Abstract
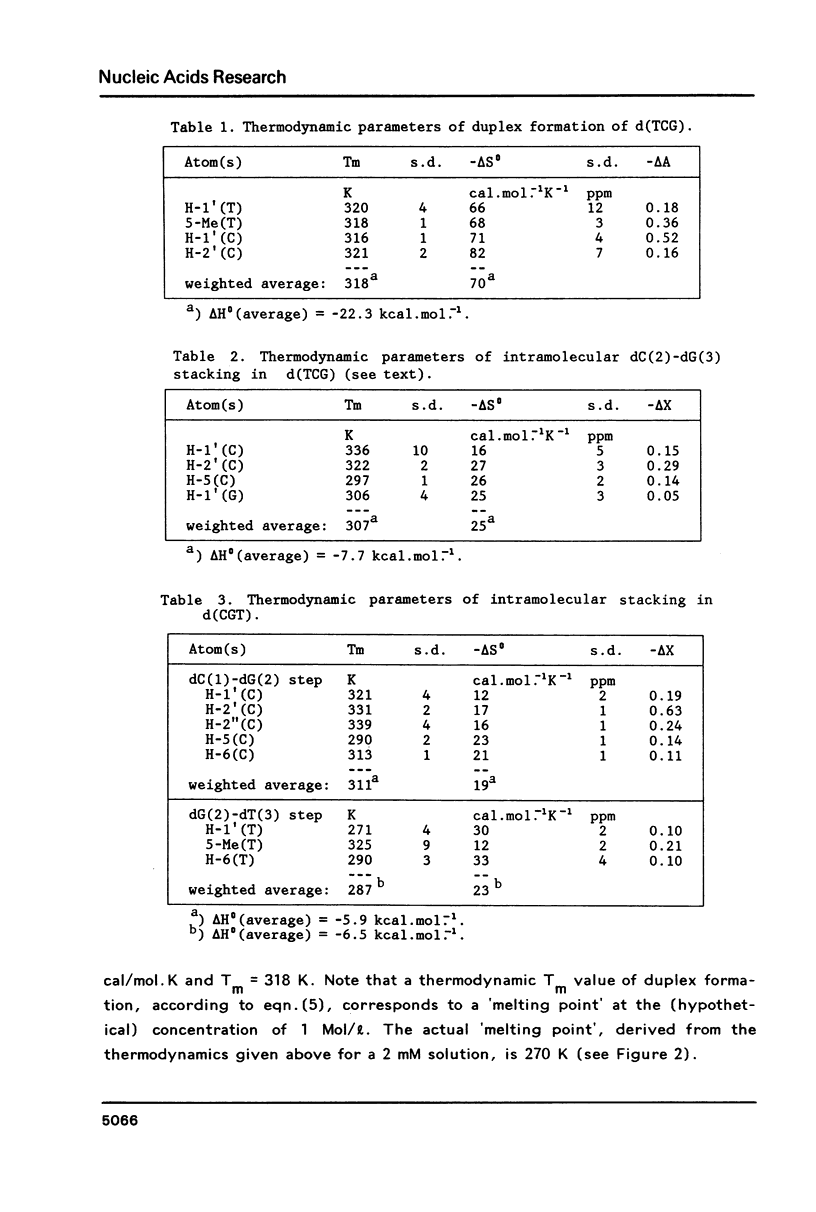
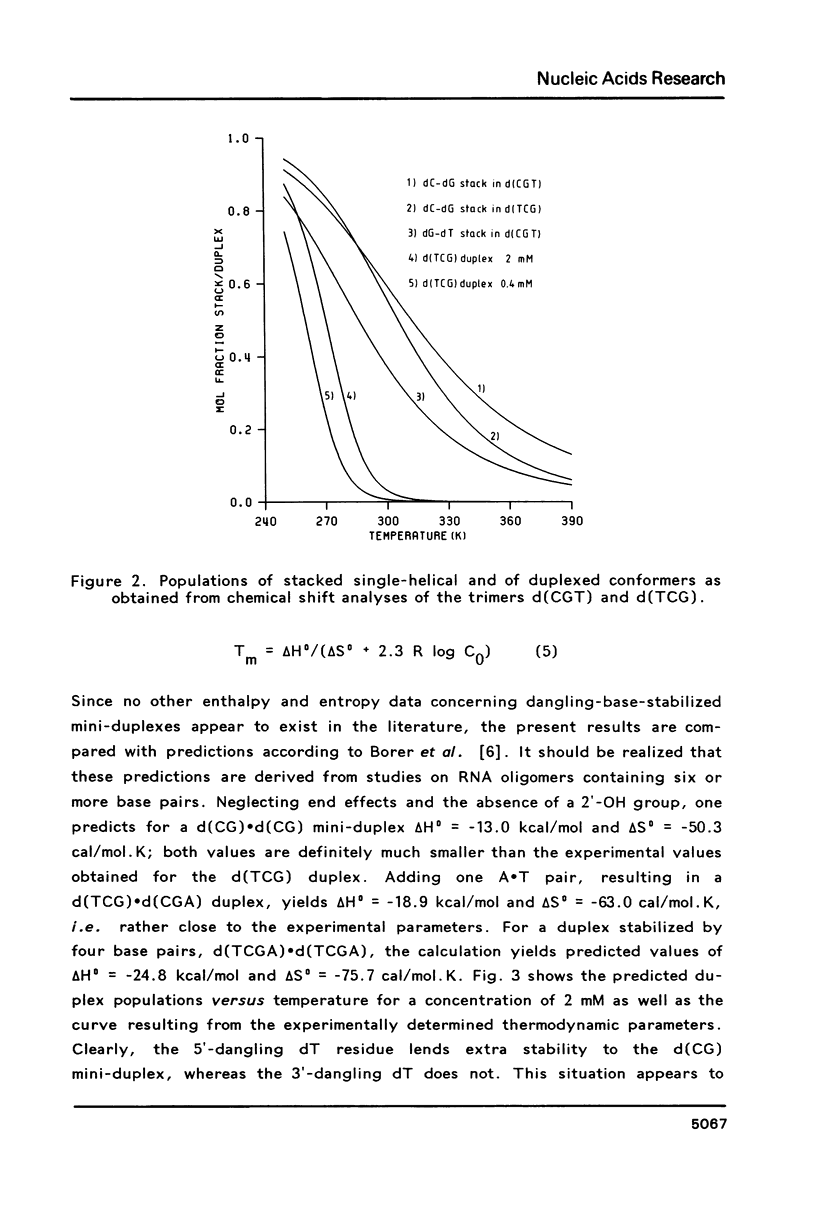
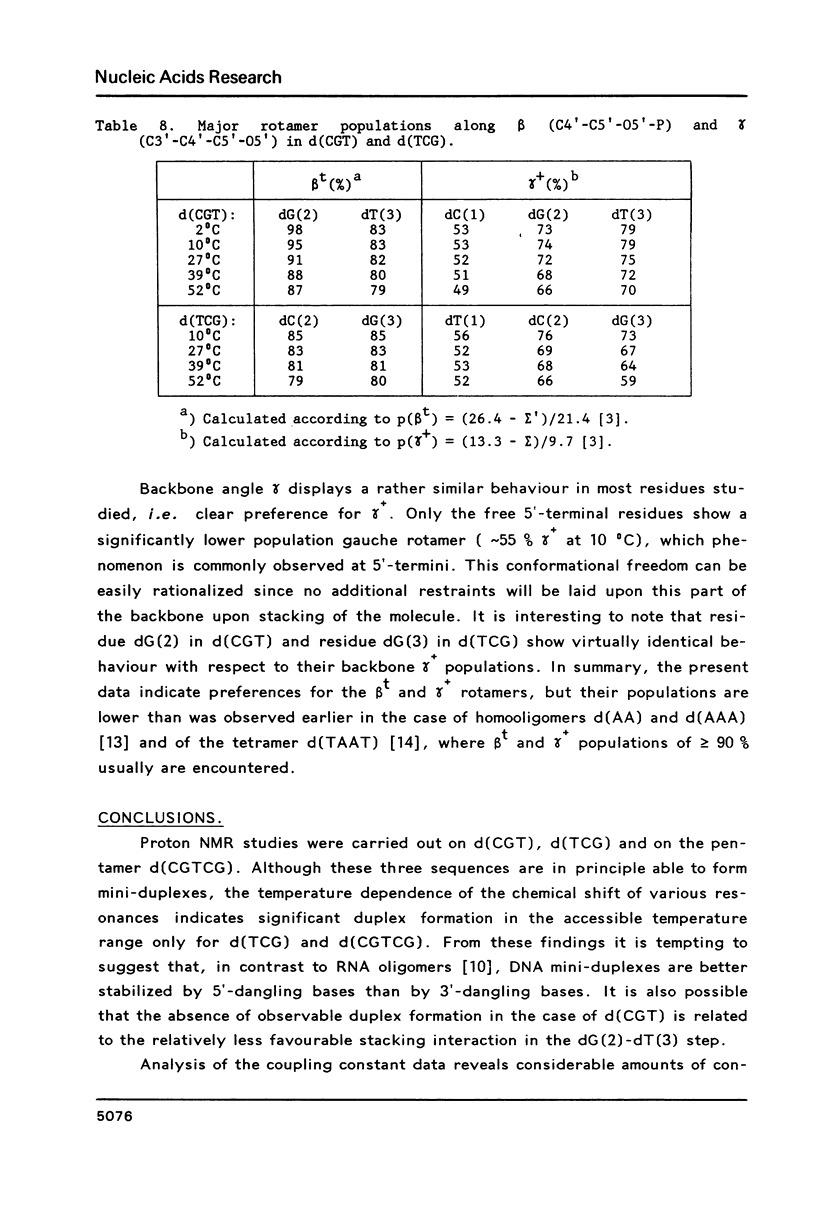
Proton NMR studies of d(CGT), d(TCG) and d(CGTCG) were carried out at 300 and 500 MHz. The temperature and concentration dependence of the chemical shifts of various resonances indicates duplex formation only in the cases of d(TCG) and d(CGTCG). It is concluded that d(TCG) forms a mini-duplex stabilized by a 5'-dangling thymine base. Thermodynamic parameters of the duplex-to-coil equilibrium of the d(TCG) duplex are: delta H0 = -22.3 kcal/mol and delta S0 = -70 cal/mol. K, which correspond to approximately 40% duplex formation at 0 degrees C in a 2 mM nucleotide solution. Comparison of these data with thermodynamic parameters given earlier [Borer, P.N., Dengler, B., Tinoco, I. and Uhlenbeck, O.C. (1974) J. Mol. Biol. 86, 843-853] leads to the conclusion that the dangling base stabilization observed here is approximately equivalent to the stabilization caused by one or two additional A . T base pairs. The chemical shift behaviour of various resonances in d(CGTCG) indicates duplex formation without looping out of the thymine bases. The T X T mismatch does not seem to disturb the helical structure to a large extent. Analysis of the vicinal proton-proton coupling constants of the three compounds yielded geometrical data for the sugar rings. The data are interpreted in terms of N and S pseudorotational ranges. It is shown that a distinct conformation-transmission effect is exerted by the guanosine residues in a 5'----3' direction.
Full text
PDF














Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altona C., Sundaralingam M. Conformational analysis of the sugar ring in nucleosides and nucleotides. Improved method for the interpretation of proton magnetic resonance coupling constants. J Am Chem Soc. 1973 Apr 4;95(7):2333–2344. doi: 10.1021/ja00788a038. [DOI] [PubMed] [Google Scholar]
- Borer P. N., Dengler B., Tinoco I., Jr, Uhlenbeck O. C. Stability of ribonucleic acid double-stranded helices. J Mol Biol. 1974 Jul 15;86(4):843–853. doi: 10.1016/0022-2836(74)90357-x. [DOI] [PubMed] [Google Scholar]
- Brahms J., Aubertin A. M., Dirheimer G., Grunberg-Manago M. Studies of trinucleotide conformations. Role of guanine residues in an oligonucleotide chain. Biochemistry. 1969 Aug;8(8):3269–3278. doi: 10.1021/bi00836a021. [DOI] [PubMed] [Google Scholar]
- Cheng D. M., Kan L. S., Leutzinger E. E., Jayaraman K., Miller P. S., Ts'o P. O. Conformational study of two short pentadeoxyribonucleotides, d-CpCpApApG and d-CpTpTpGpG, and their fragments by proton nuclear magnetic resonance. Biochemistry. 1982 Feb 16;21(4):621–630. doi: 10.1021/bi00533a004. [DOI] [PubMed] [Google Scholar]
- Grosjean H. J., de Henau S., Crothers D. M. On the physical basis for ambiguity in genetic coding interactions. Proc Natl Acad Sci U S A. 1978 Feb;75(2):610–614. doi: 10.1073/pnas.75.2.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasnoot C. A., den Hartog J. H., de Rooij J. F., van Boom J. H., Altona C. Loopstructures in synthetic oligodeoxynucleotides. Nucleic Acids Res. 1980 Jan 11;8(1):169–181. doi: 10.1093/nar/8.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartel A. J., Lankhorst P. P., Altona C. Thermodynamics of stacking and of self-association of the dinucleoside monophosphate m2(6)A-U from proton NMR chemical shifts: differential concentration temperature profile method. Eur J Biochem. 1982 Dec 15;129(2):343–357. doi: 10.1111/j.1432-1033.1982.tb07057.x. [DOI] [PubMed] [Google Scholar]
- Jaskunas S. R., Cantor C. R., Tinoco I., Jr Association of complementary oligoribonucleotides in aqueous solution. Biochemistry. 1968 Sep;7(9):3164–3178. doi: 10.1021/bi00849a020. [DOI] [PubMed] [Google Scholar]
- Mellema J. R., Haasnoot C. A., Van Boom J. H., Altona C. Complete assignment and conformational analysis of a deoxyribotetranucleotide. d(TAAT). A 360 and 500 Mhz NMR study. Biochim Biophys Acta. 1981 Sep 28;655(2):256–264. doi: 10.1016/0005-2787(81)90016-2. [DOI] [PubMed] [Google Scholar]
- Neilson T., Romaniuk P. J., Alkema D., Hughes D. W., Everett J. R., Bell R. A. The effects of base sequence and dangling bases on the stability of short ribonucleic acid duplexes. Nucleic Acids Symp Ser. 1980;(7):293–311. [PubMed] [Google Scholar]
- Olsthoorn C. S., Bostelaar L. J., Van Boom J. H., Altona C. Conformational characteristics of the trinucleoside diphosphate dApdApdA and its constituents from nuclear magnetic resonance and circular dichroism studies. Extrapolation to the stacked conformers. Eur J Biochem. 1980 Nov;112(1):95–110. doi: 10.1111/j.1432-1033.1980.tb04991.x. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Marky L. A., Rice J. A., Broka C., Itakura K., Breslauer K. J. Extra adenosine stacks into the self-complementary d(CGCAGAATTCGCG) duplex in solution. Biochemistry. 1982 Feb 2;21(3):445–451. doi: 10.1021/bi00532a004. [DOI] [PubMed] [Google Scholar]
- Tibanyenda N., De Bruin S. H., Haasnoot C. A., van der Marel G. A., van Boom J. H., Hilbers C. W. The effect of single base-pair mismatches on the duplex stability of d(T-A-T-T-A-A-T-A-T-C-A-A-G-T-T-G) . d(C-A-A-C-T-T-G-A-T-A-T-T-A-A-T-A). Eur J Biochem. 1984 Feb 15;139(1):19–27. doi: 10.1111/j.1432-1033.1984.tb07970.x. [DOI] [PubMed] [Google Scholar]


