Abstract
Recent studies emphasize the importance of mRNA splicing in human genetic disease, as 20–30% of all disease-causing mutations are predicted to result in mRNA splicing defects. The plasticity of the mRNA splicing reaction has made these mutations attractive candidates for the development of therapeutics. Familial dysautonomia (FD) is a severe neurodegenerative disorder, and all patients have an intronic IVS20+6T>C splice site mutation in the IKBKAP gene, which results in tissue-specific skipping of exon 20 and a corresponding reduction in ikappaB kinase complex associated protein (IKAP) levels. We created transgenic mouse lines using a human IKBKAP bacterial artificial chromosome (BAC) into which we inserted the IKBKAP splice mutation (FD BAC) and have shown that the transgenic mice exhibit the same tissue-specific aberrant splicing patterns as seen in FD patients. We have previously demonstrated that the plant cytokinin kinetin can significantly improve the production of wild-type IKBKAP transcripts in FD lymphoblast cell lines by improving exon inclusion. In this study, we tested the ability of kinetin to alter IKBKAP splicing in the transgenic mice carrying the FD BAC and show that it corrects IKBKAP splicing in all major tissues assayed, including the brain. The amount of wild-type IKBKAP mRNA and IKAP protein was significantly higher in the kinetin-treated mice. These exciting results prove that treatment of FD, as well as other mechanistically related splicing disorders, with kinetin holds great promise as a potential therapeutic aimed at increasing normal protein levels, which may, in turn, slow disease progression.
INTRODUCTION
The completion of the Human Genome Project and the discovery that the genome has far fewer genes than expected has highlighted the importance of alternative splicing in transcript and proteome complexity (1). Recent studies estimate that 35–74% of all human genes are alternatively spliced, and it is therefore no surprise that a significant number of disease-causing mutations alter mRNA splicing (2,3). Characterization of these mutations has shown how essential the mRNA splicing machinery is to normal physiological processes (4,5). It is not uncommon for genes with splice mutations to transcribe both wild-type and mutant versions of the message from the same allele, thereby producing the normal protein at reduced levels (6,7). Differences in the level of normal protein produced have been correlated to disease severity in some cases (8). For example, mutations in the ATP7A gene can cause severe Menkes disease or less severe occipital horn syndrome (OHS), depending on how the mutation affects the splicing machinery (9). Menkes disease is caused by mutations in the invariant donor splice site of exon 6, which leads to complete exon skipping, while OHS is the result of a point mutation at the same donor splice site that leads to incomplete exon skipping and decreased production of normal protein. Since minor shifts in splicing efficiencies have been shown to modify disease phenotype and severity, therapeutic approaches targeted at correcting aberrant splicing are attractive options. Several strategies, including antisense oligonucleotides, RNA-mediated mechanisms and pharmacologic agents, have been employed successfully to restore normal splicing patterns and increase protein production in several disorders, including spinal muscular atrophy, cystic fibrosis and Duchenne muscular dystrophy (10,11).
Familial dysautonomia (FD), a congenital sensory neuropathy with a widespread sensory and autonomic dysfunction, is a devastating genetic disorder caused by aberrant mRNA splicing of the IKBKAP gene (12). FD is an autosomal recessive disorder with a high carrier frequency ranging from 1 in 27 in the general Ashkenazi Jewish (AJ) population to 1 in 18 in Ashkenazi Jews of Polish descent (13,14). Despite the fact that FD is recessive, all patients produce normal ikappaB kinase complex associated protein (IKAP) from the mutant alleles. The levels of normal protein produced depend on the amount of correct mRNA splicing, which varies from tissue to tissue. This altered mRNA splicing causes normal IKAP protein levels to drop below the tissue-specific threshold required for normal development and neuronal differentiation. Even with early diagnosis and a significantly improved treatment regime that has been developed over the past decade, only 50% of the patients survive to 40 years of age. Adult FD patients have marked neurodegeneration and a high incidence of sudden death caused by autonomic nervous system failure. Increasing normal protein above the required threshold early in life may reduce or prevent this neurodegeneration, and therefore modification of mRNA splicing in these patients to increase functional protein levels is a viable therapeutic strategy (15,16).
In 2001, two mutations in the IKBKAP gene were shown to cause FD in AJ families (17,18). Of the three mutations in IKBKAP identified to date, at least one copy of an intronic IVS20+6T>C point mutation is present in all FD patients, with 99.5% being homozygous for this mutation (18,19). This point mutation weakens the splicing efficiency and causes frequent skipping of exon 20 in the IKBKAP transcript (20). Importantly, although FD is a recessive disease, homozygous mutant cells express both wild-type (WT) and mutant (MU) IKBKAP message, since the point mutation does not completely block inclusion of exon 20 during mRNA splicing. We have shown that the ratio of WT:MU IKBKAP transcripts varies in different human tissues, with the lowest ratio observed in the central and peripheral nervous tissues (21).
IKAP/ELP1, the protein encoded by IKBKAP, was initially identified as a scaffold protein involved in cytokine signaling, a function called into question in subsequent studies (22,23). IKAP is homologous to elongator protein 1 (Elp1) in Saccharomyces cerevisiae, and is now known to be the scaffolding component of the Elongator complex, which is required for efficient RNA-polymerase-II-mediated transcriptional elongation (24,25). IKAP has also been associated with other cellular functions in addition to its role in transcription, although some of these putative functions remain controversial (26). Studies in yeast suggest that IKAP plays a role in exocytosis and tRNA modification, whereas data in mammalian cells implicate IKAP as a scaffold protein involved in cytoplasmic JNK activation in response to extracellular stress (27–29). A recent microarray study also implicates IKAP in oligodendrocyte differentiation and myelin formation; however, this has not been demonstrated functionally (30). RNA interference studies show that the depletion of IKAP/ELP1 in mammalian cells impacts the transcriptional elongation of a number of genes, including several that are involved in cell migration and adhesion (31).
We have recently generated an Ikbkap knockout mouse model and show that, as predicted from our previous studies of the human disease, complete loss of Ikap leads to embryonic lethality in mice. Examination of transcriptional elongation using chromatin immunoprecipitation illustrates the importance of Ikap in neural and vascular development during murine embryogenesis (32). There is little doubt that deficits in cellular motility and neurodevelopmental processes resulting from reduced IKAP levels play a role in the neuropathology of FD; therefore, any strategy to boost IKAP production could reduce the severity or limit the progression of disease. To this end, we have identified the plant cytokinin kinetin as a potential modulator of IKBKAP splicing in FD cell lines. This natural compound dramatically improves the ratio of WT:MU IKBKAP transcripts and the levels of IKAP protein in FD cell lines (33). Kinetin has also been shown to affect other aberrant splicing events resulting from splice mutations in the gene causing neurofibromatosis type I demonstrating the potential of using small molecules to target particular types of mRNA splicing mutations (34,35).
In this study, we evaluate for the first time the in vivo efficacy of dietary kinetin to modify mRNA splicing using a humanized transgenic mouse line that carries the complete human IKBKAP locus with the major FD mutation. Human IKBKAP is appropriately expressed from the transgene, and the tissue-specific splicing pattern seen in FD patients is modeled perfectly in these mice (36). Effects of kinetin on IKBKAP mRNA splicing and IKAP protein levels in the transgenic mice were evaluated in various tissues following daily treatment with kinetin. To further verify the efficacy of kinetin directly in neurons, primary neuronal cultures were derived from transgenic murine embryos and treated with kinetin. The findings were uniform across both the transgenic mouse model and primary neuronal cultures: administration of kinetin improves IKBKAP splicing and IKAP protein levels in all tissues. These exciting results demonstrate for the first time that oral treatment with kinetin can modify mRNA splicing in vivo. Further, as shown here, kinetin can cross the blood-brain barrier and modify splicing in brain, further emphasizing the potential efficacy of kinetin in human FD patients.
RESULTS
We have generated several FD and WT humanized IKBKAP transgenic mouse lines, using the FD and WT bacterial artificial chromosome (BAC) constructs previously created in our laboratory. Detailed characterization of the new transgenic lines based on breeding success, transmission stability, BAC insertion (copy) number and consistency of transgene expression was performed as described previously (36). For the current study, we used an FD transgenic line containing one copy of the FD BAC (designated TgFD1).
Kinetin diet studies in transgenic mice
Recent toxicity and pharmacokinetic testing of kinetin in rodents has shown that kinetin is safe and well tolerated as an oral treatment (37). Importantly, the pharmacokinetic studies indicate that kinetin passes the blood-brain barrier and that kinetin levels in serum reach the effective concentrations we previously used in our FD lymphoblast cell culture studies (33,34). Based on our previous studies in cultured cells, our goal was to achieve serum and tissue kinetin concentrations >10 µm. The mice were fed 400 mg/kg of kinetin daily via oral gavage for a period of 30 days. There were no visible phenotypic changes observed in any of the kinetin- or vehicle-treated (control) mice and no significant changes in their body weight during the length of the experiment. All experimental subjects were alive for the duration of the trial. Following treatment, mice were sacrificed and tissues were collected for RNA isolation and LC/MS analysis. Kinetin levels in serum from the kinetin-tested mice ranged from 12.6 to 40.51 µm with an average of 31.1 µm, well above the effective concentration of kinetin in cell culture. In a parallel experiment, we administered kinetin to the transgenic mice in a formulated food diet for 30 days. The mouse chow is formulated to dose each mouse ∼400 mg/kg/day, based on the calculated average daily consumption of our mice. Kinetin levels detected in serum collected from kinetin-chow-fed mice were also above the desired concentration, with an average serum kinetin concentration of 40.3 µm. The kinetin levels achieved in brain and liver tissues confirmed our earlier studies that kinetin crosses the blood-brain barrier and is also absorbed by somatic tissues (Fig. 1B). To assess IKBKAP mRNA splicing, RT–PCR assays were performed on RNA extracted from whole blood, brain, heart, liver, lung, spleen, eye and kidney tissues. The percent exon 20 inclusion increased significantly in all tissues tested in the kinetin-treated mice when compared with the vehicle-controls (Fig. 1). RT–PCR analysis, as shown in Figure 1, shows an increase in percent exon inclusion in IKBKAP transcripts of the brain, liver, kidney, blood, spleen, lung, heart and eye samples in kinetin-treated TgFD1 mice. To further confirm that kinetin improves the inclusion of exon 20 in IKBKAP transcripts, we performed real-time RT–PCR analysis on total RNA from the brain, eye, heart, kidney and liver samples. The results indicate that kinetin increases the inclusion of exon 20 in IKBKAP transcripts as WT IKBKAP expression is significantly higher in the kinetin-treated mice compared with the vehicle-controls (Fig. 2). To determine whether higher WT IKBKAP expression levels translate into greater production of human IKAP protein, we performed western blots on the brain and liver tissues of the kinetin-fed transgenic mice. The levels of IKAP were significantly elevated in the kinetin-treated mice, when compared with the vehicle-control littermates (Fig. 3). These exciting results show, for the first time, that mRNA splicing can be modified in vivo using orally delivered kinetin, thus setting the stage for clinical trials in FD patients.
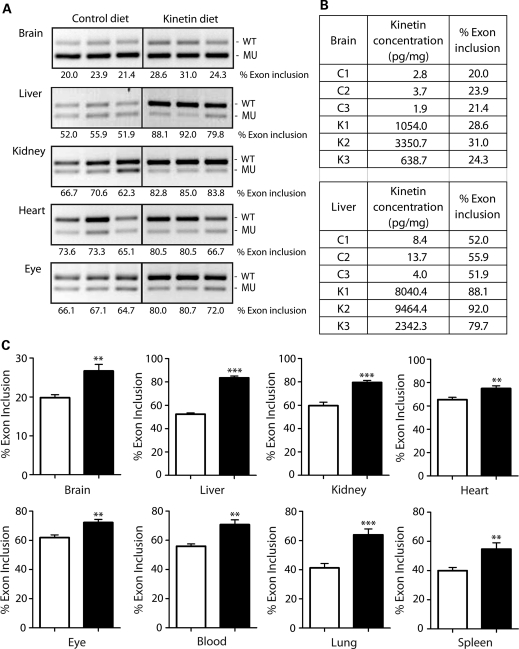
Figure 1.
Kinetin modifies IKBKAP splicing in TgFD1 transgenic mice. (A) Representative images of RT–PCR analysis of IKBKAP exon 20 splicing in different tissues from TgFD1 transgenic mice. Expression of the FD IKBKAP mRNA produces both the WT (including exon 20) and MU (excluding exon 20). Percent exon 20 inclusion (shown below each panel) was calculated using the integrated density values (IDV) obtained for each WT and MU band. An increase in the percent exon 20 inclusion is observed in all tissues tested in the kinetin-fed mice compared with the vehicle controls. (B) Table shows the kinetin concentrations and the percent exon 20 inclusion in control (C1, C2, C3) and kinetin-fed (K1, K2, K3) mice, respectively. (C) Plotted bar charts show the quantification of the percent exon 20 inclusion in multiple tissues in kinetin-fed mice and control mice. Black and white bars represent the kinetin-fed and vehicle control mice, respectively. Error bars represent SEM (n= 8). Significant differences observed between the kinetin-fed and vehicle control mice are represented by ** (P< 0.01) and (*** P< 0.0005); Student's t-test.
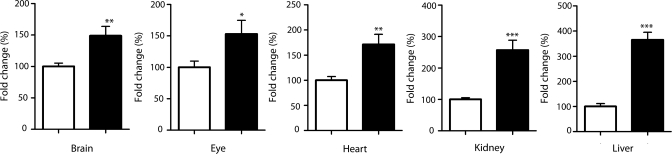
Figure 2.
Kinetin improves IKBKAP expression in transgenic mice. Bar charts represent the relative amount of WT IKBKAP transcripts in multiple tissues in the transgenic mice. Black and white bars represent the kinetin-fed and vehicle control mice, respectively. Internal normalization was performed, using mouse gapdh gene as a reference. Error bars represent SEM (n= 8). Significant differences are represented by *(P< 0.05), ** (P< 0.01) and *** (P< 0.0005); Student's t-test.
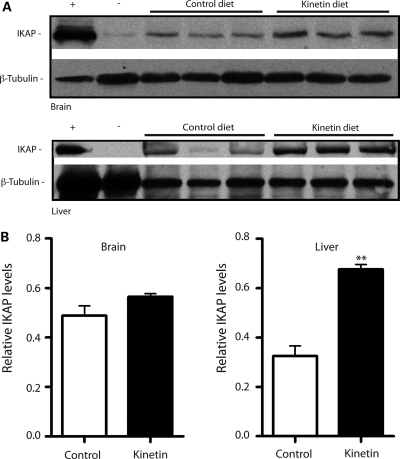
Figure 3.
Kinetin increases IKAP levels in transgenic mice. (A) Western blots of brain and liver tissues in TgFD1 transgenic mice following treatment with kinetin diet for a period of 30 days. Top and bottom panels show the blot probed with an IKAP antibody and β-tubulin antibody, respectively. Human brain and C57BL6 mouse liver tissues were used as positive (+) and negative (−) controls, respectively. (B) Bar charts show the densitometric analysis on western blots of brain and liver tissues after kinetin diet for a period of 30 days. White and black bars represent control and kinetin-fed mice, respectively. Error bars represent SEM (n= 3). Significant differences from control are represented by ** (P< 0.01); Student's t-test.
Kinetin treatment in neuronal cells
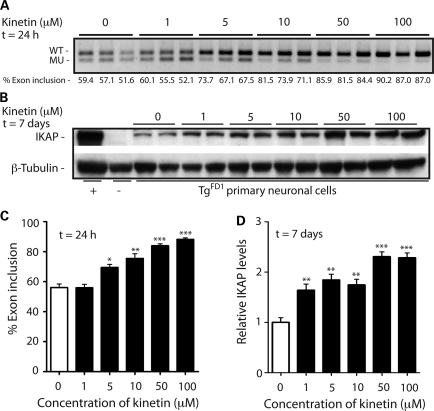
FD is a neurodegenerative disease, and the markedly reduced expression of IKAP protein in the neurons leads to FD pathophysiology. Hence, any improvement in WT IKBKAP mRNA expression by reducing aberrant IKBKAP splicing in the neurons could be beneficial. The generation of primary neuronal cells from transgenic mice carrying the human IKBKAP locus with the major IVS20+6T>C point mutation gives us a model to further study the effect of kinetin on IKBKAP splicing in a neuronal cell population. To this end, we generated neuronal cultures from the cerebrum of E17 TgFD1-positive embryos. Seven days after generation of the TgFD1-positive neuronal cultures, the cells were treated with varying concentrations of kinetin for a period of 1 or 7 days. The IKBKAP-splicing RT–PCR assays performed on RNA from the cells treated for 24 h showed significant improvement in the percent exon 20 inclusion in IKBKAP transcripts in kinetin-treated cells at concentrations as low as 5 µm (Fig. 4A and C). For the range of kinetin dosage employed in our studies, we observed a dose-dependent effect; with an increase in concentrations of kinetin, there was an increase in percent exon 20 inclusion. Western blot analysis on the kinetin-treated cells shows significantly higher IKAP levels in the kinetin-treated cells compared with the untreated controls (Fig. 4B and D). We observed higher production of human IKAP protein in the kinetin-treated cells even at concentrations of 1 µm kinetin. Taken together, our results show that kinetin crosses the blood-brain barrier and can modify splicing and increase IKAP protein in vivo, which might reduce neuronal degeneration in FD patients.
Figure 4.
Dose-dependent effect of kinetin on IKBKAP splicing and IKAP levels in TgFD1 neuronal cultures. (A) RT-PCR analysis of three TgFD1 embryos using primers specific to human IKBKAP, following treatment with 1, 5, 10, 50 and 100 µm kinetin respectively for 24 h. The percent exon 20 inclusion (shown below each lane) was determined using the IDV obtained for the WT and MU band. (B) Western blot, probed with an IKAP antibody (top panel), shows an increase in human IKAP protein production in primary neuronal cells following treatment with varying concentrations of kinetin for 7 days. The lower panel represents the same blot probed with β-tubulin antibody as a protein-loading control. Human brain and C57BL6 mouse liver were used as positive (+) and negative (−) controls, respectively. (C) Plotted bar charts show the quantification of percent exon 20 inclusion in three TgFD1 embryos, following kinetin treatment for 24 h. White and black bars represent the untreated and kinetin-treated primary neuronal cells, respectively. Error bars represent SEM (n= 3). Significant differences observed between the untreated and kinetin-treated primary neuronal cells are represented by * (P< 0.05), ** (P< 0.01) and *** (P< 0.0005); Student's t-test. (D) Bar charts show the densitometric analysis on western blots of neuronal cells treated with 1, 5, 10, 50 and 100 µm kinetin, respectively, for 7 days. White and black bars represent untreated and kinetin-treated primary neuronal cells, respectively. Error bars represent SEM (n= 4). Significant differences from control are represented by *(P< 0.05), ** (P< 0.01) and *** (P< 0.0005).
DISCUSSION
All FD patients described to date carry the major intronic mutation that leads to mis-splicing of IKBKAP mRNA and reduced levels of IKAP protein. The absence of exon 20 in the mutant transcript causes a frameshift that introduces a stop codon in the mRNA coding region of exon 21, leading to premature translational termination and degradation of the mutant RNA by nonsense-mediated decay (38). We and others have failed to detect a truncated protein product in FD patient tissues or cell lines, suggesting that the disease results from a reduction in functional IKAP protein and not aberrant activity of a truncated protein. This is further supported by genetics. FD is a recessive disease, and carrier parents have no phenotype even though they carry the mutant allele and produce low levels of mutant mRNA. If there were dominant-negative effects of a truncated IKAP protein, we would not expect to see a classic autosomal-recessive inheritance pattern. The fact that all patients have at least one copy of the major mutation, and therefore produce low levels of normal IKAP protein, suggests that FD results from IKAP levels falling below some critical threshold for neuronal survival. Our studies in Ikbkap knock-out mice support this as a complete loss of Ikap results in early embryonic lethality (32). The future challenge, therefore, will be to determine how reduction of IKAP leads to disruption of cellular functions and disease development.
This paradigm of protein reduction rather than complete loss provided us with a straightforward approach for developing potential therapeutics. Since FD patients already produce normal mRNA and protein, our approach was to identify compounds that can modify the splicing machinery and boost the production of functional IKAP protein. Kinetin, identified in our initial drug screen for small molecule modifiers of IKBKAP splicing, is a potent modulator of both in vitro and in vivo IKBKAP splicing as shown in this study (33). To date, three other compounds have been proposed as potential therapeutics for FD. Epigallocatechin gallate, an antioxidant present in green tea, was reported to modify IKBKAP splicing, and tocotrienols were reported to increase total IKBKAP transcript production in FD cell lines (39,40). However, two recent studies have failed to show that either of these two compounds has a significant effect on IKBKAP levels (41,42). Very recently, phosphatidylserine (PS), a food supplement, has also been shown to increase total IKBKAP mRNA levels in FD cells, but the effect of PS on increasing total IKBKAP is not as pronounced as that of kinetin (41).
The precise mechanism by which kinetin stimulates the splicing machinery is not known, but our previous studies indicate that the sequence of the splice donor site of exon 20, in particular the last three base pairs of exon 20, are required for activity. IKBKAP exon 20 ends with a CAA, rather than the consensus sequence CAG, and testing of kinetin on many exon 20 deletion constructs showed that the CAA sequence at the end of exon 20 was required for kinetin's activity (34). Interestingly, kinetin's ability to modify aberrant splicing events is not limited to IKBKAP. Recent studies have shown that kinetin can improve exon inclusion in neurofibromin 1 transcripts when splice mutations cause exon skipping, demonstrating the potential of kinetin and similar compounds to alter mRNA splicing and increase normal protein production in diseases caused by mechanistically similar splice mutations (34,35).
Our current study shows, for the first time, that oral dosing of kinetin can alter mRNA splicing and increase IKAP protein production in brain and somatic tissues. Examination of Figures 1 and 2 shows that although the increase in full-length IKBKAP mRNA is significant in all tissues, it does vary. This could perhaps be explained by the absolute level of kinetin in the tissues. The tables in Figure 1 show that there is some correlation between kinetin concentration and percent exon inclusion; however, our sample size was not large enough to determine whether this is a significant correlation across all tissues. We do not yet know whether increasing functional IKAP protein in FD patients will prove therapeutic, but, as described above, there are many models that demonstrate dramatic phenotypic benefits even with modest increases in protein levels. Recently, Studer and co-workers (42) created iPSCs using fibroblasts derived from FD patients and differentiated them into various cell types, including peripheral neurons. They confirmed that kinetin treatment shifted splicing and increased the level of WT IKBKAP mRNA. Interestingly, they were also able to show that kinetin treatment led to a higher percentage of differentiating neurons and an increase in expression of key peripheral neuronal genes, demonstrating positive down-stream effects of increased IKAP production. Further, our clinical studies have shown that oral administration of kinetin to adult FD carriers improves IKBKAP splicing in blood and that these changes in IKBKAP splicing are dose dependent (37,43). Kinetin activity has further been confirmed in a new model system where it improved exon 20 inclusion and restored IKAP levels in olfactory ecto-mesenchymal stem cells derived from FD patients (44). Taken together with the current study, in which we demonstrate the ability of kinetin to cross the blood-brain barrier, improve splicing and increase WT IKBKAP mRNA and IKAP protein levels in neural and somatic tissues, the stage is set for a clinical trial in FD patients. Reduction in functional IKAP protein resulting from aberrant mRNA splicing is responsible for the widespread sensory and autonomic dysfunction seen in FD. Even though FD is present at birth, there is continued neurodegeneration throughout life that eventually leads to severe ataxia, dementia and sudden death. Our studies to date suggest that kinetin treatment will improve IKBKAP splicing in patients and lead to increased IKAP production, which may, over time, slow or stop the progressive neurodegeneration that characterizes this devastating disease.
MATERIALS AND METHODS
Generation of new transgenic mice
Transgenic mice were generated as previously described in Hims et al. (36). Briefly, BAC DNA was isolated from BAC transformed bacterial culture using anion exchange columns (Nucleobond, BD Biosciences). Transgenic mice were produced at NIMH using standard protocols. BAC DNA (1 ng/µl) was injected in the pronucleus of embryos harvested from superovulated and mated C57BL6 female mice (Taconic and in-house). Embryos that advanced to the two-cell stage were transplanted into the oviducts of pseudopregnant recipient C57BL6 females. Tail biopsies were collected from 18- to 21-day-old pups for genotyping analysis. All animals were housed at Massachusetts General Hospital (Boston, MA, USA) and were treated in accordance to the NIH guidelines. All experimental protocols were approved by the Subcommittee on Research Animal Care at the Massachusetts General Hospital.
Genotype and copy number analysis in transgenic mice
The genotypes of animals and embryos were determined by PCR analysis of genomic DNA extracted from tail biopsies using Gentra Puregene Genomic Purification Kit (Qiagen, Germantown, MD, USA) and DirectPCR Lysis Reagent (Viagen Biotech Inc., Los Angeles, CA, USA). The primers used for determining the human-specific IKBKAP transgenes and copy number estimation were TgProbe1F (5′-GCCATTGTACTGTTTGCGACT) and TgProbe1R (5′-TGAGTGTCACGATTCTTTCTGC). PCR was performed using genomic DNA and 30 amplification cycles (at 94°C for 30 s, at 58°C for 30 s and at 72°C for 30 s). Copy number estimation using quantitative real-time PCR was performed with 60 ng of purified genomic DNA using primers TgProbe1F and TgProbe1R as described previously (36).
Administration of kinetin via oral gavage
Carboxymethyl cellulose sodium (CMC) was purchased from Spectrum Chemical Mfg. Corp., CA, USA. Kinetin was provided by the Dysautonomia Foundation, Inc. (New York, NY, USA). They contracted for kinetin to be produced by Taizhou Mingsheng Chemical Co., Ltd. and testing in our laboratory in various cell assays showed similar activity and efficacy to the kinetin purchased from Sigma. Stock suspension of finely powdered kinetin (40 mg/ml) was prepared in 0.5% CMC. Five transgenic mice were fed a daily dose of 400 mg/kg of kinetin suspension solution using a 20 Gauge feeding needle (Fine Science Tools Inc., CA, USA) for a period of 30 days. Five control mice were fed daily 0.5% CMC for the same duration. The mice were given food and water ad libitum, and changes in body weights were monitored on a daily basis.
Kinetin administration via specially formulated diet
Three transgenic mice were fed special AIN-76A rodent diet containing kinetin at a concentration of 4.286 g/kg of mouse chow (Research Diets, Inc., NJ, USA) for a period of 30 days. In the control group, three transgenic mice were fed AIN-76A diet without kinetin. The mice were given water ad libitum, and changes in body weights were monitored on a weekly basis.
Establishment of neuronal cultures from transgenic mice and kinetin treatment
Transgenic (TgFD1) mice were mated with pure C57BL/6 mice, and embryos were dissected from the mother via c-section at 17 days post-coitum (dpc). The neuronal cultures were established as described previously (45,46). Briefly, the cerebrum was dissected into ice-cold PBS and sequentially treated with 0.5% trypsin, 0.1% trypsin inhibitor and DNAse I for a period of 1 min each at 37°C. The neuronal tissue was broken up in neurobasal media (GIBCO) containing 2% B27 supplement (GIBCO) by repeated passage through a P1000 and P200 pipets, respectively. The dissociated cells were then plated at density of 1 × 106 cells/ml and were cultured in 37°C incubator with 5% CO2. The cells were maintained in the neurobasal media supplemented with 2% B27 supplement and 1% penicillin/streptomycin for the duration of the experiment. Seven days after the generation of the neuronal cultures, the cells were treated with 1, 5, 10, 50 and 100 µm kinetin for a period of 1 or 7 days. Following the treatment with kinetin, the cells were harvested for RNA and western blot studies.
RNA isolation from murine tissue and neuronal cells
Mice were euthanized following the treatment studies, and the brain, liver, lung, spleen, eye, kidney and heart tissues were removed. Both tissue and primary neuronal cultures were homogenized in ice-cold TRI reagent (Molecular Research Center, Inc., Cincinnati, OH, USA), using a TissueLyser (Qiagen) and the homogenates were stored at −80°C. Total RNA was extracted using the TRI reagent procedure provided by the manufacturer. Total RNA from blood samples was isolated using RiboPure™-Blood kit (Ambion, Inc., TX, USA) as per the manufacturer's protocol. The yield, purity and quality of the total RNA for each sample were determined using a Nanodrop ND-1000 spectrophotometer.
RT–PCR analysis of spliced IKBKAP isoforms
Reverse transcription was performed using 500 ng of total RNA, oligo(dT) primer and Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA, USA). PCR was performed using cDNA equivalent to 100 ng of starting RNA and run through 30 amplification cycles (at 94°C for 30 s, at 58°C for 30 s and at 72°C for 30 s). Primer sequences were as follows: human IKBKAP primers Exon 19F (5′-CCTGAGCAGCAATCATGTG), Exon 23R (5′-TACATGGTCTTCGTGACATC), mouse-specific primers Mm-sp-Ex19F (5′-AGTGGCAGTCATGAGGCCAG) and Mm-sp-Ex22R (5′-GTGACATCTTCTTCCCTGAG). PCR products were separated on 1.5% agarose gels and stained with ethidium bromide. The relative amount of WT and MU IKBKAP spliced isoforms in a single PCR were determined using Alpha 2000™ Image Analyzer (BioRad, Hercules, CA, USA) and AlphaEase FC software (Alpha Innotech, San Leandro, CA, USA), using the integrated density value (IDV) for each band as described previously, and the relative proportion of the WT isoform detected in a sample was calculated as a percentage of the total number of WT and MU isoforms (percent exon inclusion) (21).
Quantitative RT–PCR
WT IKBKAP mRNA expression levels were estimated in various tissues of the transgenic mice by quantitative RT–PCR using a BioRad iCycler, iQ SYBR-Green supermix (BioRad), and IKBKAP primers hELP1-Ex20F (5′-GCTCAGATTCGGAAGTGGTT) and hELP1-Ex22R (5′-TGAAGGTTTCCACATTTCCA), which are specific to the WT-spliced isoform of human IKBKAP. Mouse gapdh gene was used as an internal normalization control to correct for the amount of cDNA used in each reaction. Quantitative PCR was performed under the following PCR conditions: 1 cycle at 50°C for 15 min; 1 cycle at 95°C for 3 min; and 40 cycles at 95°C for 10 s, and at 59°C for 45 s.
Protein isolation and western blot analysis of tissues from transgenic mice and neuronal cultures
Brain and liver tissues from the transgenic mice were disrupted using a TissueLyser (Qiagen), followed by sonication in ice-cold radio-immunoprecipitation assay (RIPA) buffer (Boston Bioproducts, Ashland, MA, USA) supplemented with protease inhibitor cocktail (Roche, Mannheim, Germany). For the neuronal cultures, the cells were isolated and suspended in ice-cold RIPA buffer (Boston Bioproducts) supplemented with protease inhibitor cocktail (Roche). A 20 µg of total protein extract was separated on NuPage 4–12% Bis–Tris Gel (Invitrogen) and transferred to nitrocellulose transfer membrane (Thermo Scientific) using XCell SureLock Mini-Cell and XCell II Blot Module kits (Invitrogen), as per the manufacturer's protocol. The membrane was then probed with a primary antibody against IKAP (1:2250; Sigma), β-tubulin (1:4000; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) followed by HRP-conjugated secondary antibody, and visualized using ECL system (GE Healthcare UK, Ltd.) on X-ray film (Eastman Kodak, Rochester, NY, USA).
Statistical analysis
Comparisons between the kinetin treatment and control groups were performed using Student's t-test using GraphPad Prism software (GraphPad Software, La Jolla, CA, USA). Average means are represented as columns and SEM is represented as error bars. The P values and number of independent biological replicates (n) are indicated in the figure legends.
FUNDING
This work was supported by a grant from the Dysautonomia Foundation, Inc, and NIH (R01 NS36326 and R21 NS R21NS058318) to S.A.S. R.S.S. was supported by a Massachusetts General Hospital ECOR Fund for Medical Discovery Award.
ACKNOWLEDGEMENTS
We are grateful to Dr Felicia Axelrod and Dr Horacio Kaufmann of the Dysautonomia Treatment and Evaluation Center at New York University Medical School for their longstanding collaboration and helpful discussions; Dr James Gusella for his helpful discussions; Lijuan Liu for technical assistance and Dr David Schoenfeld for assistance with the statistical analysis.
Conflict of Interest statement. D.K. is the founder and employee of BRI Biopharmaceutical Research Inc.
REFERENCES
- 1.Blencowe B.J. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 2.Johnson J.M., Castle J., Garrett-Engele P., Kan Z., Loerch P.M., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 3.Modrek B., Lee C. A genomic view of alternative splicing. Nat. Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- 4.Cartegni L., Chew S.L., Krainer A.R. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 5.Faustino N.A., Cooper T.A. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- 6.Beck S., Penque D., Garcia S., Gomes A., Farinha C., Mata L., Gulbenkian S., Gil-Ferreira K., Duarte A., Pacheco P., et al. Cystic fibrosis patients with the 3272–26A→G mutation have mild disease, leaky alternative mRNA splicing, and CFTR protein at the cell membrane. Hum. Mutat. 1999;14:133–144. doi: 10.1002/(SICI)1098-1004(1999)14:2<133::AID-HUMU5>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 7.Svenson I.K., Ashley-Koch A.E., Gaskell P.C., Riney T.J., Cumming W.J., Kingston H.M., Hogan E.L., Boustany R.M., Vance J.M., Nance M.A., et al. Identification and expression analysis of spastin gene mutations in hereditary spastic paraplegia. Am. J. Hum. Genet. 2001;68:1077–1085. doi: 10.1086/320111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nissim-Rafinia M., Kerem B. The splicing machinery is a genetic modifier of disease severity. Trends Genet. 2005;21:480–483. doi: 10.1016/j.tig.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 9.Moller L.B., Tumer Z., Lund C., Petersen C., Cole T., Hanusch R., Seidel J., Jensen L.R., Horn N. Similar splice-site mutations of the ATP7A gene lead to different phenotypes: classical Menkes disease or occipital horn syndrome. Am. J. Hum. Genet. 2000;66:1211–1220. doi: 10.1086/302857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Blanco M.A., Baraniak A.P., Lasda E.L. Alternative splicing in disease and therapy. Nat. Biotechnol. 2004;22:535–546. doi: 10.1038/nbt964. [DOI] [PubMed] [Google Scholar]
- 11.Yeo G.W. Splicing regulators: targets and drugs. Genome Biol. 2005;6:240. doi: 10.1186/gb-2005-6-12-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Axelrod F.B. Familial dysautonomia. Muscle Nerve. 2004;29:352–363. doi: 10.1002/mus.10499. [DOI] [PubMed] [Google Scholar]
- 13.Lehavi O., Aizenstein O., Bercovich D., Pavzner D., Shomrat R., Orr-Urtreger A., Yaron Y. Screening for familial dysautonomia in Israel: evidence for higher carrier rate among Polish Ashkenazi Jews. Genet. Test. 2003;7:139–142. doi: 10.1089/109065703322146830. [DOI] [PubMed] [Google Scholar]
- 14.Maayan C., Kaplan E., Shachar S., Peleg O., Godfrey S. Incidence of familial dysautonomia in Israel 1977–1981. Clin. Genet. 1987;32:106–108. doi: 10.1111/j.1399-0004.1987.tb03334.x. [DOI] [PubMed] [Google Scholar]
- 15.Gold-von Simson G., Axelrod F.B. Familial dysautonomia: update and recent advances. Curr. Probl. Pediatr. Adolesc. Health Care. 2006;36:218–237. doi: 10.1016/j.cppeds.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 16.Ngai J., Kreynin I., Kim J.T., Axelrod F.B. Anesthesia management of familial dysautonomia. Paediatr. Anaesth. 2006;16:611–620. doi: 10.1111/j.1460-9592.2006.01947.x. [DOI] [PubMed] [Google Scholar]
- 17.Anderson S.L., Coli R., Daly I.W., Kichula E.A., Rork M.J., Volpi S.A., Ekstein J., Rubin B.Y. Familial dysautonomia is caused by mutations of the IKAP gene. Am. J. Hum. Genet. 2001;68:753–758. doi: 10.1086/318808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Slaugenhaupt S.A., Blumenfeld A., Gill S.P., Leyne M., Mull J., Cuajungco M.P., Liebert C.B., Chadwick B., Idelson M., Reznik L., et al. Tissue-specific expression of a splicing mutation in the IKBKAP gene causes familial dysautonomia. Am. J. Hum. Genet. 2001;68:598–605. doi: 10.1086/318810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leyne M., Mull J., Gill S.P., Cuajungco M.P., Oddoux C., Blumenfeld A., Maayan C., Gusella J.F., Axelrod F.B., Slaugenhaupt S.A. Identification of the first non-Jewish mutation in familial Dysautonomia. Am. J. Med. Genet. A. 2003;118A:305–308. doi: 10.1002/ajmg.a.20052. [DOI] [PubMed] [Google Scholar]
- 20.Ibrahim E.C., Hims M.M., Shomron N., Burge C.B., Slaugenhaupt S.A., Reed R. Weak definition of IKBKAP exon 20 leads to aberrant splicing in familial dysautonomia. Hum. Mutat. 2007;28:41–53. doi: 10.1002/humu.20401. [DOI] [PubMed] [Google Scholar]
- 21.Cuajungco M.P., Leyne M., Mull J., Gill S.P., Lu W., Zagzag D., Axelrod F.B., Maayan C., Gusella J.F., Slaugenhaupt S.A. Tissue-specific reduction in splicing efficiency of IKBKAP due to the major mutation associated with familial dysautonomia. Am. J. Hum. Genet. 2003;72:749–758. doi: 10.1086/368263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohen L., Henzel W.J., Baeuerle P.A. IKAP is a scaffold protein of the IkappaB kinase complex. Nature. 1998;395:292–296. doi: 10.1038/26254. [DOI] [PubMed] [Google Scholar]
- 23.Krappmann D., Hatada E.N., Tegethoff S., Li J., Klippel A., Giese K., Baeuerle P.A., Scheidereit C. The I kappa B kinase (IKK) complex is tripartite and contains IKK gamma but not IKAP as a regular component. J. Biol. Chem. 2000;275:29779–29787. doi: 10.1074/jbc.M003902200. [DOI] [PubMed] [Google Scholar]
- 24.Hawkes N.A., Otero G., Winkler G.S., Marshall N., Dahmus M.E., Krappmann D., Scheidereit C., Thomas C.L., Schiavo G., Erdjument-Bromage H., et al. Purification and characterization of the human elongator complex. J. Biol. Chem. 2002;277:3047–3052. doi: 10.1074/jbc.M110445200. [DOI] [PubMed] [Google Scholar]
- 25.Otero G., Fellows J., Li Y., de Bizemont T., Dirac A.M., Gustafsson C.M., Erdjument-Bromage H., Tempst P., Svejstrup J.Q. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell. 1999;3:109–118. doi: 10.1016/s1097-2765(00)80179-3. [DOI] [PubMed] [Google Scholar]
- 26.Svejstrup J.Q. Elongator complex: how many roles does it play? Curr. Opin. Cell Biol. 2007;19:331–336. doi: 10.1016/j.ceb.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 27.Esberg A., Huang B., Johansson M.J., Bystrom A.S. Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol. Cell. 2006;24:139–148. doi: 10.1016/j.molcel.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 28.Holmberg C., Katz S., Lerdrup M., Herdegen T., Jaattela M., Aronheim A., Kallunki T. A novel specific role for I kappa B kinase complex-associated protein in cytosolic stress signaling. J. Biol. Chem. 2002;277:31918–31928. doi: 10.1074/jbc.M200719200. [DOI] [PubMed] [Google Scholar]
- 29.Rahl P.B., Chen C.Z., Collins R.N. Elp1p, the yeast homolog of the FD disease syndrome protein, negatively regulates exocytosis independently of transcriptional elongation. Mol. Cell. 2005;17:841–853. doi: 10.1016/j.molcel.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 30.Cheishvili D., Maayan C., Smith Y., Ast G., Razin A. IKAP/hELP1 deficiency in the cerebrum of familial dysautonomia patients results in down regulation of genes involved in oligodendrocyte differentiation and in myelination. Hum. Mol. Genet. 2007;16:2097–2104. doi: 10.1093/hmg/ddm157. [DOI] [PubMed] [Google Scholar]
- 31.Close P., Hawkes N., Cornez I., Creppe C., Lambert C.A., Rogister B., Siebenlist U., Merville M.P., Slaugenhaupt S.A., Bours V., et al. Transcription impairment and cell migration defects in elongator-depleted cells: implication for familial dysautonomia. Mol. Cell. 2006;22:521–531. doi: 10.1016/j.molcel.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 32.Chen Y.T., Hims M.M., Shetty R.S., Mull J., Liu L., Leyne M., Slaugenhaupt S.A. Loss of mouse Ikbkap, a subunit of elongator, leads to transcriptional deficits and embryonic lethality that can be rescued by human IKBKAP. Mol. Cell. Biol. 2009;29:736–744. doi: 10.1128/MCB.01313-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Slaugenhaupt S.A., Mull J., Leyne M., Cuajungco M.P., Gill S.P., Hims M.M., Quintero F., Axelrod F.B., Gusella J.F. Rescue of a human mRNA splicing defect by the plant cytokinin kinetin. Hum. Mol. Genet. 2004;13:429–436. doi: 10.1093/hmg/ddh046. [DOI] [PubMed] [Google Scholar]
- 34.Hims M.M., Ibrahim E.C., Leyne M., Mull J., Liu L., Lazaro C., Shetty R.S., Gill S., Gusella J.F., Reed R., et al. Therapeutic potential and mechanism of kinetin as a treatment for the human splicing disease familial dysautonomia. J. Mol. Med. 2007;85:149–161. doi: 10.1007/s00109-006-0137-2. [DOI] [PubMed] [Google Scholar]
- 35.Pros E., Fernandez-Rodriguez J., Benito L., Ravella A., Capella G., Blanco I., Serra E., Lazaro C. Modulation of aberrant NF1 pre-mRNA splicing by kinetin treatment. Eur. J. Hum. Genet. 2009;18:614–617. doi: 10.1038/ejhg.2009.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hims M.M., Shetty R.S., Pickel J., Mull J., Leyne M., Liu L., Gusella J.F., Slaugenhaupt S.A. A humanized IKBKAP transgenic mouse models a tissue-specific human splicing defect. Genomics. 2007;90:389–396. doi: 10.1016/j.ygeno.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gold-von Simson G., Goldberg J.D., Rolnitzky L.M., Mull J., Leyne M., Voustianiouk A., Slaugenhaupt S.A., Axelrod F.B. Kinetin in familial dysautonomia carriers: implications for a new therapeutic strategy targeting mRNA splicing. Pediatr. Res. 2009;65:341–346. doi: 10.1203/PDR.0b013e318194fd52. [DOI] [PubMed] [Google Scholar]
- 38.Byers P.H. Killing the messenger: new insights into nonsense-mediated mRNA decay. J. Clin. Invest. 2002;109:3–6. doi: 10.1172/JCI14841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anderson S.L., Qiu J., Rubin B.Y. EGCG corrects aberrant splicing of IKAP mRNA in cells from patients with familial dysautonomia. Biochem. Biophys. Res. Commun. 2003;310:627–633. doi: 10.1016/j.bbrc.2003.09.019. [DOI] [PubMed] [Google Scholar]
- 40.Anderson S.L., Qiu J., Rubin B.Y. Tocotrienols induce IKBKAP expression: a possible therapy for familial dysautonomia. Biochem. Biophys. Res. Commun. 2003;306:303–309. doi: 10.1016/s0006-291x(03)00971-9. [DOI] [PubMed] [Google Scholar]
- 41.Keren H., Donyo M., Zeevi D., Maayan C., Pupko T., Ast G. Phosphatidylserine increases IKBKAP levels in familial dysautonomia cells. PLoS One. 2010;5:e15884. doi: 10.1371/journal.pone.0015884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee G., Papapetrou E.P., Kim H., Chambers S.M., Tomishima M.J., Fasano C.A., Ganat Y.M., Menon J., Shimizu F., Viale A., et al. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature. 2009;461:402–406. doi: 10.1038/nature08320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gold-von Simson G., Leyne M., Mull J., Rolnitzky L.M., Goldberg J.D., Berlin D., Axelrod F.B., Slaugenhaupt S.A. IKBKAP mRNA in peripheral blood leukocytes: a molecular marker of gene expression and splicing in familial dysautonomia. Pediatr. Res. 2008;63:186–190. doi: 10.1203/PDR.0b013e31815ef74b. [DOI] [PubMed] [Google Scholar]
- 44.Boone N., Loriod B., Bergon A., Sbai O., Formisano-Treziny C., Gabert J., Khrestchatisky M., Nguyen C., Feron F., Axelrod F.B., et al. Olfactory stem cells, a new cellular model for studying molecular mechanisms underlying familial dysautonomia. PLoS One. 2010;5:e15590. doi: 10.1371/journal.pone.0015590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lesuisse C., Martin L.J. Immature and mature cortical neurons engage different apoptotic mechanisms involving caspase-3 and the mitogen-activated protein kinase pathway. J. Cereb. Blood Flow Metab. 2002;22:935–950. doi: 10.1097/00004647-200208000-00005. [DOI] [PubMed] [Google Scholar]
- 46.Reis S.A., Oostra B.A., Willemsen R. Isolation of mouse neuritic mRNAs. J. Mol. Histol. 2006;37:79–86. doi: 10.1007/s10735-006-9036-7. [DOI] [PubMed] [Google Scholar]