Abstract
Genome-wide and candidate-gene association studies of bladder cancer have identified 10 susceptibility loci thus far. We conducted a meta-analysis of two previously published genome-wide scans (4501 cases and 6076 controls of European background) and followed up the most significant association signals [17 single nucleotide polymorphisms (SNPs) in 10 genomic regions] in 1382 cases and 2201 controls from four studies. A combined analysis adjusted for study center, age, sex, and smoking status identified a novel susceptibility locus that mapped to a region of 18q12.3, marked by rs7238033 (P = 8.7 × 10–9; allelic odds ratio 1.20 with 95% CI: 1.13–1.28) and two highly correlated SNPs, rs10775480/rs10853535 (r2= 1.00; P = 8.9 × 10–9; allelic odds ratio 1.16 with 95% CI: 1.10–1.22). The signal localizes to the solute carrier family 14 member 1 gene, SLC14A1, a urea transporter that regulates cellular osmotic pressure. In the kidney, SLC14A1 regulates urine volume and concentration whereas in erythrocytes it determines the Kidd blood groups. Our findings suggest that genetic variation in SLC14A1 could provide new etiological insights into bladder carcinogenesis.
INTRODUCTION
The risk for developing urinary bladder cancer, the fourth most common incident cancer in men, is strongly related to cigarette smoking and occupational exposure to aromatic amines (1). Family history is associated with an ∼2-fold risk in cancer (2), and the genetic contribution of common and uncommon alleles to bladder cancer risk has been pursued in candidate-gene studies and more recently, genome-wide association studies (GWAS). Together, the two approaches have yielded 10 distinct loci. It is notable that three of the discovered loci contain carcinogen-metabolizing genes: the N-acetyltransferase 2 (NAT2) (3), a common gene deletion of glutathione S-transferase Mu 1 (GSTM1) (4–7) and the UDP-glucuronosyltransferase UGT1A gene locus on chromosome 2q37.1 discovered by a recent GWAS (7). Cigarette smoking modifies risk associations with NAT2 and GSTM1. While the NAT2 slow acetylation status alone appears to increase the risk of bladder cancer in cigarette smokers, the GSTM1 null genotype shows stronger associations with the risk in never smokers (7).
The remaining seven susceptibility loci for bladder cancer are scattered across the genome and are currently under active investigation to understand the biological basis of the contribution of common genetic variants to bladder cancer risk. These include common single nucleotide polymorphism (SNP) markers in regions that harbor plausible candidate genes for further study: 3q28 (8) (TP63), 4p16.3 (TMEM129, TACC3-FGFR3) (9), 8q24.21 (8), 8q24.3 (10) (PSCA), 5p15.33 (TERT-CLPTM1L) (7,11), 22q13.1 (7) and 19q12 (7) (CCNE1).
GWAS of bladder cancer have included primarily cases with urothelial (transitional cell) carcinomas, which represent ∼95% of malignant bladder tumors occurring in industrialized countries (1). Most (80%) urothelial carcinomas are low grade and non-invasive at presentation (TaG1/TaG2) but have high recurrence rate, thus requiring regular screening and interventions. These tumors are clinically and molecularly distinct from the more aggressive high-grade, non-muscle invasive (TaG3/T1G2/T1G3) and muscle invasive (T2-3) tumors. Distinct tumor types are postulated to develop through different pathways, suggesting heterogeneous etiologic factors, both genetic and environmental (12,13). Smoking and occupational exposure to aromatic amines has similar associations with tumors of different grade and stage (14); however, some loci discovered though GWAS have been reported to be differentially associated according to stage and grade. While loci on chromosomes 8q24.21, 4p16.3 (TMEM129, TACC3-FGFR3) and 5p15.33 (TERT-CLPTM1L) are more strongly associated with tumors of low grade/low risk of progression (7–9), loci on chromosomes 22q13.1 and 19q12 (CCNE1) are more strongly associated with the risk of high-grade/high-risk tumors (7).
Meta-analysis of existing GWAS data offers the opportunity to discover additional loci based on current projections for the number of independent regions harboring common variants associated with bladder cancer risk (15). In this study, we conducted a meta-analysis of two previously published GWAS, followed by a validation stage in further studies to discover additional susceptibility loci.
RESULTS
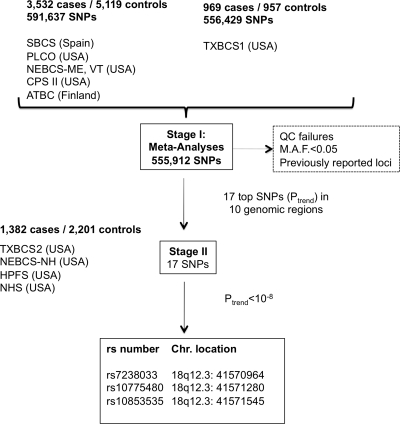
The study design for the meta-analysis (stage I) and follow up (stage II) is summarized in Figure 1, and study populations are described in Supplementary Material, Table S1.
Figure 1.
Study design of meta-analysis and follow up stages in GWAS of urinary bladder cancer. See Supplementary Material, Table S1 for details of study designs and sample sizes.
In stage I, a meta-analysis of genome-wide scan data from two previously published GWAS (7,10) was conducted in 4501 cases and 6076 controls of European background using 555 912 SNPs (common to all subjects scanned on the HumanHap 500, 610-Quad and 1M Illumina Infinium arrays). The quantile–quantile (Q–Q) plot showed minimal evidence for inflation of the test statistics when compared with the expected distribution (corrected λ1000 subjects = 1.002), which suggests that there is no substantial hidden population substructure or differential genotype calling between cases and controls (16) (Supplementary Material, Fig. S1). A Manhattan plot displays the results of the combined GWAS meta-analysis in stage I (Supplementary Material, Fig. S2). Seven distinct genomic loci were notable for P-value <1.5 × 10−5 in meta-analyses of allelic odds ratio estimates derived from logistic regression analysis adjusted for study center, age, sex and smoking status. We repeated the meta-analyses among current smokers only and identified three additional loci with P-value < 2 × 10−6 that were advanced in the replication effort, resulting on a total of 10 genomic regions being evaluated in stage II.
Based on results from stage I meta-analyses, SNPs in linkage disequilibrium with the 10 known loci or those with minor allele frequency (MAF) <5% in the controls were excluded from further analysis. Seventeen SNPs were selected based on rank P-value for follow up in stage II, which included highly correlated SNPs. These included the most significant SNPs in the most promising seven regions, namely 7p12.1 with two SNPs, 18q12.3 with three SNPs, 19q13.33 with three SNPs, 6q23.2 with three SNPs, as well as one SNP in each of the following regions: 4p16.1, 10q25.3 and 20p13. We also selected three additional variants that were highly significant in current smokers (6p21.32, 6p24.3, 9q12.3). The 17 SNPs were genotyped in each of the following regions: 1382 cases and 2201 controls from two case–control studies and two prospective cohorts in the USA (Fig. 1).
We used fixed effects meta-analyses based on estimates of allelic odds ratios for each study, adjusted by study center, age, sex and smoking status (current, former or never), to obtain combined (stages I and II) and stage-specific estimates. Analyses of combined estimates identified one locus on chromosome 18q12.3 reaching genome-wide level of significance (P < 5 × 10−8, Table 1, Supplementary Material, Table S2). The strongest signal in the 18q12.3 locus (P = 8.9 × 10−9) was for rs10775480 tagged by rs10853535 (pairwise r2= 1.0 in HapMap CEU parents). We used data on rs10775480 for all studies, except for Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial (PLCO) and Spanish Bladder Cancer Study (SBCS) in which rs10853535 was used because rs10775480 was not successfully called in the PLCO and was not included on the HumanHap 1 M array used in SBCS. Three other SNPs in the region showed associations with risk: rs7238033 had a similar signal as rs10775480/rs10853535, but it was genotyped in 8 of 10 studies included in the primary GWAS, whereas two surrogate SNPs rs11082469/rs11877720 (pairwise r2= 1.0) had a weaker signal. These associations were not independent of rs10775480/rs10853535, therefore further analyses of the 18q12.3 locus by tumor characteristics, gene–gene and gene–smoking interactions (shown below) focused on rs10775480/rs10853535. In an analysis of tumor subtypes, data suggested a possible association of the 18q12.3 locus with tumors of higher grade but the finding is not statistically significant (Pinteraction= 0.071) (Supplementary Material, Table S3). We found no evidence of modification of the 18q12.3 locus risk association by smoking status (Supplementary Material, Table S4).
Table 1.
Association of SNPs in 18q12.3 (SLC14A1) region with the risk for urinary bladder cancer
| SNP | Stage | n | Cases | Controls | MAF | Allelic OR | 95% CI |
P-value | I2 | P-het | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs7238033[C/T]* | Combined | 8 | 3992 | 4977 | 0.43 | 1.20 | 1.13 | 1.28 | 8.72E − 09 | 0.0 | 0.829 |
| Stage I | 4 | 2685 | 3150 | 0.43 | 1.19 | 1.11 | 1.29 | 6.55E − 06 | 0.0 | 0.603 | |
| Stage II | 4 | 1307 | 1827 | 0.44 | 1.22 | 1.09 | 1.36 | 3.31E − 04 | 0.0 | 0.641 | |
| rs10775480[C/T] | Combined | 10 | 5801 | 7894 | 0.43 | 1.16 | 1.10 | 1.22 | 8.95E − 09 | 0.0 | 0.535 |
| rs10853535[T/C]** | Stage I | 6 | 4499 | 6068 | 0.43 | 1.15 | 1.08 | 1.22 | 4.41E − 06 | 0.0 | 0.453 |
| Stage II | 4 | 1302 | 1826 | 0.44 | 1.22 | 1.09 | 1.36 | 3.16E − 04 | 0.0 | 0.519 | |
| rs11082469[A/G] | Combined | 10 | 5792 | 7821 | 0.49 | 0.89 | 0.85 | 0.94 | 1.84E − 05 | 33.5 | 0.150 |
| rs11877720[A/G]** | Stage I | 6 | 4487 | 5987 | 0.49 | 0.91 | 0.86 | 0.96 | 1.11E − 03 | 45.0 | 0.105 |
| Stage II | 4 | 1305 | 1834 | 0.49 | 0.85 | 0.76 | 0.95 | 3.00E − 03 | 0.0 | 0.399 | |
Results from meta-analyses of allelic odds ratios adjusted by study center, age, gender and smoking status, obtained from up to 10 case–control and cohort studies (Stage I: SBCS, PLCO, NEBCS-ME,VT, CPSII, ATBC, TXBCS1; Stage II: NBCS-NH, HPFS, NHS, TXBCS2).
*Data on rs7238033 was not available in PLCO and SBCS. The effect estimates for rs7238033 and rs10775480 in the subset of three studies with data on both SNPs were very similar (P = 3.4 × 10−4 for rs7238033 based on 2193 cases and 1716 controls and P = 2.6 × 10−4 for rs10775480 based on 2184 cases and 1705 controls). Inclusion of both SNP simultaneously in the same model resulted in loss of significance for both SNPs.
**These SNPs were used as surrogates in PLCO and SBCS because of no availability of data on rs10775480 and rs11082469 from these studies. The pairwise r2 values based on HapMap data (release 28) for the surrogate pairs are r2 = 1.00. The pairwise r2 values between rs10775480/rs10853535 and rs11082469/rs11877720 are r2 = 0.66.
We explored whether the allelic relative risk for the new locus on 18q12.3 varied by genotypes for each of the 10 previously identified loci (Supplementary Material, Table S5). Genotype-specific estimates and P-values for multiplicative gene × gene interactions were obtained by including main effects and an interaction term between the 18q12.3 locus and each locus in a series of multivariate logistic regression models adjusted by study center, sex, age and smoking status. Our preliminary analyses suggest that the association at 18q12.3 is limited to subjects with the AA genotype (87%) of the UGTA1 locus (rs11892031) encoding a UGTA1 family of proteins that facilitate solubility and removal of carcinogens though urine (Pinteraction = 0.018; Table 2); however, given the limited power to detect gene × gene interactions and the possibility that this could represent a false positive finding, further work is needed to confirm this interaction. We did not observe notable departures from multiplicative joint effects for other susceptibility regions.
Table 2.
Association of rs10775480[T]/rs10853535[C]* in 18q12.3 (SLC14A1) with the risk for urinary bladder cancer, stratified by rs11892031 (UGT1A) and rs1495741 (NAT2) genotypes
| Genotypes | Cases | Controls | OR | 95% CI |
P-value | |
|---|---|---|---|---|---|---|
| rs11892031 (UGTA1) | ||||||
| AA | 5004 | 6607 | 1.19 | 1.12 | 1.25 | 6.7E − 10 |
| AC | 753 | 1200 | 1.02 | 0.89 | 1.16 | 0.793 |
| CC | 22 | 57 | 0.82 | 0.42 | 1.60 | 0.553 |
| AC/CC | 775 | 1257 | 1.01 | 0.88 | 1.15 | 0.938 |
| Total | 5779 | 7864 | Pint**= | 0.018 | ||
| rs1495741 (NAT2) | ||||||
| AA | 3649 | 4652 | 1.13 | 1.06 | 1.21 | 1.8E − 04 |
| AG | 1863 | 2749 | 1.20 | 1.10 | 1.31 | 2.7E − 05 |
| GG | 241 | 374 | 1.33 | 1.04 | 1.70 | 2.5E − 02 |
| GG/AG | 2104 | 3123 | 1.22 | 1.12 | 1.32 | 2.6E − 06 |
| Total | 5753 | 7775 | Pint**= | 0.116 | ||
Results from logistic regression models to estimate allelic odds ratios adjusted by study center, age, gender and smoking status, including data from 10 case–control and cohort studies (Stage I: SBCS, PLCO, NEBCS-ME,VT, CPSII, ATBC, TXBCS1; Stage II: NBCS-NH, HPFS, NHS, TXBCS2).
*Surrogate SNPs in PLCO and SBCS: rs10775480 and rs10853535; r2 = 1.08.
**P-value from test for interaction with rs10775480[T]/rs10853535[C].
DISCUSSION
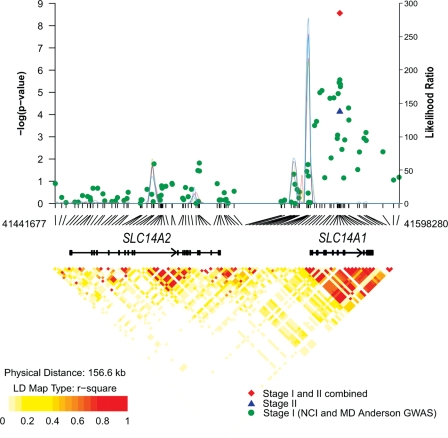
Our combined analysis of 5883 individuals with bladder cancer and 8277 controls has identified a new genomic region on 18q12.3 associated with urinary bladder cancer risk. The 18q12.3 locus contains two related genes from the solute carrier family 14 transporters, member 1 (SLC14A1) and member 2 (SLC14A2) separated by ∼50 kb. SLC14A2 is the main renal tubular urea transporter responsible for renal urinary concentration. However, our GWAS meta-analysis did not reveal a notable association signal in SLC14A2 and we confirmed the mapping of a recombination breakpoint between SLC14A1 and SLC14A2 (Fig. 2), thus pointing towards the former as a plausible candidate gene for study.
Figure 2.
Association results, recombination and linkage disequilibrium plots for chromosome 18q12.3. Association results of stage I (green circles), stage II (blue triangle) and combined data from stages I and II (red diamond) for log-additive models are shown in the top panel with −log10 P values (left y-axis). Overlaid on the top panel is the LR statistics (right y-axis) to estimate putative recombination hotspot across the region based on five sets of 100 randomly selected control samples (connected lines in various colors). Pairwise r2 values based on control populations are displayed at the bottom for all SNPs included in the GWAS analysis. Genomic coordinates are based on NCBI Human Genome Build 36.3.
SLC14A1 functions as a urea transporter in the kidney and erythrocytes, and is a determinant for the Kidd blood group and regulates urinary concentration, specifically the capacity to filter urea (17). A non-synonymous variant rs1058396 at amino acid 280 distinguishes the two Kidd blood groups Jk(A) and Jk(B); Asp-280 encodes Jk(A) and Asn-280 encodes Jk(B). Notably, rs1058396 is in strong linkage disequilibrium with the signals discovered in our meta-analysis (r2 = 0.64, 0.71 and 0.93 for rs10775480, rs7238033 and rs11082469, respectively). It has been suggested that a second non-synonymous variant, rs2298720 (Glu44Lys) defines a weaker version of the JkA group, Jk(a)W (18). The three variants analyzed in our meta-analysis and follow-up samples adequately tag the Kidd blood groups as shown in haplotype analyses presented in Supplementary Material, Figure S3: the risk allele corresponds to the Jk(A) group, whereas the protective allele corresponds to the Jk(B) and Jk(A)W groups.
In <0.1% of the population, erythrocytes in individuals lacking both Jk(A) and Jk(B) antigens are resistant to lysis by 2 m urea (19,20). These individuals are unable to adequately concentrate urine, producing larger volumes of diluted urine (21). A similar phenotype observed in a knock-out mouse model of SLC14A1, demonstrated ∼50% reduction in urea concentrating capacity and 1.5-fold greater daily output of urine of lower osmolarity (22). There is no clear evidence of different renal function in individuals with common Jk(A), Jk(B) and Jk(A)W blood groups, but this should be explored further in bladder cancer patients and controls. Variations in urine volumes and concentration could modify exposure of bladder epithelium to carcinogens in the urine. In this regard, our study raises the possibility that common genetic variation could alter the function or expression of the Kidd blood group antigen and contribute to the risk for urinary bladder cancer. Fine-mapping studies of this region should provide an optimal set of variants for functional studies to pursue the role of the Kidd blood group as a urine transporter in the carcinogenesis of urinary bladder cancer. It is also possible that the common variants act through a different mechanism yet to be determined and not related to the Kidd blood group antigen.
The addition of the 18q12.3 locus to the catalog of conclusively associated regions with urinary bladder represents a small, but important step in defining a comprehensive set of variants that could be used in generating a risk model in combination with other risk factors for bladder cancer (i.e. smoking, occupational and environmental exposures and family history). At the same time, the discovery of novel genomic regions provides the foundation for biological insights into the genesis of urinary bladder cancer that ultimately could lead to improved preventive, diagnostic and/or therapeutic approaches to this challenging cancer.
MATERIAL AND METHODS
Study populations
Participants were drawn from 10 prospective cohort and case–control studies (Supplementary Material, Table S1).
Stage I meta-analyses
NCI GWAS
Case and controls in the National Cancer Institute (NCI) GWAS were derived from two case–control studies [SBCS and the Maine and Vermont components of the New England Bladder Cancer Study (NEBCS-ME, VT)], and three prospective cohorts [PLCO, The American Cancer Society Cancer Prevention Study II Nutrition Cohort (CPS II) and Alpha-Tocopherol, Beta-Carotene Cancer Prevention Study (ATBC)]. Cases were defined as histologically confirmed primary carcinoma of the urinary bladder including carcinoma in situ (ICD-0-2 topography codes C67.0–C67.9 or ICD9 codes 188.1–188.9). Analyses were restricted to individuals of European background (<15% non-European admixture) as determined by population structure analyses with STRUCTURE (23). There were no age, gender or cancer-stage restrictions. Each participating study obtained informed consent from study participants and approval from its Institutional Review Board (IRB) for this study. Participating studies obtained institutional certification permitting data sharing in accordance with the NIH Policy for Sharing of Data Obtained in NIH Supported or Conducted GWAS.
MD Anderson GWAS
Cases and controls for the primary scan were derived from the Texas Bladder Cancer Study 1 (TXBCS 1), a hospital-based bladder cancer case–control study (10). Cases were defined as histologically confirmed and previously untreated incident bladder cancer cases recruited from the University of Texas MD Anderson Cancer Center and Baylor College of Medicine between 1999 and 2007. There were no age, gender, cancer-stage restrictions. All included subjects were self-identified as European background. Informed consent was obtained from all study participants before the collection of epidemiological data and blood samples by trained M.D. Anderson staff interviewers. The study was approved by the IRB of MD Anderson Cancer Center, Baylor College of Medicine and the Kelsey-Seybold Clinic.
Stage II follow up
Cases and control in stage II were derived from two case–control studies [Texas Bladder Cancer Study 2 (TXBCS 2), the New Hampshire component of the New England Bladder Cancer Study (NEBCS-NH)], and two prospective cohorts [Nurse's Health Study (NHS) and Health Professionals Follow up Study (HPFS)]. Each participating study obtained informed consent from study participants and approval from its IRB for this study. There were no age, gender, ethnicity or cancer-stage restrictions for TXCBS1, and analyses were restricted to individuals of European background as determined by self-report for NEBCS-NH, NHS and HPFS.
Genotyping and quality control
Genotyping, quality control and assessment of population structure for studies in stage I has been previously described [see Rothman et al. for NCI GWAS (7) and Wu et al. for MD Anderson GWAS (10)]. The meta-analysis was performed on the common set of SNPs called in both the MD Anderson and NCI GWAS sets. All samples had been scanned with Illumina Infinium Arrays.
We estimate the inflation of the test statistic, λ, adjusted to a sample size of 1000 cases/1000 controls as per the method of de Bakker et al. (24): λ(corrected) = 1 + (λ− 1) × [ncase−1 + ncont−1]/[2 × 10−3]. The corrected estimated λ1000 is 1.002 while the uncorrected λ is 1.014 (Supplementary Material, Fig. S1).
The final participant count for stage I analysis was 4501 cases and 6076 controls. The number of SNPs available for association analysis in all studies after quality control metrics applied was 556 429 for MD Anderson and 591 637 for NCI. Of these SNPs, 555 912 overlapped exactly between the Infinium HumanHap 1 M chip and 610Quad/550k data and were used for this meta-analysis.
TaqMan custom genotyping assays (ABI, Foster City, CA, USA) were designed and optimized for the 17 SNPs genotyped in stage II studies. In an analysis of 1000 samples, the comparison of the Illumina calls with the TaqMan assays showed a concordance rate of 100%. The Illumina Infinium cluster plots for the two surrogate SNPs in the novel region in 18q12.3 (rs10775480 and rs10853535) are shown in Supplementary Material, Figure S4.
Statistical analysis
Meta-analyses in stage I were based on allelic odds ratio estimates derived from logistic regression models adjusted for study center, age (in 5-year categories), sex and smoking status (current, former or never). Each SNP genotype was coded as a count of minor alleles, with the exception of X-linked SNPs among men that were coded as 2 if the participant carried the minor allele and 0 if he carried the major allele (25). A score test with one degree of freedom was performed on all genetic parameters in each model to determine statistical significance. For the inclusion of stage II data, we conducted meta-analyses based on estimates of allelic odds ratio for each study, adjusted by study center, age, sex and smoking status (never, former, current).
Polytomous logistic regression was used to obtain estimates of effect for different tumor subtypes. Case-only analyses with tumor type as an outcome were used to test for differences in effect size across subtypes. Polytomous logistic regression models for tumor grade constrained the effect size to increase linearly across levels. Gene–gene interactions were assessed using logistic regression models adjusted by study center, age, sex and smoking status and including interaction terms. Genotype–smoking interactions were assessed using logistic regression for grouped data adjusted by study center and sex, and including interaction terms.
A haplotype-based association analysis was performed across the region of interest (shown in the Supplementary Material, Figure S3) using the PLINK (version 1.07) (26). Haplotype-specific odds ratio and P-values were estimated for each haplotype versus all others, adjusted for the effects of age, gender, study center and when appropriate, smoking status.
Data analysis and management was performed with GLU (Genotyping Library and Utilities version 1.0), a suite of tools available as an open-source application for management, storage and analysis of GWAS data, and STATA S.E. v.11.1 (College Station, TX, USA).
Estimate of recombination hotspots
SequenceLDhot (27) that uses an approximate marginal likelihood method (28) was used to compute likelihood ratio (LR) statistics for a set of putative hotspots across the region of interest. We sequentially analyzed subsets of 100 controls of European background (by pooling five controls from each study). We used Phasev2.1 to infer the haplotypes as well as background recombination rates. The analysis was repeated with five non-overlapping sets of 100 controls.
URLs
CGEMS portal: http://cgems.cancer.gov/.
CGF: http://cgf.nci.nih.gov/.
GLU: http://code.google.com/p/glu-genetics/.
EIGENSTRAT: http://genepath.med.harvard.edu/∼reich/Software.htm.
SNP500Cancer: http://snp500cancer.nci.nih.gov/.
STRUCTURE: http://pritch.bsd.uchicago.edu/structure.html.
SUPPLEMENTARY MATERIAL
FUNDING
The NCI bladder cancer GWAS was supported by the intramural research program of the National Institutes of Health, National Cancer Institute. This project has been funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
SBCS (D.T.S., N.R., S.J.C., M.G.C.)—Intramural Research Program of the National Institutes of Health, National Cancer Institute, Division of Cancer Epidemiology and Genetics and intramural contract number NCI N02-CP-11015. FIS/Spain 98/1274, FIS/Spain 00/0745, PI061614 and G03/174, Fundació Marató TV3, Red Temática Investigación Cooperativa en Cáncer (RTICC), Consolíder ONCOBIO, EU-FP7-201663; and RO1-CA089715 and CA34627.
NEBCS (D.T.S.)—Intramural research program of the National Institutes of Health, National Cancer Institute, Division of Cancer Epidemiology and Genetics and intramural contract number NCI N02-CP-01037.
PLCO (M.P.P)—the NIH Genes, Environment and Health Initiative (GEI) partly funded DNA extraction and statistical analyses (HG-06-033-NCI-01 and RO1HL091172-01), genotyping at the Johns Hopkins University Center for Inherited Disease Research (U01HG004438 and NIH HHSN268200782096C) and study coordination at the GENEVA (N.C.)—The NIH Genes, Environment and Health Initiative (GEI) partly funded DNA extraction and statistical analyses (HG-06-033-NCI-01 and RO1HL091172-01), genotyping at the Johns Hopkins University Center for Inherited Disease Research (U01HG004438 and NIH HHSN268200782096C) and study coordination at the GENEVA Coordination Center (U01 HG004446) for EAGLE and part of PLCO studies. Genotyping for the remaining part of PLCO and all ATBC and CPS-II samples were supported by the Intramural Research Program of the National Institutes of Health, NCI, Division of Cancer Epidemiology and Genetics. The PLCO is supported by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics and supported by contracts from the Division of Cancer Prevention, National Cancer Institute, National Institutes of Health.
ATBC (D.A.)—this research was supported in part by the Intramural Research Program of the NIH and the National Cancer Institute. Additionally, this research was supported by U.S. Public Health Service contracts N01-CN-45165, N01-RC-45035 and N01-RC-37004 from the National Cancer Institute, Department of Health and Human Services.
TXBCS1 & TXBCS2—U01 CA 127615 (X.W.), R01 CA 74880 (X.W.) and P50 CA 91846 (X.W., C.P.D.), UT MD Anderson Cancer Centre Research Trust (X.W.), Centre for Translational and Public Health Genomics at MD Anderson Cancer Centre.
NHS & HPFS (I.D.V.)—CA055075 and CA087969.
AUTHOR CONTRIBUTIONS
M.G.-C., Y.Y., N.R., J.F.F., D.T.S., S.J.C. and X.W. organized and designed the study. S.J.C., K.B.J., A.H., Z.W., Y.-P.F., L.P.-O., L.B., X.W. conducted and supervised genotyping of samples. M.G.-C., Y.Y., N.R., N.C., K.B.J., Y.-P.F., L.P.-O., J.G., J.L., D.T.S., S.J.C. and X.W. contributed to the design and execution of statistical analysis. M.G.-C., Y.Y., N.R., J.D.F., D.T.S., S.J.C. and X.W. wrote the first draft of the manuscript. M.G.-C., Y.Y., N.R., J.D.F., N.M., C.P.D., N.C., L.P.-O., Z.W., J.L., F.X.R., K.B.J., D.B., M.T., I.DeV., D.A., M.P.P., M.K., A.M.K., S.P.L., H.B.G., J.G., X.P., A.H., Y.-P.F., L.B., M.Y., W.T., A.T., C.S., A.C., R.G.-C., J.Ll., A.J., M.S., M.R.K., A.S., G.A., R.G., A.B., E.J.J., W.R.D., S.M.G., S.J.W., J.V., D.J.H., N.C., M.T.L., J.F.F., D.T.S., S.J.C. and X.W. conducted the epidemiologic studies and contributed samples to the bladder cancer GWAS and/or replication.
All authors contributed to the writing of the manuscript.
Supplementary Material
ACKNOWLEDGEMENTS
Leslie Carroll (Information Management Services, Silver Spring, MD, USA); Gemma Castaño-Vinyals (Institut Municipal d'Investigació Mèdica, Barcelona, Spain); Fernando Fernández (Institut Municipal d'Investigació Mèdica, Barcelona, Spain); Paul Hurwitz (Westat, Inc., Rockville, MD, USA); Charles Lawrence (Westat, Inc., Rockville, MD, USA); Marta Lopez-Brea (Marqués de Valdecilla University Hospital, Santander, Cantabria, Spain); Anna McIntosh (Westat, Inc., Rockville, MD, USA); Angeles Panadero (Hospital Ciudad de Coria, Coria (Cáceres), Spain); Fernando Rivera (Marqués de Valdecilla University Hospital, Santander, Cantabria, Spain); Robert Saal (Westat, Rockville, MD, USA); Maria Sala (Institut Municipal d'Investigació Mèdica, Barcelona, Spain); Kirk Snyder (Information Management Services, Inc., Silver Spring, MD, USA); Anne Taylor (Information Management Services, Inc., Silver Spring, MD, USA); Montserrat Torà (Institut Municipal d'Investigació Mèdica, Barcelona, Spain); Jane Wang (Information Management Services, Silver Spring, MD, USA).
REFERENCES
- 1.Silverman D., Devesa S.S., Moore L.E., Rothman N. Bladder Cancer. In: Schottenfeld D., Fraumeni J.F. Jr, editors. Cancer Epidemiology and Prevention. New York, NY: Oxford University Press; 2006. pp. 1101–1127. [Google Scholar]
- 2.Murta-Nascimento C., Silverman D.T., Kogevinas M., Garcia-Closas M., Rothman N., Tardon A., Garcia-Closas R., Serra C., Carrato A., Villanueva C., et al. Risk of bladder cancer associated with family history of cancer: do low-penetrance polymorphisms account for the increase in risk? Cancer Epidemiol. Biomarkers Prev. 2007;16:1595–1600. doi: 10.1158/1055-9965.EPI-06-0743. [DOI] [PubMed] [Google Scholar]
- 3.Lower G.M., Jr, Nilsson T., Nelson C.E., Wolf H., Gamsky T.E., Bryan G.T. N-acetyltransferase phenotype and risk in urinary bladder cancer: approaches in molecular epidemiology. Preliminary results in Sweden and Denmark. Environ. Health Perspect. 1979;29:71–79. doi: 10.1289/ehp.792971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell D.A., Taylor J.A., Paulson D.F., Robertson C.N., Mohler J.L., Lucier G.W. Genetic risk and carcinogen exposure: a common inherited defect of the carcinogen-metabolism gene glutathione S-transferase M1 (GSTM1) that increases susceptibility to bladder cancer. J. Natl Cancer Inst. 1993;85:1159–1164. doi: 10.1093/jnci/85.14.1159. [DOI] [PubMed] [Google Scholar]
- 5.Garcia-Closas M., Malats N., Silverman D., Dosemeci M., Kogevinas M., Hein D.W., Tardon A., Serra C., Carrato A., Garcia-Closas R., et al. NAT2 slow acetylation, GSTM1 null genotype, and risk of bladder cancer: results from the Spanish Bladder Cancer Study and meta-analyses. Lancet. 2005;366:649–659. doi: 10.1016/S0140-6736(05)67137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rothman N., Garcia-Closas M., Hein D.W. Commentary: reflections on G. M. Lower and colleagues’ 1979 study associating slow acetylator phenotype with urinary bladder cancer: meta-analysis, historical refinements of the hypothesis, and lessons learned. Int. J. Epidemiol. 2007;36:23–28. doi: 10.1093/ije/dym026. [DOI] [PubMed] [Google Scholar]
- 7.Rothman N., Garcia-Closas M., Chatterjee N., Malats N., Wu X., Figueroa J.D., Real F.X., Van Den Berg D., Matullo G., Baris D., et al. A multi-stage genome-wide association study of bladder cancer identifies multiple susceptibility loci. Nat. Genet. 2010;42:978–984. doi: 10.1038/ng.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kiemeney L.A., Thorlacius S., Sulem P., Geller F., Aben K.K., Stacey S.N., Gudmundsson J., Jakobsdottir M., Bergthorsson J.T., Sigurdsson A., et al. Sequence variant on 8q24 confers susceptibility to urinary bladder cancer. Nat. Genet. 2008;40:1307–1312. doi: 10.1038/ng.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiemeney L.A., Sulem P., Besenbacher S., Vermeulen S.H., Sigurdsson A., Thorleifsson G., Gudbjartsson D.F., Stacey S.N., Gudmundsson J., Zanon C., et al. A sequence variant at 4p16.3 confers susceptibility to urinary bladder cancer. Nat. Genet. 2010;42:415–419. doi: 10.1038/ng.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu X., Ye Y., Kiemeney L.A., Sulem P., Rafnar T., Matullo G., Seminara D., Yoshida T., Saeki N., Andrew A.S., et al. Genetic variation in the prostate stem cell antigen gene PSCA confers susceptibility to urinary bladder cancer. Nat. Genet. 2009;41:991–995. doi: 10.1038/ng.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rafnar T., Sulem P., Stacey S.N., Geller F., Gudmundsson J., Sigurdsson A., Jakobsdottir M., Helgadottir H., Thorlacius S., Aben K.K., et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat. Genet. 2009;41:221–227. doi: 10.1038/ng.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goebell P.J., Knowles M.A. Bladder cancer or bladder cancers? Genetically distinct malignant conditions of the urothelium. Urol. Oncol. 2010;28:409–428. doi: 10.1016/j.urolonc.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 13.Guey L.T., Garcia-Closas M., Murta-Nascimento C., Lloreta J., Palencia L., Kogevinas M., Rothman N., Vellalta G., Calle M.L., Marenne G., et al. Genetic susceptibility to distinct bladder cancer subphenotypes. Eur. Urol. 2010;57:283–292. doi: 10.1016/j.eururo.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sturgeon S.R., Hartge P., Silverman D.T., Kantor A.F., Linehan W.M., Lynch C., Hoover R.N. Associations between bladder cancer risk factors and tumor stage and grade at diagnosis. Epidemiology. 1994;5:218–225. doi: 10.1097/00001648-199403000-00012. [DOI] [PubMed] [Google Scholar]
- 15.Park J.H., Wacholder S., Gail M.H., Peters U., Jacobs K.B., Chanock S.J., Chatterjee N. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat. Genet. 2010;42:570–575. doi: 10.1038/ng.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freedman M.L., Reich D., Penney K.L., McDonald G.J., Mignault A.A., Patterson N., Gabriel S.B., Topol E.J., Smoller J.W., Pato C.N., et al. Assessing the impact of population stratification on genetic association studies. Nat. Genet. 2004;36:388–393. doi: 10.1038/ng1333. [DOI] [PubMed] [Google Scholar]
- 17.Shayakul C., Hediger M.A. The SLC14 gene family of urea transporters. Pflugers Arch. 2004;447:603–609. doi: 10.1007/s00424-003-1124-x. [DOI] [PubMed] [Google Scholar]
- 18.Wester E.S., Storry J.R., Olsson M.L. Characterization of Jk(a + (weak)): a new blood group phenotype associated with an altered JK*01 allele. Transfusion. 2011;51:380–392. doi: 10.1111/j.1537-2995.2010.02795.x. [DOI] [PubMed] [Google Scholar]
- 19.Heaton D.C., McLoughlin K. Jk(a-b-) red blood cells resist urea lysis. Transfusion. 1982;22:70–71. doi: 10.1046/j.1537-2995.1982.22182154224.x. [DOI] [PubMed] [Google Scholar]
- 20.Frohlich O., Macey R.I., Edwards-Moulds J., Gargus J.J., Gunn R.B. Urea transport deficiency in Jk(a-b-) erythrocytes. Am. J. Physiol. 1991;260:C778–C783. doi: 10.1152/ajpcell.1991.260.4.C778. [DOI] [PubMed] [Google Scholar]
- 21.Sands J.M., Gargus J.J., Frohlich O., Gunn R.B., Kokko J.P. Urinary concentrating ability in patients with Jk(a-b-) blood type who lack carrier-mediated urea transport. J. Am. Soc. Nephrol. 1992;2:1689–1696. doi: 10.1681/ASN.V2121689. [DOI] [PubMed] [Google Scholar]
- 22.Yang B., Bankir L., Gillespie A., Epstein C.J., Verkman A.S. Urea-selective concentrating defect in transgenic mice lacking urea transporter UT-B. J. Biol. Chem. 2002;277:10633–10637. doi: 10.1074/jbc.M200207200. [DOI] [PubMed] [Google Scholar]
- 23.Pritchard J.K., Stephens M., Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Bakker P.I., Ferreira M.A., Jia X., Neale B.M., Raychaudhuri S., Voight B.F. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum. Mol. Genet. 2008;17:R122–R128. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clayton D. Testing for association on the X chromosome. Biostatistics. 2008;9:593–600. doi: 10.1093/biostatistics/kxn007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fearnhead P. SequenceLDhot: detecting recombination hotspots. Bioinformatics. 2006;22:3061–3066. doi: 10.1093/bioinformatics/btl540. [DOI] [PubMed] [Google Scholar]
- 28.Fearnhead P., Harding R.M., Schneider J.A., Myers S., Donnelly P. Application of coalescent methods to reveal fine-scale rate variation and recombination hotspots. Genetics. 2004;167:2067–2081. doi: 10.1534/genetics.103.021584. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.