Abstract
Three genome-wide association studies in Europe and the USA have reported eight urinary bladder cancer (UBC) susceptibility loci. Using extended case and control series and 1000 Genomes imputations of 5 340 737 single-nucleotide polymorphisms (SNPs), we searched for additional loci in the European GWAS. The discovery sample set consisted of 1631 cases and 3822 controls from the Netherlands and 603 cases and 37 781 controls from Iceland. For follow-up, we used 3790 cases and 7507 controls from 13 sample sets of European and Iranian ancestry. Based on the discovery analysis, we followed up signals in the urea transporter (UT) gene SLC14A. The strongest signal at this locus was represented by a SNP in intron 3, rs17674580, that reached genome-wide significance in the overall analysis of the discovery and follow-up groups: odds ratio = 1.17, P = 7.6 × 10−11. SLC14A1 codes for UTs that define the Kidd blood group and are crucial for the maintenance of a constant urea concentration gradient in the renal medulla and, through this, the kidney's ability to concentrate urine. It is speculated that rs17674580, or other sequence variants in LD with it, indirectly modifies UBC risk by affecting urine production. If confirmed, this would support the ‘urogenous contact hypothesis’ that urine production and voiding frequency modify the risk of UBC.
INTRODUCTION
Globally each year, almost 400 000 new patients are diagnosed with urinary bladder cancer (UBC) and more than 150 000 patients die from the disease (1,2). Most patients with UBC are treated with conservative surgery but they experience an extremely high risk of frequent recurrences. Because of that, bladder cancer is the most expensive cancer in many Western communities (3). Both in the USA and western Europe, 1 in every 25 men and 1 in 80 women will develop bladder cancer sometime during life (1). This 3:1 male/female ratio is largely explained by historical differences in smoking habits and occupational exposure to carcinogens, the most important risk factors for UBC. Bladder cancer has historically not been perceived as a disease with a strong genetic background, even though high-risk UBC families have been identified (4). The risk of UBC is increased almost 2-fold for first-degree relatives of UBC cases but this clustering may largely be explained by low-penetrance genetic polymorphisms (5–8). Candidate gene association studies have consistently shown that NAT2 slow acetylator and GSTM1 null genotypes increase UBC risk (9). Dozens of other suggestions from such studies have not been replicated. Recently, three genome-wide association studies (GWAS) have identified eight additional UBC risk loci (10–13) (Table 1). All of these loci have been extensively replicated (12).
Table 1.
Extensively replicated UBC susceptibility loci
| Locus | Gene region | SNP | Risk allelea | Allelic OR | Risk allele frequency | Reference | Replicated by |
|---|---|---|---|---|---|---|---|
| 8q24.21 | MYC | rs9642880 | T | 1.22 | 0.45 | (10) | (12,13,39,50,51) |
| 3q28 | TP63 | rs710521 | A | 1.19 | 0.73 | (10) | (12,13,37) |
| 5p15.33 | TERT | rs2736098 | A | 1.12b | 0.26 | (52) | (53) |
| 5p15.33 | CLPTM1L | rs401681 | C | 1.07b | 0.54 | (52) | (12,53) |
| 8q24.3 | PSCA | rs2294008 | T | 1.15 | 0.46 | (13) | (12,54) |
| 4p16.3 | TACC3-FGFR3 | rs798766 | T | 1.24 | 0.19 | (11) | (12,55) |
| 22q13.1 | CBX6, APOBEC3A | rs1014971 | T | 1.14 | 0.62 | (12) | (12) included data from (11,13) |
| 19q12 | CCNE1 | rs8102137 | C | 1.13 | 0.33 | (12) | (12) included data from (11,13) |
| 2q37.1 | UGT1A | rs11892031 | A | 1.19 | 0.92 | (12) | (12) included data from (11,13) |
| 8p22 | NAT2 | rs1495741 | A | 1.15 | 0.80 | c | (12) |
| 1p13.3 | GSTM1 | — | Null | 1.47 | 0.51 | c | (12) |
aFor GSTM1, these do not refer to the risk allele but to the risk genotype.
bReported ORs are mutually adjusted for rs401681 and rs2736098.
cThese susceptibility loci have frequently been reported by candidate gene association studies, using different assays.
Here, we report on a new UBC susceptibility locus discovered from an extension of the European UBC GWAS.
RESULTS
To search for variants that affect the risk of UBC, we imputed 1000 Genomes project's single-nucleotide polymorphisms (SNPs) into our two discovery data sets, composed of 603 Icelandic cases and 37 781 Icelandic controls and 1631 Dutch cases and 3822 Dutch controls, genotyped on the HumanHap300 or HumanCNV370-duo BeadChips. For the Icelandic data set, the imputation was done with version 3 of the 1000 Genomes data set, whereas version 2 was used for the Dutch data set (see Materials and Methods). After excluding SNPs that failed quality control in either of the data sets and SNPs with minor allele frequency <0.01, 5 340 737 SNPs present in both data sets were included in a combined analysis of the two sets. After excluding SNPs that showed associations with significant heterogeneity between the two populations and a single SNP, rs10094872, located at the previously reported bladder cancer locus at 8q24 (10), no variants reached genome-wide significance, here defined conservatively as P < 1 × 10−8 (=0.05/5 million SNPs).
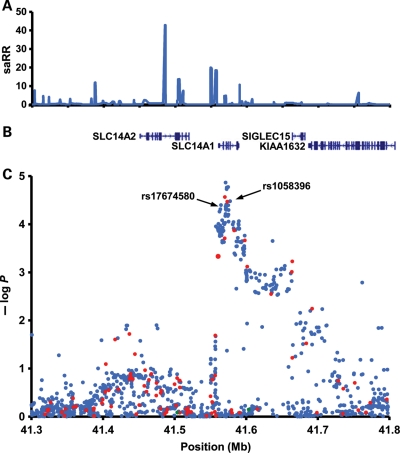
Assessing missense variants in protein-coding genes, we noted that among the top variants were two SNPs located in the solute carrier family 14, member 1 (SLC14A1 or UT-B) gene, a member of the SLC14A family of urea transporters (UTs) crucial to the kidney's ability to concentrate urine. The gene is also expressed on red blood cells where different isoforms, defined by two alleles Jka and Jkb, form the Kidd blood group system (14). The G allele of marker rs1058396 (rs1058396[G], SLC14A1 D280N) showed an odds ratio (OR) of 1.16 and a P-value of 3.4 × 10−5, whereas rs11877062[C] (SLC14A1 R4W) had an OR of 1.15 and a P-value of 9.8 × 10−5. The SNP that showed the stronger association with UBC, rs1058396[G], causes the amino acid change D280N which defines the two alleles of the Kidd blood group system (15). Since the two missense variants are highly correlated (r2= 0.88 based on the 566 chromosomes in the 1000 Genomes version 3 training set), subsequent follow-up studies focused on rs1058396 (D280N). Before proceeding to such follow-up studies, however, we scrutinized the association results between UBC and all tested SNPs in a 0.5 Mb region centered on rs1058396 (Fig. 1). Apart from rs1058396 (D280N) and rs11877062 (R4W), a total of 35 markers had P-values lower than 1.0 × 10−4 (Supplementary Material, Table S1) and of those, 18 were strongly correlated with the coding SNPs (r2> 0.8). For follow-up, we also selected the SNP rs17674580 as a representative of a group of 10 correlated SNPs that did not correlate well with rs1058396 (r2 ≈ 0.5). rs17674580 is located in an intron of SLC14A1, ∼12 kb centromeric to rs1058396 (D280N) (Fig. 1). The LD block where both rs17674580 and rs1058396 (D280N) are located contains only the SLC14A1 gene (Fig. 1). After adjusting for either rs1058396 or rs17674580, only one of the other 35 SNPs associated with UBC with nominal significance and none after adjusting for the number of SNPs (Supplementary Material, Table S1).
Figure 1.
A schematic view of the structure and association results in the UBC-associated region on chromosome 18q12.3. (A) Estimated recombination rates (saRR) in cM/Mb from the HapMap (release 22) Phase II data. (B) Location of known genes in the region. (C) Schematic view of the association with bladder cancer for all SNPs tested in the region for the initial scan (Iceland and the Netherlands). The y-axis indicates the –log 10 P-value. Red dots indicate SNPs directly genotyped in both discovery series; blue dots indicate SNPs imputed in both discovery series; green dots indicate SNPs directly genotyped in the Icelandic series but imputed in the Dutch series.
We genotyped both rs17674580 and rs1058396 (D280N) in 13 additional UBC case–control sample sets from Iceland, Italy, the UK, Spain, Sweden, Belgium, Germany, Eastern Europe and Iran (Table 2). Both SNPs replicated in the follow-up groups combined and both reached GW significance in the overall analysis of the discovery and follow-up groups with an OR of 1.17 and a P-value of 7.6 × 10−11 for rs17674580[T] and an OR of 1.14 and a P-value of 2.9 × 10−9 for rs1058396[G]. We did not observe significant heterogeneity of the ORs between studies [Phet = 0.10 and 0.54, I2 = 33.0 and 0 for rs17674580 and rs1058396 (D280N), respectively]. Relative to the non-carriers, the ORs for heterozygous and homozygous carriers of the rs17674580[T] were estimated to be 1.17 and 1.37, respectively.
Table 2.
Association of rs1058396[G] and rs17674580[T] on 18q12.3 with UBC
| Study population (n cases/n controls) | rs1058396[G] |
rs17674580[T] |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Frequency |
OR | 95% CI | P-value | Info | Frequency |
OR | 95% CI | P-value | Info | |||
| Cases | Controls | Cases | Controls | |||||||||
| Discovery groups (GWA) | ||||||||||||
| Iceland (603/37 781)a | 0.531 | 0.487 | 1.20 | 1.06–1.34 | 2.5 × 10−3 | 0.986 | 0.381 | 0.334 | 1.24 | 1.10–1.40 | 5.0 × 10−4 | 0.992 |
| The Netherlands (1631/3822)a | 0.548 | 0.515 | 1.14 | 1.05–1.24 | 1.6 × 10−3 | 1000 | 0.392 | 0.366 | 1.13 | 1.03–1.23 | 6.8 × 10−3 | 0.987 |
| Follow-up groups | ||||||||||||
| Iceland II (178/2055)b | 0.55 | 0.48 | 1.29 | 1.02–1.63 | 0.031 | 0.41 | 0.36 | 1.23 | 0.98–1.54 | 0.07 | ||
| Iceland III (350/3500)b | 0.50 | 0.48 | 1.18 | 1.05–1.34 | 0.0071 | 0.36 | 0.34 | 1.26 | 1.11–1.43 | 3.1 × 10−4 | ||
| UK (724/535) | 0.56 | 0.55 | 1.04 | 0.89–1.22 | 0.65 | 0.38 | 0.38 | 1.01 | 0.85–1.19 | 0.93 | ||
| Italy—Torino (328/389) | 0.51 | 0.51 | 1.00 | 0.81–1.23 | 0.97 | 0.40 | 0.35 | 1.22 | 0.98–1.53 | 0.072 | ||
| Italy—Brescia (181/192) | 0.52 | 0.46 | 1.26 | 0.95–1.68 | 0.11 | 0.38 | 0.34 | 1.21 | 0.89–1.64 | 0.22 | ||
| Belgium (191/378) | 0.48 | 0.52 | 0.87 | 0.68–1.21 | 0.27 | 0.32 | 0.38 | 0.78 | 0.60–1.02 | 0.064 | ||
| Eastern Europe (213/526) | 0.52 | 0.48 | 1.19 | 0.94–1.49 | 0.15 | 0.37 | 0.34 | 1.17 | 0.92–1.48 | 0.20 | ||
| Sweden (343/1264) | 0.52 | 0.51 | 1.07 | 0.90–1.27 | 0.46 | 0.36 | 0.34 | 1.12 | 0.94–1.34 | 0.20 | ||
| Spain (238/881) | 0.56 | 0.52 | 1.20 | 0.97–1.48 | 0.085 | 0.37 | 0.35 | 1.07 | 0.87–1.32 | 0.52 | ||
| Iran (269/246) | 0.57 | 0.54 | 1.09 | 0.85–1.39 | 0.51 | 0.36 | 0.33 | 1.15 | 0.88–1.49 | 0.31 | ||
| Germany—LW (213/198) | 0.55 | 0.50 | 1.23 | 0.93–1.62 | 0.14 | 0.40 | 0.33 | 1.35 | 1.01–1.80 | 0.044 | ||
| Germany—Dortmund (196/239) | 0.55 | 0.49 | 1.29 | 0.99–1.69 | 0.059 | 0.41 | 0.32 | 1.45 | 1.09–1.92 | 9.5 × 10−3 | ||
| Germany—Neuss (216/104) | 0.56 | 0.50 | 1.29 | 0.92–1.79 | 0.14 | 0.42 | 0.33 | 1.45 | 1.03–2.06 | 0.035 | ||
| GWA (2234/41 603)c | 1.16 | 1.08–1.24 | 3.4 × 10−5 | 1.17 | 1.08–1.25 | 4.9 × 10−5 | ||||||
| Follow-up groups (3790/7507)c | 1.13 | 1.07–1.20 | 2.5 × 10−5 | 1.16 | 1.10–1.23 | 5.8 × 10−7 | ||||||
| All combined (6024/49 110)c | 1.14 | 1.09–1.19 | 2.9 × 10−9 | 1.17 | 1.11–1.22 | 7.6 × 10−11 | ||||||
All P-values shown are two-sided. Shown are the corresponding numbers of cases and controls (n), allelic frequencies of variants in affected and control individuals, the allelic odds-ratio (OR) with P-values based on the multiplicative model.
aResults presented for Iceland and the Netherlands were individually adjusted by the method of genomic control (see Materials and Methods).
bIceland II consists of cases and controls genotyped by single-SNP assay, Iceland III consists of cases and controls genotyped in silico, using genotype information on relatives.
cFor the combined study populations, the control frequency was the average, unweighted control frequency of the individual populations, whereas the OR and the P-values were estimated using the Mantel–Haenszel model.
We examined the joint effect of rs17674580[T] and rs1058396[G] using samples directly genotyped by a single-track assay for both SNPs (Table 3). After adjusting for rs1058396[G] (D280N), the association of rs17674580[T] remained significant (OR = 1.1, 95% CI 1.03–1.18, P= 0.0061), whereas the association between rs1058396[G] (D280N) and UBC almost disappeared after adjustment for rs17674580[T].
Table 3.
Joint analysis of rs17674580[T] and rs1058396[G] in UBC
| Population | rs17674580[T] adjusted for rs1058396[G] |
rs1058396[G] adjusted for rs17674580[T] |
||||
|---|---|---|---|---|---|---|
| OR | 95% CI | P-value | OR | 95% CI | P-value | |
| The Netherlands | 1.04 | 0.92–1.18 | 0.52 | 1.11 | 0.98–1.25 | 0.094 |
| Iceland | 1.17 | 0.99–1.37 | 0.063 | 1.08 | 0.92–1.27 | 0.36 |
| Belgium | 0.80 | 0.55–1.17 | 0.25 | 1.00 | 0.70–1.43 | 1.00 |
| Germany—Lutherstadt–Wittenberg | 1.34 | 0.88–2.02 | 0.17 | 0.99 | 0.67–1.49 | 0.98 |
| Germany—Dortmund | 1.42 | 0.95–2.13 | 0.08 | 1.03 | 0.70–1.52 | 0.89 |
| Eastern Europe | 1.17 | 0.82–1.67 | 0.39 | 1.04 | 0.73–1.47 | 0.83 |
| Iceland II | 1.13 | 0.79–1.62 | 0.52 | 1.19 | 0.83–1.70 | 0.34 |
| Iran | 1.14 | 0.81–1.61 | 0.46 | 0.99 | 0.71–1.38 | 0.97 |
| Italy—Brescia | 0.99 | 0.63–1.55 | 0.97 | 1.30 | 0.85–1.99 | 0.23 |
| Italy—Torino | 1.33 | 0.97–1.83 | 0.07 | 0.85 | 0.63–1.16 | 0.30 |
| Germany—Neuss | 1.39 | 0.85–2.27 | 0.19 | 1.07 | 0.67–1.71 | 0.77 |
| Spain | 1.02 | 0.75–1.38 | 0.91 | 1.15 | 0.85–1.54 | 0.36 |
| Sweden | 1.18 | 0.91–1.54 | 0.20 | 0.94 | 0.73–1.21 | 0.63 |
| UK | 0.96 | 0.77–1.21 | 0.76 | 1.07 | 0.86–1.34 | 0.53 |
| Combined | 1.1 | 1.03–1.18 | 0.0061 | 1.06 | 1.00–1.14 | 0.069 |
| Phet = 0.62, I2 = 0.0 | Phet = 0.97, I2 = 0.0 | |||||
We have previously reported a UBC risk variant at the FGFR3/TACC3 locus at 4p16 that shows significantly stronger association with UBC with a predicted ‘low risk’ of progression (tumors confined to the bladder mucosa and not poorly differentiated) than with UBC with a predicted ‘high risk’ of progression (tumor invasion in or beyond the lamina propria or poorly differentiated). Using the same classification of cases, we found that the frequency of rs17674580[T] did not differ between patients with tumors of different aggressiveness (combined OR high versus low risk = 1.07, P= 0.17, Table 4). By regressing on the age at diagnosis for 4750 cases, we found that rs17674580[T] did not associate with age at diagnosis of UBC (effect = −0.25 years per allele; P= 0.27, Phet = 0.33).
Table 4.
Association of rs17674580[T] with risk of progression
| Study population (n high risk/n low risk) | rs17674580[T] |
||||||
|---|---|---|---|---|---|---|---|
| Frequency |
OR | 95% CI | P-value | ||||
| High risk | Low risk | ||||||
| Iceland (59/121) | 0.34 | 0.42 | 0.72 | 0.44–1.15 | 0.17 | ||
| The Netherlands (467/609) | 0.41 | 0.39 | 1.10 | 0.92–1.31 | 0.29 | ||
| UK (405/274) | 0.38 | 0.37 | 1.05 | 0.84–1.31 | 0.66 | ||
| Italy—Torino (112/152) | 0.40 | 0.40 | 0.99 | 0.70–1.40 | 0.95 | ||
| Italy—Brescia (126/53) | 0.39 | 0.36 | 1.14 | 0.71–1.82 | 0.59 | ||
| Eastern Europe (100/34) | 0.41 | 0.34 | 1.33 | 0.75–2.36 | 0.33 | ||
| Sweden (193/142) | 0.38 | 0.35 | 1.11 | 0.80–1.52 | 0.53 | ||
| Spain (135/28) | 0.36 | 0.30 | 1.27 | 0.68–2.34 | 0.45 | ||
| Germany—Lutherstadt–Wittenberg (142/65) | 0.41 | 0.38 | 1.12 | 0.73–1.71 | 0.60 | ||
| Germany—Dortmund (74/118) | 0.43 | 0.39 | 1.14 | 0.75–1.73 | 0.54 | ||
| Germany—Neuss (108/98) | 0.46 | 0.39 | 1.34 | 0.90–1.98 | 0.15 | ||
| All combined (1921/1694)a | 0.40 | 0.37 | 1.07 | 0.97–1.18 | 0.17 | Phet = 0.68 | I2 = 0.0 |
aFor the combined study populations, the control frequency was the average, unweighted control frequency of the individual populations, whereas the OR and the P-values were estimated using the Mantel–Haenszel model.
Since smoking is a strong risk factor for UBC and variants in the sequence of the genome that affect smoking behavior have been identified, we tested whether either rs17674580[T] or rs1058396[G] (D280N) was associated with smoking behavior, using genotypes and information on smoking from 17 617 Icelanders who have a history of smoking (16). Neither rs17674580[T] nor rs1058396[G] (D280N) associated with smoking quantity as measured by number of cigarettes smoked per day [P= 0.94 and 0.88 for rs17674580[T] and rs1058396[G] (D280N), respectively]. Finally, we tested for association between rs17674580[T] and serum urea in an Icelandic sample set that had urea measurements (n = 3641) (17). No significant association between blood urea and rs17674580 was observed (P= 0.68).
DISCUSSION
Urea is produced in the liver as the main nitrogenous end product of protein catabolism. Depending on the diet, ∼20–30 g of urea is produced per day and passed into the blood. It is filtered in the kidney and either excreted or absorbed and concentrated in the renal medulla in order to negate the osmotic effect of urea in the urine and thereby maintain body fluid balance (18). Specific proteins, called facilitative UTs, mediate urea flux across cellular membranes, necessary for the process of concentrating urine and salvaging nitrogen. These proteins are derived from two distinct genes, Solute Carrier Family 14, member 1; SLC14A1 (or UT-B) and SLC14A2 (or UT-A). The genes occur in tandem on chromosome 18q12.1–q21.1 and are probably the result of duplication of a primordial UT gene (19). UT-A and UT-B transporters have a similar basic structure and function but there are also a number of significant differences. Whereas UT-A transporters are mainly found in the kidney, UT-B expression is mainly found on the erythrocyte plasma membrane and, to a lesser extent, in the descending vasa recta of the kidney, in the brain, ear, testis, intestine and urinary bladder (18). Unlike UT-A, UT-Bs transport water in addition to urea, have a higher transport rate, are inhibited by mercurial compounds and are less acutely regulated (14).
Until now, no overt pathologies associated with defects in urea transport are known. Several mutations in the UT-B gene lead to expression of defective UT-B proteins and a lack of urea transport in erythrocytes. People with such UT-B mutations have red blood cells that are devoid of the Kidd blood group antigen (i.e. JKnull) but further than that there is no clear phenotypic effect. Only when JKnull individuals are dehydrated, they appear to have a mild (∼20%) reduction in maximal urinary concentrating capacity (20). In agreement with this finding, studies in UT-B knockout mice show a 45-fold reduction in red blood cell urea permeability, 50% increase in urine output and 30% decrease in urine osmolality compared with WT mice (21). The UT-B null mice also gain less weight, which suggests that intestinal UT-B plays an important role in weight gain. This observation may also be caused by an adverse effect on general health but the mice displayed no obvious signs of ill-health.
The UT-B (SLC14A1) gene consists of 11 exons covering 30 kb. The gene is known to produce two isoforms via alternative splicing: UT-B1, which consists of 389 amino acids, and UT-B2, which consists of 445 amino acids. Both transporters consist of 10 transmembrane-spanning domains, a large extracellular loop containing an N-glycosylation site, plus intracellular N and C termini (14). rs1058396 (the ‘Kidd blood group polymorphism’) is a G-to-A transition at nucleotide 838, located in exon 8 of the gene. It results in an asp280-to-asn (D280N) amino acid substitution and an MnlI RFLP (15). Whereas it was this variant that made us focus on this new bladder cancer locus, several other variants at this locus show similar association with bladder cancer in our discovery analysis. Moreover, analysis of one of those variants, rs17674580, a variant that is only moderately correlated with rs1058396, indicates that rs1058396 cannot fully account for the observed association of this locus with bladder cancer. When analyzing those two variants jointly, it appeared that rs17674580 can account for the observed association with rs1058396, although the converse is not true.
We do not have any evidence for functionality of rs17674580 or markers in strong LD with it. It has been reported that variants in the UT-A (SLC14A2) gene, residing in the LD block next to the block containing UT-B (SLC14A1), associate with blood urea nitrogen levels in a Japanese population (22). In our study, neither rs17674580 nor other variants in the LD block containing SLC14A1 associated with serum urea levels. The UT-B (SLC14A1) gene is expressed in the urinary bladder but no difference in expression between normal and cancerous urothelial tissue has been observed (http://www.ncbi.nlm.nih.gov/geoprofiles) (23,24). Also, as far as we have known, there are no data available on expression differences in urothelium by rs1058396 or rs17674580 genotype. It is not certain that SLC14A1 variants have a direct effect on the urothelium. Possibly, the association with UBC is indirect through urine concentration or frequency of urination. It has long been suggested that the amount of daily fluid intake may have an effect on the risk of bladder cancer. The so-called urogenous contact hypothesis states that high fluid intake may reduce exposure to carcinogens by diluting the urine and reducing the contact time through increased micturition (25). Large prospective epidemiological studies have yielded inconsistent results on this topic (26,27) which may be related to the enormous methodological difficulties in studying this relationship in an observational setting. In an experimental model, however, it was clearly shown that beagle dogs administered 4-aminobiphenyl have decreased levels of urothelial DNA adducts with increased voiding frequency, supporting the urogenous contact hypothesis (28). Consequently, SLC14A1 variants that influence the urea concentration gradient at the level of the descending vasa recta blood vessels in the renal medulla may modify the risk of UBC through an effect on urine production. Unfortunately, we were not able to evaluate this because data on daily fluid intake and voiding frequency were not available in our study.
In summary, we have discovered an association between genetic variants at the SLC14A1 locus at chromosome 18q and the risk of UBC. The gene codes for UTs that define the Kidd blood group and help maintain constant urea concentration gradient in the renal medulla. It is speculated that one or more sequence variants at this locus indirectly modify UBC risk by affecting the urine production. If this is correct, our finding supports the urogenous contact hypothesis that urine production and voiding frequency modify the risk of bladder carcinogens.
MATERIALS AND METHODS
Study subjects from Iceland
Records of all UBC diagnoses were obtained from the Icelandic Cancer Registry (ICR) (http://www.krabbameinsskra.is). The ICR contains all cancer diagnoses in Iceland from 1 January 1955. The ICR contained records of 1845 Icelandic UBC patients diagnosed until 31 December 2009. Recruitment of UBC cases was initiated in 2001 and included all prevalent cases as well as newly diagnosed cases from that time. Patients were recruited by trained nurses on behalf of the patients' treating physicians, through special recruitment clinics. Participants in the study donated a blood sample and answered a lifestyle questionnaire. A total of 603 patients (77% males; diagnosed from December 1964 to December 2009) were included in a genome-wide SNP genotyping effort, using the Infinium II assay method and either the Sentrix HumanHap 300 or HumanCNV370-duo BeadChip (Illumina). The median age at diagnosis for all consenting cases was 67.8 years (range 20–95 years) compared with 68.5 years for all UBC patients in the ICR. The 37 781 controls (41% males; mean age 61 years; SD = 21) used in this study consisted of individuals from other ongoing GWAS at deCODE and represent >15% of the adult population of Iceland. No individual disease group is represented by >10% of the total control group. Cancer patients (prostate, breast, colorectal and lung) were analyzed separately, and the frequency of the sequence variants studied did not differ from other controls. Samples from prostate, breast, colorectal and lung cancer patients as well as individuals used for the analysis of smoking variables come from other ongoing project at deCODE Genetics. The study was approved by the Data Protection Authority of Iceland and the National Bioethics Committee. Written informed consent was obtained from all patients, relatives and controls. Personal identifiers associated with medical information and blood samples were encrypted with a third-party encryption system for which the Data Protection Authority maintains the code.
In addition to the chip-genotyped case–control series that was used for the discovery phase of the study (Iceland I), we selected two follow-up (replication) case–control sample sets. The first of these (Iceland II) consists of 178 UBC cases and 2055 controls who were recruited after 2007 in the same way as the group ‘Iceland I’ but who were not whole-genome genotyped. This group was genotyped using single-SNP assays. The second follow-up group (Iceland III) was generated using information on 950 UBC cases without DNA who had close relatives that had been genotyped. These cases were assigned in silico genotypes, corresponding to an effective sample size of approximately 350 cases and 3500 controls who were also in silico genotyped [see the section In silico genotyping (Iceland III)].
Study subjects from the Netherlands
The Dutch patients were recruited for the Nijmegen Bladder Cancer Study (NBCS: http://dceg.cancer.gov/icbc/membership.html). The NBCS identified patients through the population-based regional cancer registry held by the Comprehensive Cancer Centre East, Nijmegen, that serves a region of 1.3 million inhabitants in the eastern part of the Netherlands (www.ikcnet.nl). Patients diagnosed between 1995 and 2006 under the age of 75 years were selected and their vital status and current addresses updated through the hospital information systems of the seven community hospitals and one university hospital (Radboud University Nijmegen Medical Centre, RUNMC) that are covered by the cancer registry. All patients still alive on 1 August 2007 were invited to the study by the Comprehensive Cancer Center on behalf of the patients' treating physicians. A second group of patients, diagnosed between 2007 and 2008, was invited in 2009. In case of consent, patients were sent a lifestyle questionnaire to fill out and blood samples were collected by Thrombosis Service centers which hold offices in all the communities in the region. In total, 1940 patients were invited to participate. Of all the invitees, 1275 gave informed consent (66%): 1185 filled out the questionnaire (61%) and 1219 (63%) provided a blood sample. The number of participating patients was increased with a non-overlapping series of 439 bladder cancer patients who were recruited previously for a study on gene–environment interactions in three hospitals (RUNMC, Canisius Wilhelmina Hospital, Nijmegen, and Streekziekenhuis Midden-Twente, Hengelo, the Netherlands). Ultimately, completed questionnaires and blood samples were available for 1429 and 1691 patients, respectively. All the patients selected for the analyses (n = 1631) were of self-reported European descent. The median age at diagnosis was 62 (range 25–93) years. Eighty-two percent of the participants were males. Data on tumor stage and grade were obtained through the cancer registry.
The control group (n = 3822) came from different sources. A total of 1918 cancer-free control individuals (46% males) were recruited within a project entitled ‘Nijmegen biomedical study’ (NBS). The details of this study were reported previously (29). Briefly, this is a population-based survey conducted by the Department of Epidemiology and Biostatistics and the Department of Clinical Chemistry of the RUNMC, in which 9371 individuals participated from a total of 22 500 age- and sex-stratified, randomly selected inhabitants of Nijmegen. Control individuals from the NBS were invited to participate in a study on gene–environment interactions in multifactorial diseases such as cancer. They were all of self-reported European descent and fully informed about the goals and the procedures of the study. The study protocols of the NBCS and the NBS were approved by the Institutional Review Board of the RUNMC and all study subjects gave written informed consent. The NBS control group was extended with a Dutch control group provided by collaborators from the University of Utrecht that had been chip-genotyped in conjunction with other studies and have been described in a previous publication (30). Briefly, these controls were recruited from two sources: (i) 613 control individuals were unrelated, healthy volunteers who accompanied non-amyotrophic lateral sclerosis (ALS) patients to the University Medical Center Utrecht neurology outpatient clinic, (ii) 1358 individuals were included from a GWAS on schizophrenia. All were of Dutch descent, with at least three out of four grandparents of Dutch ancestry. Of the 3881 chip-typed controls, 3822 passed all quality control filters and were used for analysis.
Study subjects from the UK
Details of the Leeds Bladder Cancer Study have been reported previously (31). In brief, patients from the urology department of St James's University Hospital, Leeds, were recruited from August 2002 to March 2006. All patients undergoing cystoscopy or transurethral resection of a bladder tumor who had previously been found, or were subsequently shown, to have urothelial cell carcinoma of the bladder were included. Exclusion criterion was significant mental impairment or a blood transfusion in the past month. All non-Caucasians were excluded from the study, leaving 764 patients. The median age at diagnosis of the patients was 73 years (range 30–101). Seventy-one percent of the patients were male and 36% of all the patients had tumors with low risk of progression (pTaG1/2). The controls were recruited from the otolaryngology outpatients and ophthalmology inpatient and outpatient departments at St James's Hospital, Leeds, from August 2002 to March 2006. All controls of appropriate age for frequency-matching with the cases were approached and recruited. As for the cases, exclusion criteria for the controls were significant mental impairment or a blood transfusion in the past month. Also, controls were excluded if they had symptoms suggestive of bladder cancer, such as hematuria; 2.8% of the controls were non-Caucasian, leaving 530 Caucasian controls for the study, and 71% of the controls were male. Data were collected by a health questionnaire on smoking habits and smoking history (non-, ex- or current smoker, smoking dose in pack-years), occupational exposure history (to plastics, rubber, laboratories, printing, dyes and paints, diesel fumes), family history of bladder cancer, ethnicity and place of birth and places of birth of parents. The participation rate of cases was ∼99%, and that among the controls was ∼80%. Ethical approval for the study was obtained from Leeds (East) Local Research Ethics Committee, project number 02/192.
Study subjects from Torino, Italy
The sources of cases for the Torino bladder cancer study were two urology departments of the main hospital in Torino, the San Giovanni Battista Hospital (32). Cases were all Caucasian men, aged 40–75 years (median 63 years) and living in the Torino metropolitan area. They were newly diagnosed between 1994 and 2006 with a histologically confirmed, invasive or in situ, bladder cancer. Of all the patients with information on stage and grade, 56% were at low risk of progression (pTaG1/2). The sources of controls are urology, medical and surgical departments of the same hospital in Torino. All controls are Caucasian men resident in the Torino metropolitan area. They were diagnosed and treated between 1994 and 2006 for benign diseases (such as prostatic hyperplasia, cystitis, hernias, heart failure, asthma and benign ear diseases). Controls with cancer, liver or renal diseases and smoking-related conditions were excluded. The median age of the controls was 57 years (range 40–74). Data were collected by a professional interviewer who used a structured questionnaire to interview both cases and controls face-to-face. Data collected included demographics (age, sex, ethnicity, region and education) and smoking. For cases, additional data were collected on tumor histology, tumor site, size, stage, grade and treatment of the primary tumor. The participation rates were 90% for cases and 75% for controls, resulting in 328 cases and 389 controls. Ethical approval for the study was obtained from Comitato Etico Interaziendale, A.O.U. San Giovanni Battista/C.T.O./Maria Adelaide.
Study subjects from Brescia, Italy
The Brescia bladder cancer study is a hospital-based case–control study. The study was reported in detail previously (33). In short, the catchment area of the cases and controls was the Province of Brescia, a highly industrialized area in Northern Italy (mainly metal and mechanical industry, construction, transport, textiles) but also with relevant agricultural areas. Cases and controls were enrolled in 1997–2000 from the two main city hospitals. The total number of eligible subjects was 216 cases and 220 controls. The participation rate (enrolled/eligible) was 93% (n = 201) for cases and 97% (n = 214) for controls. Only males were included. All cases and controls had Italian nationality and were of Caucasian ethnicity. All cases had to be residents of the Province of Brescia, aged between 20 and 80, and newly diagnosed with histologically confirmed bladder cancer. The median age of the patients was 63 years (range 22–80); 29% of all the patients with known stage and grade had tumors of low risk of progression (pTaG1/2). Controls were patients admitted for various urological non-neoplastic diseases and were frequency-matched to cases on age, hospital and period of admission. The study was formally approved by the ethical committee of the hospital where the majority of subjects were recruited. A written informed consent was obtained from all participants. Data were collected from clinical charts (tumor histology, site, grade, stage, treatments, etc.) and by means of face-to-face interviews during hospital admission, using a standardized semi-structured questionnaire. The questionnaire included data on demographics (age, ethnicity, region, education, residence, etc.) and smoking. ISCO and ISIC codes and expert assessments were used for occupational coding. Blood samples were collected from cases and controls for genotyping and DNA adduct analyses.
Study subjects from Belgium
The Belgian study has been reported in detail (34). In brief, cases were selected from the Limburg Cancer Registry (LIKAR) and were approached through urologists and general practitioners. All cases were diagnosed with histologically confirmed urothelial cell carcinoma of the bladder between 1999 and 2004, and were Caucasian inhabitants of the Belgian province of Limburg. The median age of the patients was 68 years, and 86% of all the patients were males. For the recruitment of controls, a request was made to the ‘Kruispuntbank’ of the social security for simple random sampling, stratified by municipality and socio-economic status, among all citizens >50 years of age of the province. The median age of the controls was 64 years; 59% of the controls were males. Three trained interviewers visited cases and controls at home. Information was collected through a structured interview and a standardized food frequency questionnaire. In addition, biological samples were collected. Data collected included medical history, lifetime smoking history, family history of bladder cancer and a lifetime occupational history. Informed consent was obtained from all participants and the study was approved by the ethical review board of the Medical School of the Catholic University of Leuven, Belgium.
Study subjects from Eastern Europe
The details of this study have been described previously (35). Cases and controls were recruited as part of a study designed to evaluate the risk of various cancers due to environmental arsenic exposure in Hungary, Romania and Slovakia between 2002 and 2004. The recruitment was carried out in the counties of Bacs, Bekes, Csongrad and Jasz-Nagykun-Szolnok in Hungary; Bihor and Arad in Romania; and Banska Bystrica and Nitra in Slovakia. The cases (n = 214) and controls (n = 533) selected were of Hungarian, Romanian and Slovak nationalities. Bladder cancer patients were invited on the basis of histopathological examinations by pathologists. Hospital-based controls were included in the study, subject to fulfillment of a set of criteria. All general hospitals in the study areas were involved in the process of control recruitment. The controls were frequency-matched with cases for age, gender, country of residence and ethnicity. Controls included general surgery, orthopedic and trauma patients aged 30–79 years. Patients with malignant tumors, diabetes and cardiovascular diseases were excluded as controls. The median age of the bladder cancer patients was 65 years (range 36–90). Eighty-three percent of the patients were males. The median age for the controls was 61 years (range 28–83). Fifty-one percent of the controls were males. The participation rates among cases and controls were ∼70%. Of all the patients with known stage and grade information, 28% had a low-risk tumor (pTaG1/2). Clinicians took venous blood and other biological samples from cases and controls after consent forms had been signed. Cases and controls recruited to the study were interviewed by trained personnel and completed a general lifestyle questionnaire. Ethnic background for cases and controls was recorded along with other characteristics of the study population. Local ethical boards approved the study plan and design.
Study subjects from Sweden
The Swedish patients came from a population-based study of UBC patients diagnosed in the Stockholm region in 1995–1996 (36). Blood samples from 352 patients were available out of a collection of 538 patients with primary urothelial carcinoma of the bladder. The average age at onset for these patients was 69 years (range 32–97 years) and 67% of the patients were males. Clinical data, including age at onset, grade and stage of tumor, were prospectively obtained from hospitals and urology units in the region. The control samples came from blood donors in the Stockholm region and were from cancer-free individuals of both genders. The regional ethical committee approved of the study and all participants gave informed consent.
Study subjects from Spain
The Spanish study patients were recruited from the urology and oncology departments of Zaragoza Hospital between September 2007 and June 2009. A total of 246 patients with histologically proven urothelial cell carcinoma of the bladder were enrolled (response 77%). Clinical information including age at onset, grade and stage was obtained from medical records. The median age at diagnosis for the patients was 65 years (range 27–94) and 87% were males. The 890 Spanish control individuals were part of a larger collection of control samples obtained from individuals who had attended the University Hospital in Zaragoza, Spain, for diseases other than cancer between November 2001 and May 2007. The controls were of both genders and median age was 52 years (range 11–87). Controls were questioned to rule out prior cancers before drawing the blood sample. All patients and controls were of self-reported European descent. Study protocols were approved by the Institutional Review Board of Zaragoza University Hospital. All subjects gave written informed consent.
Study subjects from Germany
The study subjects from Germany came from three different studies.
The Neuss bladder cancer study. Details of the bladder cancer cases of this study have been published previously (37). The ongoing case–control series consists of 217 bladder cancer cases and 105 controls from the Department of Urology, Lukasklinik Neuss, Germany, located ∼20 km from the Ruhr area. The median age at diagnosis was 72.9 (range 26.1–93.4) years. Seventy-eight percent of the participants were males. Data on tumor stage and grade were obtained through the cancer registry. Forty-five percent of the patients had a low-risk tumor (pTaG1/2). The 105 control individuals (64% males) (median age 42.4, range 18.0–89.0) were cancer-free. Data were collected from June 2009 to July 2010. The local ethics committee approved the study plan and design.
The Dortmund bladder cancer study. Details of the bladder cancer cases of this study have been published previously (37). The case–control series consists of 197 patients with a confirmed bladder cancer from the Department of Urology, St.-Josefs-Hospital Dortmund-Hörde, located in the Ruhr area, an area of former coal, iron and steel industries and 240 controls from the same Department of Urology, admitted for treatment of benign urological diseases, enrolled from July 2009 to July 2010. The median age at diagnosis was 71.2 (range 35.0–89.2) years. Seventy-five percent of the participants were males, and 60% of the patients had tumors with low risk of progression (pTaG1/2). The 240 control individuals (77% males) were cancer-free and frequency-matched for age with the cases (median age 70.7, range 21.7–100). The local ethics committee approved the study plan and design.
The Lutherstadt Wittenberg (LW) study. Details of the bladder cancer cases of this study have been reported previously (37–39). In brief, 221 patients with a confirmed bladder cancer from the Department of Urology, Paul Gerhardt Foundation, Lutherstadt Wittenberg, Germany, were included. Patients were enrolled from December 1995 to January 1999. Exclusion criterion was a missing written informed consent into the study. The median age of the patients was 65 years (range 20–91); 86% of the patients were males. A total of 214 controls were from the same Department of Urology, but were admitted for treatment of benign urological diseases. Exclusion criterion was a malignant disease in the medical history or a missing written informed consent. The median age of the controls was 68 years (range 29–91); 84% of the controls were males. Data were collected from July 2000 to May 2005. All cases and controls were Caucasians, which was confirmed by questionnaire-based documentation of nationality. Cases and controls were matched for age. Data collected in cases and controls include age, gender, a complete documentation of occupational activities performed at least for 6 months, documentation of work places with known bladder cancer risk over the entire working life, exposures to known or suspected occupational bladder carcinogens, lifetime smoking habits, family history of bladder cancer, numbers of urinary infections treated by drugs during the previous 10 years, place of birth and places of residency for >10 years. For bladder cancer cases, data on tumor staging, grading and treatment were taken from the records. First diagnosis of bladder cancer was recorded from July 1979 to January 1999. The local ethics committee approved the study plan and design.
Study subjects from Iran
Samples from Iranian bladder cancer patients were obtained from the Biobank of Shiraz Institute for Cancer Research (http://icr.ir/groups/biobank.html). The majority of the patients were residents of the province of Fars (of which Shiraz is the capital city) in the south of Iran. The people of Fars are Persian with little racial heterogeneity. However, since hospitals affiliated with Shiraz University of Medical Sciences are referral hospitals for large parts of southern Iran, inclusion of patients from other ethnic groups cannot be ruled out. The DNA collection of all cancer patients in the Biobank of Shiraz Institute for Cancer Research is an ongoing process that started in 1999. For the present study, 267 patients (229 males and 38 females) with histologically verified bladder cancer were available. The mean ages at diagnosis were 63.6 and 60.1 years, respectively. For 3% of all cases, age could not be determined. The majority of the patients had urothelial cell carcinoma (n = 239; 89.5%); the remaining had squamous cell carcinoma, adenocarcinoma, rhabdomyosarcoma or unspecified/unknown morphology. The control group consisted of 222 male and 25 female healthy Iranian subjects without apparent cancer or autoimmune diseases and a negative first-degree family history of cancer. The controls were recruited among healthy blood donors at the Fars blood transfusion center. The mean ages were 54.0 years for males and 53.5 years for females. DNA of the controls was also obtained from the Biobank of Shiraz Institute for Cancer Research. Informed consent was obtained from all patients and controls. The biobanking procedures have been approved by Shiraz University of Medical Sciences Ethics Committee.
Illumina genome-wide genotyping
The Dutch and Icelandic case and control samples were assayed with the Illumina HumanHap300 or HumanHapCNV370 bead chips (Illumina, San Diego, CA, USA), containing 317 503 and 370 404 haplotype tagging SNPs derived from phase I of the International HapMap project, respectively. SNPs were excluded if they had: (i) a yield <95% in cases or controls; (ii) a minor allele frequency <1% in the population; or (iii) showed significant deviation from Hardy–Weinberg equilibrium in the controls (P < 0.001). Any samples with a call rate <98% were excluded from the analysis.
SNP imputations
For the Dutch data set, we used 292 650 autosomal SNPs present on both chip types to impute an additional 7 543 837 ungenotyped SNPs using the IMPUTE v2.1 software (40,41) and a training set consisting of the combined 1000 Genomes low-coverage pilot haplotypes (released June 2010, 120 chromosomes) and the HapMap3 haplotypes (released February 2009, 1920 chromosomes) downloaded from http://mathgen.stats.ox.ac.uk/impute/impute_v2.html#filtered_1kg_hm3_haps. Individuals with <96% yield for the 292 650 SNPs and SNPs with minor allele frequency <0.01, which were not in Hardy–Weinberg equilibrium (P < 10−5) or with a different frequency for the two chip types used (P < 10−5), were excluded from the imputation. For the Icelandic data set, we used 289 572 autosomal SNPs to impute an additional 11 283 069 ungenotyped SNPs using as training set the 566 phased European haplotypes of the August 2010 release of the 1000 Genomes project, downloaded from http://mathgen.stats.ox.ac.uk/impute/impute_v2.html#August_haps. The imputation method used for the Icelandic data set has been described previously (42). In short, it is an adaption of the methodology as used in the IMPUTE software but taking advantage of the long-range phasing of the whole Icelandic data set. As for the Dutch data set, individuals with low yield and SNPs that failed quality control were excluded prior to imputation.
Genome-wide analysis
For both the Icelandic and the Dutch data set, we used SNPTEST v2.1.1 (41) to test each SNP for association with bladder cancer, assuming additive genetic effect and using missing data likelihood score test (the method score option) to account for imputation uncertainty. Individuals with genotype yield <98% and SNPs with information (proper_info) <0.4 in the analysis of either data set were excluded. When combining the two data sets, we restricted the analysis to SNPs that were present in both data sets and that had MAF >1%. As the August 2010 release of the 1000 Genomes is mapped to NCBI Genome build 37, the location of those SNPs were mapped to build 36 using the UCSC LiftOver tool (http://genome.ucsc.edu/cgi-bin/hgLiftOver). A total of 5 340 737 SNPs that passed quality criteria and that were identified in both data sets were included when the results for the two data sets were combined.
Prior to combining the two data sets, the results were adjusted using the method of genomic control by dividing the χ2 statistics by 1.071 for the Dutch data set and by 1.011 for the Icelandic data set. For the combined analysis, we assumed a fixed effect model implemented in the program METAL (http://www.sph.umich.edu/csg/abecasis/Metal/) and weighted the contribution of each data set with the corresponding standard error. Heterogeneity in the effect estimate for each SNP was calculated by including the corresponding option in METAL.
Single-SNP genotyping
Single-SNP genotyping for all samples was carried out at deCODE Genetics in Reykjavik, Iceland, applying the same platform to all populations studied. All single-SNP genotyping was carried out using the Centaurus (Nanogen) platform (43). The quality of each Centaurus SNP assay was evaluated by genotyping each assay on the CEU samples and comparing the results with the HapMap data (44). All assays had mismatch rate <0.5%. Additionally, all markers were re-genotyped on >10% of samples typed with the Illumina platform, resulting in an observed mismatch in <0.5% of samples.
In silico genotyping (Iceland III)
In addition to imputing rs1058396 and rs17674580 into chip-genotyped individuals, we also performed a second imputation step where genotypes were imputed into relatives of chip-genotyped individuals, creating in silico genotypes. The inputs into the second imputation step are the fully phased (in particular, every allele has been assigned a parent of origin) imputed and chip-typed genotypes of the available chip-typed individuals. The algorithm used to perform the second imputation step consists of:
For each ungenotyped individual (the proband), find all chip-genotyped individuals within two meioses of the individual. The six possible types of two-meioses relatives of the proband are (ignoring more complicated relationships due to pedigree loops) parents, full and half siblings, grandparents, children and grandchildren. If all pedigree paths from the proband to a genotyped relative go through other genotyped relatives, then that relative is excluded. For example, if a parent of the proband is genotyped, then the proband's grandparents through that parent are excluded. If the number of meioses in the pedigree around the proband exceeds a threshold (we used 12), then relatives are removed from the pedigree until the number of meioses falls below 12, in order to reduce computational complexity.
At every point in the genome, calculate the probability for each genotyped relative sharing with the proband based on the autosomal SNPs used for phasing. A multipoint algorithm, based on the hidden Markov-model Lander–Green multipoint linkage algorithm using fast Fourier transforms, is used to calculate these sharing probabilities (10,45,46). First, single-point sharing probabilities are calculated by dividing the genome into 0.5 cM bins and using the haplotypes over these bins as alleles. Haplotypes that are the same, except that they can differ for one SNP at most, are treated as identical. When the haplotypes in the pedigree are incompatible over a bin, then a uniform probability distribution was used for that bin. The most common causes for such incompatibilities are recombinations within the pedigree, phasing errors and genotyping errors. Note that since the input genotypes are fully phased, the single-point information is substantially more informative than for unphased genotyped persons. In particular one haplotype of the parent of a genotyped child is always known. The single-point distributions are then convolved using the multipoint algorithm to obtain multipoint sharing probabilities at the center of each bin. Genetic distances were obtained from the most recent version of the deCODE genetic map (47).
- Based on the sharing probabilities at the center of each bin, all the SNPs from the whole-genome genotyping are imputed into the proband. To impute the genotype of the paternal allele of an SNP located at x, flanked by bins with centers at xleft and xright, starting with the left bin, going through all possible sharing patterns ν, let Iν be the set of haplotypes of genotyped individuals that share identically by descent within the pedigree with the proband's paternal haplotype given the sharing pattern ν and P(ν) be the probability of ν at the left bin—this is the output from step (ii) above—and let ei be the expected allele count of the SNP for haplotype i. Then
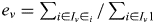 is the expected allele count of the paternal haplotype of the proband given ν and an overall estimate of the allele count given the sharing distribution at the left bin is obtained from
is the expected allele count of the paternal haplotype of the proband given ν and an overall estimate of the allele count given the sharing distribution at the left bin is obtained from 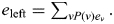 If Iν is empty, then no relative shares with the proband's paternal haplotype given ν and thus there is no information about the allele count. We therefore store the probability that some genotyped relative shared the proband's paternal haplotype,
If Iν is empty, then no relative shares with the proband's paternal haplotype given ν and thus there is no information about the allele count. We therefore store the probability that some genotyped relative shared the proband's paternal haplotype, 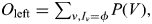 and an expected allele count, conditional on the proband's paternal haplotype being shared by at least one genotyped relative:
and an expected allele count, conditional on the proband's paternal haplotype being shared by at least one genotyped relative: 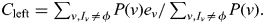 In the same way, calculate Oright and Cright. Linear interpolation is then used to get an estimate at the SNP from the two flanking bins:
In the same way, calculate Oright and Cright. Linear interpolation is then used to get an estimate at the SNP from the two flanking bins:
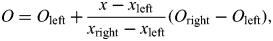
If θ is an estimate of the population frequency of the SNP, then Oc + (1 – O)θ is an estimate of the allele count for the proband's paternal haplotype. Similarly, an expected allele count can be obtained for the proband's maternal haplotype.
When performing case–control analysis with the in silico genotypes, we use logistic regression to test for association between SNPs and disease, treating disease status as the response and expected genotype counts from imputation or allele counts from direct genotyping as covariates. Testing was performed using the likelihood ratio statistic. When testing for association based on the in silico genotypes, controls were matched to cases based on the informativeness of the imputed genotypes, such that for each case C, controls of matching informativeness were chosen. Failing to match cases and controls will lead to a highly inflated genomic control factor, and in some cases may lead to spurious false-positive findings. The informativeness of each of the imputation of each one of an individual's haplotypes was estimated by taking the average of
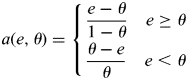 |
over all SNPs imputed for the individual, where e is the expected allele count for the haplotype at the SNP and θ is the population frequency of the SNP. Note that a(θ,θ) = 0 and a(0,θ) = a(1,θ) = 1. The mean informativeness values cluster into groups corresponding to the most common pedigree configurations used in the imputation, such as imputing from parent into child or from child into parent. Based on this clustering of imputation informativeness, we divided the haplotypes of individuals into 7 groups of varying informativeness, which created 27 groups of individuals of similar imputation informativeness; 7 groups of individuals with both haplotypes having similar informativeness, 21 groups of individuals with the two haplotypes having different informativeness, minus the one group of individuals with neither haplotype being imputed well. Within each group, we calculate the ratio of the number of controls and the number of cases, and choose the largest integer C that was less than this ratio in all the groups. For example, if in one group there are 10.3 times as many controls as cases and if in all other groups this ratio was greater, then we would set C = 10 and within each group randomly select 10 times as many controls as there are cases.
Association analysis
For association analysis of the replication data sets, we used a standard likelihood ratio statistic, implemented in the NEMO software (48) to calculate two-sided P-values for each individual allele, assuming a multiplicative model for UBC risk, i.e. the relative risk from carrying two copies of the high-risk allele (compared with none) is assumed to be the square of the relative risk from carrying one copy. Results from multiple case–control groups were combined using a Mantel–Haenszel model in which the groups were allowed to have different population frequencies for alleles and genotypes but were assumed to have common ORs. Stratified analyses were conducted by smoking and by UBC aggressiveness. For the latter, all patients for whom detailed histology information was available were classified with regard to risk of progression, based on stage and grade information. Patients with ‘low risk of progression’ were defined as having TNM stage pTa in combination with WHO 1973 differentiation grade 1 or 2 or WHO/ISUP 2004 low grade. All other tumors were classified as ‘high risk of progression’ (stage pTis or ≥ pT1 or WHO 1973 grade 3 or WHO/ISUP 2004 high grade).
Heterogeneity calculations
Heterogeneity was tested by comparing the null hypothesis of the effect being the same in all populations to the alternative hypothesis of each population having a different effect using a likelihood ratio test. I2 lies between 0 and 100% and describes the proportion of total variation in study estimates that is due to heterogeneity (49).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the following funding agencies. Collection of samples and data in Iceland and the Netherlands was funded in part by the European Commission (POLYGENE: LSHC-CT-2005) (grant number 018827) and a research investment grant of the Radboud University Nijmegen Medical Centre (RUNMC). Control samples from the University of Utrecht were genotyped with generous support from the ‘Prinses Beatrix Fonds’, VSB Fonds, H. Kersten and M. Kersten (Kersten Foundation), The Netherlands ALS Foundation, J.R. van Dijk and the Adessium Foundation. The controls from the Dutch Schizophrenia GWA study were genotyped with the support of the National Institute of Mental Health (NIH/NIMH MH078075). The Leeds Bladder Cancer Study was funded by Cancer Research UK and Yorkshire Cancer Research. Torino Bladder Cancer Case Control Study was supported by a grant to ECNIS (Environmental Cancer Risk, Nutrition and Individual Susceptibility), a network of excellence operating within the European Union 6th Framework Program, Priority 5: ‘Food Quality and Safety’ (Contract No. 513943); by a grant of the Compagnia di San Paolo—Human Genetics Foundation (HuGeF), the Italian Association for Cancer Research, Italy and the Piedmont Region Progetti di Ricerca Sanitaria Finalizzata. The Belgian bladder cancer study was funded by the Flemish government and by the health authorities of the Belgian province of Limburg. The Swedish study was funded by the Swedish Cancer Society and the Swedish Research Council. The Iranian study was supported by a grant from Shiraz Institute for Cancer Research, Shiraz, Iran (grant number ICR-87-502). C.Z. and S.B. are funded by a European Commission 7th Framework Programme FP7-MC-IAPP (grant agreement number 218071 CancerGene). J.I.M. is funded by Red Temática de Investigación Cooperativa en Cáncer (grant number RD06/0020/1054).
Supplementary Material
ACKNOWLEDGEMENTS
We thank the individuals who participated in the study and whose contribution made this work possible. We also thank the personnel at all the recruitment centers. We acknowledge the cancer registries in Iceland and the Netherlands for assistance in the ascertainment of the Icelandic and Dutch UBC patients.
Conflict of Interest statement. The authors who are affiliated with deCODE genetics are all employees of deCODE, a biotechnology company that provides genetic testing services, and some own stocks or stock options in the company.
REFERENCES
- 1.Ploeg M., Aben K.K., Kiemeney L.A. The present and future burden of urinary bladder cancer in the world. World J. Urol. 2009;27:289–293. doi: 10.1007/s00345-009-0383-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferlay J., Shin H.R., Bray F., Forman D., Mathers C., Parkin D.M. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 3.Sievert K.D., Amend B., Nagele U., Schilling D., Bedke J., Horstmann M., Hennenlotter J., Kruck S., Stenzl A. Economic aspects of bladder cancer: what are the benefits and costs? World J. Urol. 2009;27:295–300. doi: 10.1007/s00345-009-0395-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mueller C.M., Caporaso N., Greene M.H. Familial and genetic risk of transitional cell carcinoma of the urinary tract. Urol. Oncol. 2008;26:451–464. doi: 10.1016/j.urolonc.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aben K.K., Witjes J.A., Schoenberg M.P., Hulsbergen-van de Kaa C., Verbeek A.L., Kiemeney L.A. Familial aggregation of urothelial cell carcinoma. Int. J. Cancer. 2002;98:274–278. doi: 10.1002/ijc.10191. [DOI] [PubMed] [Google Scholar]
- 6.Aben K.K., Baglietto L., Baffoe-Bonnie A., Coebergh J.W., Bailey-Wilson J.E., Trink B., Verbeek A.L., Schoenberg M.P., Alfred W.J., Kiemeney L.A. Segregation analysis of urothelial cell carcinoma. Eur. J. Cancer. 2006;42:1428–1433. doi: 10.1016/j.ejca.2005.07.039. [DOI] [PubMed] [Google Scholar]
- 7.Murta-Nascimento C., Silverman D.T., Kogevinas M., Garcia-Closas M., Rothman N., Tardon A., Garcia-Closas R., Serra C., Carrato A., Villanueva C., et al. Risk of bladder cancer associated with family history of cancer: do low-penetrance polymorphisms account for the increase in risk? Cancer Epidemiol. Biomarkers Prev. 2007;16:1595–1600. doi: 10.1158/1055-9965.EPI-06-0743. [DOI] [PubMed] [Google Scholar]
- 8.Amundadottir L.T., Thorvaldsson S., Gudbjartsson D.F., Sulem P., Kristjansson K., Arnason S., Gulcher J.R., Bjornsson J., Kong A., Thorsteinsdottir U., Stefansson K. Cancer as a complex phenotype: pattern of cancer distribution within and beyond the nuclear family. PLoS Med. 2004;1:e65. doi: 10.1371/journal.pmed.0010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Closas M., Malats N., Silverman D., Dosemeci M., Kogevinas M., Hein D.W., Tardon A., Serra C., Carrato A., Garcia-Closas R., et al. NAT2 slow acetylation, GSTM1 null genotype, and risk of bladder cancer: results from the Spanish Bladder Cancer Study and meta-analyses. Lancet. 2005;366:649–659. doi: 10.1016/S0140-6736(05)67137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kiemeney L.A., Thorlacius S., Sulem P., Geller F., Aben K.K., Stacey S.N., Gudmundsson J., Jakobsdottir M., Bergthorsson J.T., Sigurdsson A., et al. Sequence variant on 8q24 confers susceptibility to urinary bladder cancer. Nat. Genet. 2008;40:1307–1312. doi: 10.1038/ng.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kiemeney L.A., Sulem P., Besenbacher S., Vermeulen S.H., Sigurdsson A., Thorleifsson G., Gudbjartsson D.F., Stacey S.N., Gudmundsson J., Zanon C., et al. A sequence variant at 4p16.3 confers susceptibility to urinary bladder cancer. Nat. Genet. 2010;42:415–419. doi: 10.1038/ng.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rothman N., Garcia-Closas M., Chatterjee N., Malats N., Wu X., Figueroa J.D., Real F.X., Van Den B.D., Matullo G., Baris D., et al. A multi-stage genome-wide association study of bladder cancer identifies multiple susceptibility loci. Nat. Genet. 2010;42:978–984. doi: 10.1038/ng.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu X., Ye Y., Kiemeney L.A., Sulem P., Rafnar T., Matullo G., Seminara D., Yoshida T., Saeki N., Andrew A.S., et al. Genetic variation in the prostate stem cell antigen gene PSCA confers susceptibility to urinary bladder cancer. Nat. Genet. 2009;41:991–995. doi: 10.1038/ng.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stewart G. The emerging physiological roles of the SLC14A family of urea transporters. Br. J. Pharmacol. 2011;41:991–995. doi: 10.1111/j.1476-5381.2011.01377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olives B., Merriman M., Bailly P., Bain S., Barnett A., Todd J., Cartron J.P., Merriman T. The molecular basis of the Kidd blood group polymorphism and its lack of association with type 1 diabetes susceptibility. Hum. Mol. Genet. 1997;6:1017–1020. doi: 10.1093/hmg/6.7.1017. [DOI] [PubMed] [Google Scholar]
- 16.Thorgeirsson T.E., Geller F., Sulem P., Rafnar T., Wiste A., Magnusson K.P., Manolescu A., Thorleifsson G., Stefansson H., Ingason A., et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gudbjartsson D.F., Holm H., Indridason O.S., Thorleifsson G., Edvardsson V., Sulem P., de Vegt F., d'Ancona F.C., den Heijer M., Wetzels J.F., et al. Association of variants at UMOD with chronic kidney disease and kidney stones—role of age and comorbid diseases. PLoS Genet. 2010;6:e1001039. doi: 10.1371/journal.pgen.1001039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith C.P. Mammalian urea transporters. Exp. Physiol. 2009;94:180–185. doi: 10.1113/expphysiol.2008.043042. [DOI] [PubMed] [Google Scholar]
- 19.Fenton R.A., Hewitt J.E., Howorth A., Cottingham C.A., Smith C.P. The murine urea transporter genes Slc14a1 and Slc14a2 occur in tandem on chromosome 18. Cytogenet. Cell Genet. 1999;87:95–96. doi: 10.1159/000015401. [DOI] [PubMed] [Google Scholar]
- 20.Sands J.M., Gargus J.J., Frohlich O., Gunn R.B., Kokko J.P. Urinary concentrating ability in patients with Jk(a-b-) blood type who lack carrier-mediated urea transport. J. Am. Soc. Nephrol. 1992;2:1689–1696. doi: 10.1681/ASN.V2121689. [DOI] [PubMed] [Google Scholar]
- 21.Yang B., Bankir L., Gillespie A., Epstein C.J., Verkman A.S. Urea-selective concentrating defect in transgenic mice lacking urea transporter UT-B. J. Biol. Chem. 2002;277:10633–10637. doi: 10.1074/jbc.M200207200. [DOI] [PubMed] [Google Scholar]
- 22.Kamatani Y., Matsuda K., Okada Y., Kubo M., Hosono N., Daigo Y., Nakamura Y., Kamatani N. Genome-wide association study of hematological and biochemical traits in a Japanese population. Nat. Genet. 2010;42:210–215. doi: 10.1038/ng.531. [DOI] [PubMed] [Google Scholar]
- 23.Dyrskjot L., Thykjaer T., Kruhoffer M., Jensen J.L., Marcussen N., Hamilton-Dutoit S., Wolf H., Orntoft T.F. Identifying distinct classes of bladder carcinoma using microarrays. Nat. Genet. 2003;33:90–96. doi: 10.1038/ng1061. [DOI] [PubMed] [Google Scholar]
- 24.Dyrskjot L., Kruhoffer M., Thykjaer T., Marcussen N., Jensen J.L., Moller K., Orntoft T.F. Gene expression in the urinary bladder: a common carcinoma in situ gene expression signature exists disregarding histopathological classification. Cancer Res. 2004;64:4040–4048. doi: 10.1158/0008-5472.CAN-03-3620. [DOI] [PubMed] [Google Scholar]
- 25.Braver D.J., Modan M., Chetrit A., Lusky A., Braf Z. Drinking, micturition habits, and urine concentration as potential risk factors in urinary bladder cancer. J. Natl Cancer Inst. 1987;78:437–440. [PubMed] [Google Scholar]
- 26.Michaud D.S., Spiegelman D., Clinton S.K., Rimm E.B., Curhan G.C., Willett W.C., Giovannucci E.L. Fluid intake and the risk of bladder cancer in men. N. Engl. J. Med. 1999;340:1390–1397. doi: 10.1056/NEJM199905063401803. [DOI] [PubMed] [Google Scholar]
- 27.Ros M.M., Bas Bueno-de-Mesquita H.B., Buchner F.L., Aben K.K., Kampman E., Egevad L., Overvad K., Tjonneland A., Roswall N., Clavel-Chapelon F., et al. Fluid intake and the risk of urothelial cell carcinomas in the European Prospective Investigation into Cancer and Nutrition (EPIC) Int. J. Cancer. 2011;128:2695–2708. doi: 10.1002/ijc.25592. [DOI] [PubMed] [Google Scholar]
- 28.Kadlubar F.F., Dooley K.L., Teitel C.H., Roberts D.W., Benson R.W., Butler M.A., Bailey J.R., Young J.F., Skipper P.W., Tannenbaum S.R. Frequency of urination and its effects on metabolism, pharmacokinetics, blood hemoglobin adduct formation, and liver and urinary bladder DNA adduct levels in beagle dogs given the carcinogen 4-aminobiphenyl. Cancer Res. 1991;51:4371–4377. [PubMed] [Google Scholar]
- 29.Wetzels J.F., Kiemeney L.A., Swinkels D.W., Willems H.L., den Heijer M. Age- and gender-specific reference values of estimated GFR in Caucasians: the Nijmegen Biomedical Study. Kidney Int. 2007;72:632–637. doi: 10.1038/sj.ki.5002374. [DOI] [PubMed] [Google Scholar]
- 30.van Es M.A., Veldink J.H., Saris C.G., Blauw H.M., van Vught P.W., Birve A., Lemmens R., Schelhaas H.J., Groen E.J., Huisman M.H., et al. Genome-wide association study identifies 19p13.3 (UNC13A) and 9p21.2 as susceptibility loci for sporadic amyotrophic lateral sclerosis. Nat. Genet. 2009;41:1083–1087. doi: 10.1038/ng.442. [DOI] [PubMed] [Google Scholar]
- 31.Sak S.C., Barrett J.H., Paul A.B., Bishop D.T., Kiltie A.E. The polyAT, intronic IVS11–6 and Lys939Gln XPC polymorphisms are not associated with transitional cell carcinoma of the bladder. Br. J. Cancer. 2005;92:2262–2265. doi: 10.1038/sj.bjc.6602616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matullo G., Guarrera S., Sacerdote C., Polidoro S., Davico L., Gamberini S., Karagas M., Casetta G., Rolle L., Piazza A., Vineis P. Polymorphisms/haplotypes in DNA repair genes and smoking: a bladder cancer case-control study. Cancer Epidemiol. Biomarkers Prev. 2005;14:2569–2578. doi: 10.1158/1055-9965.EPI-05-0189. [DOI] [PubMed] [Google Scholar]
- 33.Shen M., Hung R.J., Brennan P., Malaveille C., Donato F., Placidi D., Carta A., Hautefeuille A., Boffetta P., Porru S. Polymorphisms of the DNA repair genes XRCC1, XRCC3, XPD, interaction with environmental exposures, and bladder cancer risk in a case-control study in northern Italy. Cancer Epidemiol. Biomarkers Prev. 2003;12:1234–1240. [PubMed] [Google Scholar]
- 34.Kellen E., Zeegers M., Paulussen A., Van Dongen M., Buntinx F. Fruit consumption reduces the effect of smoking on bladder cancer risk. The Belgian case control study on bladder cancer. Int. J. Cancer. 2006;118:2572–2578. doi: 10.1002/ijc.21714. [DOI] [PubMed] [Google Scholar]
- 35.Thirumaran R.K., Bermejo J.L., Rudnai P., Gurzau E., Koppova K., Goessler W., Vahter M., Leonardi G.S., Clemens F., Fletcher T., et al. Single nucleotide polymorphisms in DNA repair genes and basal cell carcinoma of skin. Carcinogenesis. 2006;27:1676–1681. doi: 10.1093/carcin/bgi381. [DOI] [PubMed] [Google Scholar]
- 36.Larsson P., Wijkstrom H., Thorstenson A., Adolfsson J., Norming U., Wiklund P., Onelov E., Steineck G. A population-based study of 538 patients with newly detected urinary bladder neoplasms followed during 5 years. Scand. J. Urol. Nephrol. 2003;37:195–201. doi: 10.1080/00365590310008037. [DOI] [PubMed] [Google Scholar]
- 37.Lehmann M.L., Selinski S., Blaszkewicz M., Orlich M., Ovsiannikov D., Moormann O., Guballa C., Kress A., Truss M.C., Gerullis H., et al. Rs710521[A] on chromosome 3q28 close to TP63 is associated with increased urinary bladder cancer risk. Arch. Toxicol. 2010;84:967–978. doi: 10.1007/s00204-010-0617-6. [DOI] [PubMed] [Google Scholar]
- 38.Golka K., Schmidt T., Seidel T., Dietrich H., Roemer H.C., Lohlein D., Reckwitz T., Sokeland J., Weistenhofer W., Blaszkewicz M., Selinski S. The influence of polymorphisms of glutathione S-transferases M1 and M3 on the development of human urothelial cancer. J. Toxicol. Environ. Health A. 2008;71:881–886. doi: 10.1080/15287390801988087. [DOI] [PubMed] [Google Scholar]
- 39.Golka K., Hermes M., Selinski S., Blaszkewicz M., Bolt H.M., Roth G., Dietrich H., Prager H.M., Ickstadt K., Hengstler J.G. Susceptibility to urinary bladder cancer: relevance of rs9642880[T], GSTM1 0/0 and occupational exposure. Pharmacogenet. Genomics. 2009;19:903–906. doi: 10.1097/FPC.0b013e328331b554. [DOI] [PubMed] [Google Scholar]
- 40.Howie B.N., Donnelly P., Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marchini J., Howie B., Myers S., McVean G., Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 42.Kong A., Steinthorsdottir V., Masson G., Thorleifsson G., Sulem P., Besenbacher S., Jonasdottir A., Sigurdsson A., Kristinsson K.T., Jonasdottir A., et al. Parental origin of sequence variants associated with complex diseases. Nature. 2009;462:868–874. doi: 10.1038/nature08625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kutyavin I.V., Milesi D., Belousov Y., Podyminogin M., Vorobiev A., Gorn V., Lukhtanov E.A., Vermeulen N.M., Mahoney W. A novel endonuclease IV post-PCR genotyping system. Nucleic Acids Res. 2006;34:e128. doi: 10.1093/nar/gkl679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.International Hapmap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kruglyak L., Lander E.S. Faster multipoint linkage analysis using Fourier transforms. J. Comput. Biol. 1998;5:1–7. doi: 10.1089/cmb.1998.5.1. [DOI] [PubMed] [Google Scholar]
- 46.Lander E.S., Green P. Construction of multilocus genetic linkage maps in humans. Proc. Natl Acad. Sci. USA. 1987;84:2363–2367. doi: 10.1073/pnas.84.8.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kong A., Thorleifsson G., Gudbjartsson D.F., Masson G., Sigurdsson A., Jonasdottir A., Walters G.B., Jonasdottir A., Gylfason A., Kristinsson K.T., et al. Fine-scale recombination rate differences between sexes, populations and individuals. Nature. 2010;467:1099–1103. doi: 10.1038/nature09525. [DOI] [PubMed] [Google Scholar]
- 48.Gretarsdottir S., Thorleifsson G., Reynisdottir S.T., Manolescu A., Jonsdottir S., Jonsdottir T., Gudmundsdottir T., Bjarnadottir S.M., Einarsson O.B., Gudjonsdottir H.M., et al. The gene encoding phosphodiesterase 4D confers risk of ischemic stroke. Nat. Genet. 2003;35:131–138. doi: 10.1038/ng1245. [DOI] [PubMed] [Google Scholar]
- 49.Higgins J.P., Thompson S.G. Quantifying heterogeneity in a meta-analysis. Stat. Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 50.Cortessis V.K., Yuan J.M., Van Den B.D., Jiang X., Gago-Dominguez M., Stern M.C., Castelao J.E., Xiang Y.B., Gao Y.T., Pike M.C., Conti D.V. Risk of urinary bladder cancer is associated with 8q24 variant rs9642880[T] in multiple racial/ethnic groups: results from the Los Angeles-Shanghai case-control study. Cancer Epidemiol. Biomarkers Prev. 2010;19:3150–3156. doi: 10.1158/1055-9965.EPI-10-0763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang M., Wang M., Zhang W., Yuan L., Fu G., Wei Q., Zhang Z. Common genetic variants on 8q24 contribute to susceptibility to bladder cancer in a Chinese population. Carcinogenesis. 2009;30:991–996. doi: 10.1093/carcin/bgp091. [DOI] [PubMed] [Google Scholar]
- 52.Rafnar T., Sulem P., Stacey S.N., Geller F., Gudmundsson J., Sigurdsson A., Jakobsdottir M., Helgadottir H., Thorlacius S., Aben K.K., et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat. Genet. 2009;41:221–227. doi: 10.1038/ng.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gago-Dominguez M., Jiang X., Conti D.V., Castelao J.E., Stern M.C., Cortessis V.K., Pike M.C., Xiang Y.B., Gao Y.T., Yuan J.M., Van Den Berg D.J. Genetic variations on chromosomes 5p15 and 15q25 and bladder cancer risk: findings from the Los Angeles-Shanghai bladder case-control study. Carcinogenesis. 2011;32:197–202. doi: 10.1093/carcin/bgq233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang S., Tang J., Wang M., Yuan L., Zhang Z. Genetic variation in PSCA and bladder cancer susceptibility in a Chinese population. Carcinogenesis. 2010;31:621–624. doi: 10.1093/carcin/bgp323. [DOI] [PubMed] [Google Scholar]
- 55.Wang M., Chu H., Yan F., Qin C., Li P., Yuan L., Yin C., Xu J., Zhang Z. Chromosome 4p16.3 variant modify bladder cancer risk in a Chinese population. Carcinogenesis. 2011;32:872–875. doi: 10.1093/carcin/bgr060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.