yuan Y, Wang JY, Xu LY, Cai R, Chen Z, Luo BY. Microrna expression changes in the hippocampi of rats subjected to global ischemia. J Clin Neurosci. 2010;17:774–778. doi: 10.1016/j.jocn.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 67.Ziu M, Fletcher L, Rana S, Jimenez DF, Digicaylioglu M. Temporal differences in microrna expression patterns in astrocytes and neurons after ischemic injury. PLoS One. 2011;6:e14724. doi: 10.1371/journal.pone.0014724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yin KJ, Deng Z, Hamblin M, Xiang Y, Huang H, Zhang J, Jiang X, Wang Y, Chen YE. Peroxisome proliferator-activated receptor delta regulation of mir-15a in ischemia-induced cerebral vascular endothelial injury. J Neurosci. 2010;30:6398–6408. doi: 10.1523/JNEUROSCI.0780-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tan JR, Koo YX, Kaur P, Liu F, Armugam A, Wong PT, Jeyaseelan K. Micrornas in stroke pathogenesis. Curr Mol Med. 2011;11:76–92. doi: 10.2174/156652411794859232. [DOI] [PubMed] [Google Scholar]
- 70.Sepramaniam S, Armugam A, Lim KY, Karolina DS, Swaminathan P, Tan JR, Jeyaseelan K. Microrna 320a functions as a novel endogenous modulator of aquaporins 1 and 4 as well as a potential therapeutic target in cerebral ischemia. J Biol Chem. 2010;285:29223–29230. doi: 10.1074/jbc.M110.144576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Buller B, Liu X, Wang X, Zhang RL, Zhang L, Hozeska-Solgot A, Chopp M, Zhang ZG. Microrna-21 protects neurons from ischemic death. FEBS J. 2010;277:4299–4307. doi: 10.1111/j.1742-4658.2010.07818.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Siegel C, Li J, Liu F, Benashski SE, McCullough LD. Mir-23a regulation of x-linked inhibitor of apoptosis (xiap) contributes to sex differences in the response to cerebral ischemia. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1102635108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Villeneuve LM, Natarajan R. The role of epigenetics in the pathology of diabetic complications. Am J Physiol Renal Physiol. 2010;299:F14–25. doi: 10.1152/ajprenal.00200.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Villeneuve LM, Kato M, Reddy MA, Wang M, Lanting L, Natarajan R. Enhanced levels of microrna-125b in vascular smooth muscle cells of diabetic db/db mice lead to increased inflammatory gene expression by targeting the histone methyltransferase suv39h1. Diabetes. 2010;59:2904–2915. doi: 10.2337/db10-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Caporali A, Meloni M, Vollenkle C, Bonci D, Sala-Newby GB, Addis R, Spinetti G, Losa S, Masson R, Baker AH, Agami R, le Sage C, Condorelli G, Madeddu P, Martelli F, Emanueli C. Deregulation of microrna-503 contributes to diabetes mellitus-induced impairment of endothelial function and reparative angiogenesis after limb ischemia. Circulation. 2011;123:282–291. doi: 10.1161/CIRCULATIONAHA.110.952325. [DOI] [PubMed] [Google Scholar]
- 76.Urbich C, Kuehbacher A, Dimmeler S. Role of micrornas in vascular diseases, inflammation, and angiogenesis. Cardiovasc Res. 2008;79:581–588. doi: 10.1093/cvr/cvn156. [DOI] [PubMed] [Google Scholar]
- 77.Suarez Y, Fernandez-Hernando C, Yu J, Gerber SA, Harrison KD, Pober JS, Iruela-Arispe ML, Merkenschlager M, Sessa WC. Dicer-dependent endothelial micrornas are necessary for postnatal angiogenesis. Proc Natl Acad Sci U S A. 2008;105:14082–14087. doi: 10.1073/pnas.0804597105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Buysschaert I, Schmidt T, Roncal C, Carmeliet P, Lambrechts D. Genetics, epigenetics and pharmaco-(epi)genomics in angiogenesis. J Cell Mol Med. 2008;12:2533–2551. doi: 10.1111/j.1582-4934.2008.00515.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang S, Olson EN. Angiomirs--key regulators of angiogenesis. Curr Opin Genet Dev. 2009;19:205–211. doi: 10.1016/j.gde.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bonauer A, Carmona G, Iwasaki M, Mione M, Koyanagi M, Fischer A, Burchfield J, Fox H, Doebele C, Ohtani K, Chavakis E, Potente M, Tjwa M, Urbich C, Zeiher AM, Dimmeler S. Microrna-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science. 2009;324:1710–1713. doi: 10.1126/science.1174381. [DOI] [PubMed] [Google Scholar]
- 81.Daubman S. Micrornas in angiogenesis and vascular smooth muscle cell function. Circ Res. 2010;106:423–425. doi: 10.1161/RES.0b013e3181d61a0d. [DOI] [PubMed] [Google Scholar]
- 82.Ghosh G, Subramanian IV, Adhikari N, Zhang X, Joshi HP, Basi D, Chandrashekhar YS, Hall JL, Roy S, Zeng Y, Ramakrishnan S. Hypoxia-induced microrna-424 expression in human endothelial cells regulates hif-alpha isoforms and promotes angiogenesis. J Clin Invest. 2010;120:4141–4154. doi: 10.1172/JCI42980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bavan L, Midwood K, Nanchahal J. Microrna epigenetics: A new avenue for wound healing research. BioDrugs. 2011;25:27–41. doi: 10.2165/11585010-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 84.Chan MC, Hilyard AC, Wu C, Davis BN, Hill NS, Lal A, Lieberman J, Lagna G, Hata A. Molecular basis for antagonism between pdgf and the tgfbeta family of signalling pathways by control of mir-24 expression. EMBO J. 2010;29:559–573. doi: 10.1038/emboj.2009.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Romm E, Nielsen JA, Kim JG, Hudson LD. Myt1 family recruits histone deacetylase to regulate neural transcription. J Neurochem. 2005;93:1444–1453. doi: 10.1111/j.1471-4159.2005.03131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kuhn AR, Schlauch K, Lao R, Halayko AJ, Gerthoffer WT, Singer CA. Microrna expression in human airway smooth muscle cells: Role of mir-25 in regulation of airway smooth muscle phenotype. Am J Respir Cell Mol Biol. 2010;42:506–513. doi: 10.1165/rcmb.2009-0123OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Neppl RL, Wang DZ. Smooth(ing) muscle differentiation by micrornas. Cell Stem Cell. 2009;5:130–132. doi: 10.1016/j.stem.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Parmacek MS. Microrna-modulated targeting of vascular smooth muscle cells. J Clin Invest. 2009;119:2526–2528. doi: 10.1172/JCI40503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang C. Microrna and vascular smooth muscle cell phenotype: New therapy for atherosclerosis? Genome Med. 2009;1:85. doi: 10.1186/gm85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zeng L, Carter AD, Childs SJ. Mir-145 directs intestinal maturation in zebrafish. Proc Natl Acad Sci U S A. 2009;106:17793–17798. doi: 10.1073/pnas.0903693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Albinsson S, Suarez Y, Skoura A, Offermanns S, Miano JM, Sessa WC. Micrornas are necessary for vascular smooth muscle growth, differentiation, and function. Arterioscler Thromb Vasc Biol. 2010;30:1118–1126. doi: 10.1161/ATVBAHA.109.200873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen J, Yin H, Jiang Y, Radhakrishnan SK, Huang ZP, Li J, Shi Z, Kilsdonk EP, Gui Y, Wang DZ, Zheng XL. Induction of microrna-1 by myocardin in smooth muscle cells inhibits cell proliferation. Arterioscler Thromb Vasc Biol. 2011;31:368–375. doi: 10.1161/ATVBAHA.110.218149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ohtani K, Dimmeler S. Control of cardiovascular differentiation by micrornas. Basic Res Cardiol. 2011;106:5–11. doi: 10.1007/s00395-010-0139-7. [DOI] [PubMed] [Google Scholar]
- 94.Vemuganti R. The micrornas and stroke: No need to be coded to be counted. Transl Stroke Res. 2010;1:158–160. doi: 10.1007/s12975-010-0030-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lim KY, Chua J, Tan J, Swaminathan P, Sepramaniam S, Armugam A, Wong PT, Jeyaseelan K. Micrornas in cerebral ischemia. Transl Stroke Res. 2010;1:287–303. doi: 10.1007/s12975-010-0035-3. [DOI] [PubMed] [Google Scholar]