Abstract
A multiplexed, ratiometric method is described that can confidently distinguish between cancerous and noncancerous epithelial prostate cells in vitro. The technique is based on bright surface-enhanced resonance Raman scattering (SERRS) biotags (SBTs) infused with unique Raman reporter molecules, and carrying cell-specific peptides. Two sets of SBTs were used. One targets the neuropilin-1 (NRP-1) receptors of cancer cells through the RPARPAR peptide. The other functions as a positive control (PC) and binds to both noncancerous and cancer cells through the HIV-derived TAT peptide. Point-by-point 2D Raman maps of the spatial distribution of the two tags were constructed with subcellular resolution from cells simultaneously incubated with the two sets of SBTs. Averaging the SERRS signal over a given cell yielded an NRP/PC ratio from which a robust quantitative measure of the overexpression of the NRP-1 by the cancer cell line was extracted. The use of a local, on-cell reference produces quantitative, statistically robust measures of overexpression independent of such sources of uncertainty as variations in the location of the focal plane, the local cell concentration, and turbidity.
Keywords: surface-enhanced Raman spectroscopy biomarker , cancer cell identification, multiplexing, silver nanoparticles
Early and rapid identification of malignant cells [ideally free-flowing in biological fluids such as urine (1) or blood (2–4)] is a central goal in cancer research. Accordingly, seeking improvements in sensitivity and accuracy of detection and quantification through, for example, the development of cell-specific diagnostic tools and biomarkers is an active enterprise. Although it is known that such cells are present in body fluids, concentrating them to levels appropriate for confident identification and quantification is still a serious challenge, despite significant advances that have been made by, for example, using microfluidics (5). Once collected, the cells must be identified as either cancerous or noncancerous within tolerable confidence limits, which may be improved by, for example, using several nonredundant biomarkers. Numerous promising identification methods have been reported using antibodies. In this article, we report a technique based on surface-enhanced Raman spectroscopy (SERS) and biotags that produce bright surface-enhanced resonance Raman scattering (SERRS) signals, comparable to or exceeding the intensities of fluorescent tags. The SERRS biotags (SBTs) used in this study exploit peptides as recognition moieties. We show that using SBTs ratiometrically can provide highly statistically significant, quantitative measures of neuropilin-1 (NRP-1) overexpression insensitive to normal causes of uncertainty in optical measurements such as variations in focal plane, cell concentration, and turbidity.
Normal Raman signals from cells and their infrared (FTIR) absorbance patterns have been previously used to differentiate between cancer and normal cells (6–9). SERS has been used as an alternative immunohistochemistry (IHC) tool (10, 11) for the detection of biomarkers in biological fluids or in vivo (12), and for cancer detection from blood (13). However, Raman and FTIR signals measured directly from cells are typically much weaker than those that are measurable with bright labels. Moreover, sample identification is often based on slight differences between highly overlapping spectra. By contrast, numerous nonoverlapping SERS and SBTs can be routinely synthesized (14) and simultaneously excited with a single, very low intensity laser source, making the determination of the relative contribution of the individual SBTs to the overall spectrum tractable. SERS analysis is also, in general, less time consuming than IHC protocols, because cell labeling is carried out in a single step, without pre- or posttreatments like washes, fixation, and permeabilization.
The central feature of SERS is its facile multiplexing capability by using premade, encapsulated nanoparticle clusters that are then infused with one of several highly Raman-active reporter molecules. The SERS spectrum of an individual SBT acts as a unique barcode that is easily differentiable in a composite SERS spectrum originating from many tags. Several synthetic challenges have been overcome in the course of this study so that their synthesis is now prescriptive. The SERS intensities achieved are comparable to fluorescence (15). We have previously shown that forerunners of the SBTs reported here are capable of direct binding to, and uptake by, live cells (16).
Importantly for biomedical applications, SERS employs tissue-penetrating lasers in the red to near-infrared range resulting in low autofluorescence; the high signal enhancement and sharp peaks make it possible to distinguish SERS and especially SERRS in biological specimens at low laser powers (16). Finally, several versions of the biotags can be incubated simultaneously to probe a variety of cell-specific markers taking advantage of the multiplexing capability of SERS.
In this paper, we describe a SERRS-based approach to discriminate between prostate cancer cells (PPC-1) and noncancerous prostate epithelial cells (RWPE-1) in vitro using cell-specific peptides. Multiplexed SBTs consisting of polymer-encapsulated silver particles each labeled with a unique Raman reporter molecule (14) display either a receptor-specific peptide, or a general cell-penetrating peptide (CPP). The SBTs are incubated with cells, SERS mapped, and the signal from the various SBTs deconvoluted to identify the cells while in suspension, simulating their capture from blood. The central idea being that tags displaying the CPP act as a cell-resident calibrant against which the cancer-indicating tags can be ratioed, thereby greatly reducing the uncertainty in the quantitative measure of the cancer-indicative marker that can result from, for example, variability in focal plane, changes in turbidity, cell concentration, or abbreviated measurement time. The use of a cell-resident reference would be particularly useful when a cancerous cell is identified on the basis of the degree of overexpression of a biomarker that may also be present (but to a lesser extent) on the noncancerous cell. The ratiometric approach, in essence, provides a quantitative local measure of the extent of expression of biomarkers associated with the cancerous modifications of a specific cell, essentially providing a basis for quantifying overexpression.
Results and Discussion
Two types of SBTs functionalized with cell-targeting peptides were synthesized. The first carries Cys-TAT (sequence CGRKKRRQRRR, cys added simply for conjugation purposes), a peptide derived from the HIV-1 TAT protein and known CPP (17, 18). TAT was used as a general targeting peptide because it binds rapidly and uniformly on the surface of cells (Fig. S1), and also enters many types through endocytosis (19). The second SBT uses the peptide Cys-RPARPAR, which specifically binds to a pocket in the biomarker protein NRP-1, (20) a receptor expressed on the outer plasma membrane of certain types of cancer cells, which is involved in angiogenesis, a hallmark of cancer growth, and tumor cell migration (21). The binding ability of the RPARPAR peptide to the NRP-1 receptor is given by the amino acid sequence that follows the so-called C-end rule (CendR): a C-terminal arginine (or lysine) in a general sequence of the type R/KXXR/K (where X indicates a generic amino acid) is fundamental for the binding activity of the peptide (20).
Cys-TAT is small and highly cationic. The exact mechanism that allows this peptide to enter cells is still uncertain; however, it is generally agreed that it relies heavily on the initial attachment to the negatively charged moieties on most membranes. When TAT is used to transport nanoparticles in cells, the main uptake mechanism has been shown to be lipid-raft macropinocytosis (22). TAT also has a cryptic CendR R/KXXR/K motif and could compete with binding of RPARPAR (20) particles. This, however, was not observed, likely because of the low concentrations of SBTs. Our group has previously achieved rapid cellular uptake of hollow gold nanoshells through the self-assembly of a TAT–lipid layer around them (23). For the present application, TAT is bound directly to a protein on the surface of the SBT.
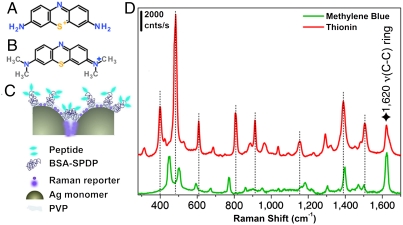
The two SBT sets each carry a Raman reporter: methylene blue (MB) (Fig. 1A) on the FAM-Cys-TAT positive control set (called PC-SBT) and thionin (Fig. 1B) on the FAM-Cys-RPARPAR carrying particles (therefore called NRP-SBT). The absorption maxima of the tags are approximately 670 nm for MB, and approximately 600 nm for thionin. Although the SERS enhancement arises primarily on account of plasmonic excitation in the silver nanoparticles, resonance Raman enhancement due to the laser wavelength’s (633 nm) resonance with the two reporter molecules likely contributes to their high overall SERRS cross-section. The structure of the Raman reporter molecules and the SERRS spectra of the SBTs are shown in Fig. 1. The number of SBTs that gives rise to each of the spectra in Fig. 1C was estimated to be approximately 4, assuming approximately 1 nM colloid concentration and a focal volume of 7 μm3 for the 10× objective. Signals arising from individual SBTs can be detected in deposited samples mapped using the 100× objective (Fig. S2). We note in passing that our purification step does leave some particles with Ag monomer cores rather than Ag dimers. These do not produce strong signals (14).
Fig. 1.
Structural description of SBT system and spectra. (A) Thionin. (B) MB. (C) Schematic of a typical silver SBT: Two Ag monomers are encapsulated in a thin polymer shell and functionalized with a modified BSA carrying several copies of a cell-specific or a universal peptide, then the SBTs are infused with a Raman reporter (either thionin or MB). (D) Corresponding SERRS spectra obtained from an ensemble solution of SBTs (approximately 4 per focal volume). The spectra have been shifted along the ordinate axis for clarity, and the dotted lines identify the bands of thionin that are nonoverlapping with bands in the MB spectrum. The star indicates a common band. Laser 633 nm, 10× objective, power at sample approximately 0.11 mW, hole 400 μm, slit 400 μm, and exposure time 1 s × 3 accumulations.
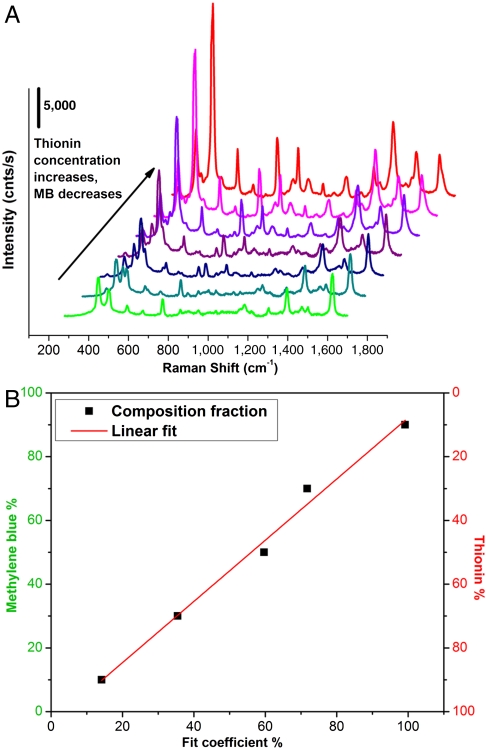
When a mixture of SBTs reside together in the laser’s focal volume a composite spectrum results, which can be deconvoluted into its individual components, allowing the relative concentrations of each contributing species to be determined (14, 24, 25) provided that reference spectra of the pure components are known. Spectra were deconvoluted by assuming the composite spectrum to be a linear combination of the individual components—a reasonable assumption for stable, noninteracting particles. Concentrations were derived from the individual spectroscopic contribution using a calibration plot, constructed by deconvoluting SERS spectra of solutions containing known concentrations of the two SBTs (PC-SBT and NRP-SBT) (Fig. 2A). After subtracting the background, the spectra were normalized to the common band at 1,620 cm-1, assigned to the ring ν(C-C) vibration mode of both thionin and MB (26, 27). The data were fit (Fig. S3) using a weighted least squares method (28). Fig. 2B shows the resulting calibration curve.
Fig. 2.
Calibration plots for the mixed-SBT deconvolution series. (A) SERRS spectra of mixtures of the two SBTs: green trace, 100% PC-SBT; red trace, 100% NRP-SBT. (B) Calibration curve derived from the relative least-squares coefficients for the two components corrected for the known concentrations of MB or thionin SBT present in the solution. This calibration curve was used to calculate the relative quantity of each tag resident on a given cell.
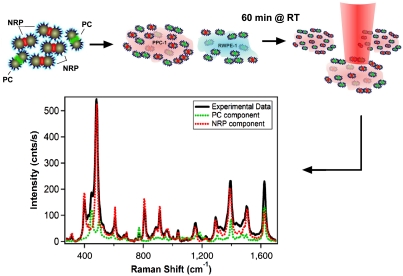
The cell lines chosen are RWPE-1 [prostate epithelium cells, non tumorigenic, from normal tissue, immortalized by HPV (29, 30)] and PPC-1 (epithelial, originated from a bone metastasis of a prostate cancer patient, expressing the biomarker NRP-1). After incubating the PC-SBT and NRP-SBT simultaneously with each cell line suspended in DMEM supplemented with 10% FBS at room temperature for 60 min, cells were placed on a microscope slide and SERRS-mapped without fixing, by scanning the focused laser beam (100× objective) in steps of 1.5 μm and recording a full SERRS spectrum at each point (Fig. 3).
Fig. 3.
Schematic of the SERRS cell mapping experiment. NRP- and PC-SBTs are synthesized, combined, and added to either noncancerous or cancer cells suspended in DMEM supplemented with 10% FBS. These are incubated for 60 min at room temperature. Cells are placed on a microscope slide, and their SERRS spectrum is acquired using microRaman. The sample is mapped along the x and y axes in 1.5-μm steps, 5-μW power at sample, 1-s exposure time. The SERRS spectrum averaged over the whole cell area is deconvoluted into the individual components.
The SERRS maps [each point comprised of a full (270–1,700 cm-1) spectrum] were analyzed using Mathematica. The spectra were fit to a linear combination of the two tags’ individual reference spectra, and included sloping baseline corrections: S(x) = a∗A(x) + b∗B(x) + c + d∗x, where a, b, c, and d are nonnegative adjustable fitting parameters; A(x) and B(x) are the reference spectra as a function of the wavenumber value x. The a and b coefficients together with the calibration plot (Fig. 2B) were used to construct the composition maps.
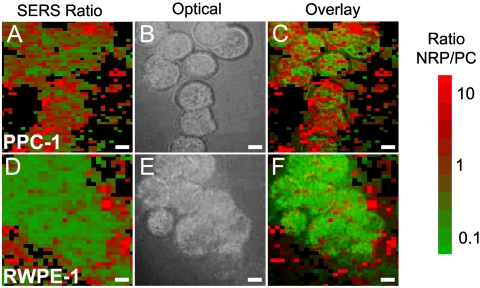
Percent composition maps are depicted in Fig. 4 A and D and colorized on the basis of the ratio of thionin (cancer marker NRP-1) to MB (control). A high value of the NRP/PC ratio is colored red; a low value of the ratio is green; intermediate values are red-green color mixtures as shown in the bar at the right of Fig. 4. Points corresponding to intensities below 100 counts per second were assumed not to contain SBTs and set to black. This threshold was based on the consideration of noise and the observation that individual SBTs deposited out of a dilute solution onto a glass slide produced well-resolved signals above 100 counts per second (Fig. S2). Bright field images of the same cell groups are shown in Fig. 4 B and E. The SERRS and bright field images are overlaid in Fig. 4 C and F to show that signals arise from the cells and not from the glass substrate. It is gratifying to note that the PPC-1 cells, which are expected to bind both peptides, are indeed stained as a mosaic with a mean ratio of approximately 1.0. By contrast, the SERRS images of the noncancerous prostate cell RWPE-1 show very little red, indicating very low NRP/PC ratios with the few red points situated primarily outside the cells (mean of approximately 0.3 including outer rim), likely indicating nonspecifically bound SBTs. The maps and their histograms (Figs. S4 and S5) agree well with the histograms of average-cell signals discussed below (Fig. 5A).
Fig. 4.
Two-dimensional mappings for cancer and normal cells. Color indicates the NRP/PC ratio extracted by deconvolution of the point-by-point SERRS spectra across the scanned area of a group of (A) PPC-1 cells and a group of (D) RWPE-1 cells. Bright field image of (B) the same PPC-1 and (E) RWPE-1 cells. (C and F) The corresponding image overlays. Color code indicates the NRP/PC ratio ranging from 0.1 (green) to 10 (red). Scale bar, 5 μm.
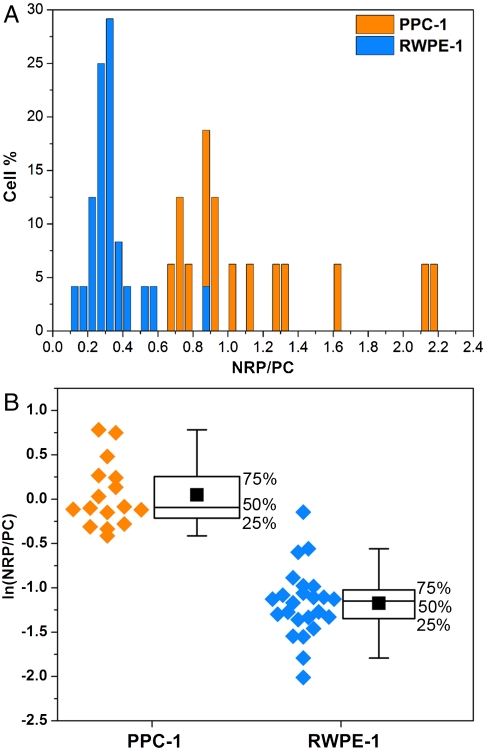
Fig. 5.
Statistics of the SERRS ratios obtained from cancer and normal cell populations. (A) The histogram represents the percentage of either RWPE-1 (blue) or PPC-1 (orange) cells that have the values of the NRP/PC ratio indicated. (B) Box plots of the NRP/PC ratio for both PPC-1 and RWPE-1 populations after application of the logarithmic transformation to render both distributions normal. The experimental data (orange dots for PPC-1 cells and blue dots for RWPE-1 cells) are depicted on the left of each box as a visual aid. The middle line of each box represents the median, the solid black square is the mean, and the whiskers define the inner upper and lower fences beyond which possible outliers are found. The boxes do not overlap, indicating statistically excellent differentiation between these two populations; t test n1 = 16 and n2 = 24, p < 0.001.
To discriminate confidently between these different types of cells, we analyzed the average spectrum arising at both the entire cell level and in maps with subcellular resolution (Figs. S6 and S7), extracting ratios of NRP/PC. Sixteen PPC-1 cells and 24 control RWPE-1 cells were analyzed, and the resulting histograms of the NRP/PC ratio are shown in Fig. 5A. Ninety-six percent of the RWPE-1 cells were found to have NRP/PC ratios below 0.6, whereas 100% of the PPC-1 were found to have ratios above this value. Interestingly, the distribution for the PPC-1 cells is skewed toward high ratio values, likely reflecting the heterogeneity of cell expression of NRP-1 and particle affinity, binding/uptake rates, or receptor recycling during prolonged incubation. We found that changing the concentration of the SBT mixture did not significantly affect the ratios.
The nonnormal distribution depicted by the histogram (Fig. 5A) was converted into a normal distribution through a logarithmic transformation. The statistical analyses are summarized in Fig. 5B. Student’s t test was also performed on transformed data. In Fig. 5B, the boxes define the interquartile range (IQR) (i.e. the range that contains the middle 50% of the data in the distribution), the black square dots are the means of the distributions, and the whiskers determine the upper and lower inner fences outside which suspected outliers (extreme values) lie. These limits are located, respectively, at Q1 (lower quartile) - 1.5 × IQR and Q3 (upper quartile)+1.5 × IQR. No extreme values were observed for the PPC-1 group, whereas the RWPE-1 group has two. One is in the upper fence at ln(NRP/PC) = -0.15 and the other in the lower fence at ln(NRP/PC) = -2.01; however, neither were rejected as outliers. The higher extreme value observed was likely due either to nonspecifically bound SBTs (Fig. S8) or glass-bound SBTs located coincidentally close to a cell. The lower extreme is likely caused by a few nonspecifically bound tags. This is clearly illustrated in Fig. S8, which shows that for a noncancerous cell the body is uniformly green (PC-dominated) with the few red points (indicating NRP contribution) at the periphery. The location of the outliers at the cells’ periphery suggests that the high degree of confidence in distinguishing between cancerous and noncancerous cells based on statistics can be further augmented through microscopic observation of the cells. RWPE-1 cells show weak but nonzero RPARPAR binding (resulting in a mean observed NRP/PC ratio of approximately 0.3, somewhat greater than zero—the expected value if all interactions were specific). This is likely due to nonspecific adsorption of NRP-SBT on the control cells’ membrane.
The statistics indicate that the ratiometric SERS multiplexing approach we used does an excellent job in distinguishing cancerous from noncancerous epithelial prostate cells. Referring to Fig. 5B, the two cell populations form distinct clusters along the ratiometric axis, well separated both visually and numerically. Applying the t test to the two populations also shows them to be highly distinct (p < 0.001), suggesting that with proper choice of tags and peptides the ratio (or rather, its natural logarithm) easily discriminates between noncancerous and cancerous cells.
Conclusions
The multiplexed, ratiometric method described was shown to successfully distinguish between cancerous and noncancerous epithelial prostate cells. The technique is based on deconvoluting SERRS spectra arising from two sets of bright SBTs simultaneously bound to the cells. One of the two sets of SBTs was functionalized to target the NRP-1 receptors of cancer cells through the RPARPAR CendR peptide. The other functions as a positive control and binds to both noncancerous and cancer cells through the HIV-derived TAT peptide. Each was infused with a unique Raman reporter molecule. Averaging the SERRS signal over a given cell yielded an NRP/PC ratio that functioned as a statistically robust quantitative measure of the overexpression of the NRP-1 by the cancer cell line. This indicative ratio is independent of variations in the location of the focal plane, the local cell concentration, and turbidity. The technique also benefits from the very low laser intensities (100 μW) needed for good signal to noise and the use of a single wavelength to excite all SBTs.
Materials and Methods
Materials.
All chemicals for the synthesis of the SBTs were purchased from Sigma except where specified. Silver nitrate (AgNO3) 99.9999%; trisodium citrate dihydrate (Fisher); hexamethylenediamine (HMD), 98%; polyvinylpyrrolidone (PVP), molecular weight 40,000; bovine serum albumin (BSA), powder; potassium chloride (Mallinckrodt Baker); thionin acetate salt (dye content approximately 90%); MB (dye content ≥82%); N-succinimidyl 3-(2-pyridyldithio)propionate (SPDP), Thermo Fisher Pierce no. 21857; Fluorescein-C-X-GRPARPAR-OH (where X is an aminohexanoic linker) designated FAM-Cys-RPARPAR was a gift from Erkki Ruoslahti’s group at Sanford–Burnham Medical Research Institute, University of California, Santa Barbara, CA; Cys-TAT (CGRKKRRQRRR-OH), Anaspec; Fluorescein-NHS (#46410, Pierce); polyoxyethylene (20) sorbitan monolaurate solution 10% in H2O (Tween 20, T20); D-PBS (Gibco), Syringe filters Millex-GV, 0.22 μm, PVDF, 13 mm, ethylene oxide sterilized.
Peptide Labeling.
Cys-TAT was labeled with NHS-Fluorescein (FAM-NHS, Pierce). FAM-NHS first was dissolved at 5 mg/mL in DMSO and stored at -20 °C. Cys-TAT was dissolved in water (Hyclone) to a concentration of 1 mg/mL and also stored at -20 °C. The two were combined, 300 μL peptide and 25 μL dye, and reacted for 3 h at room temperature and stored at -20 °C.
BSA Modification.
BSA was reacted with SPDP (Thermo Fisher Pierce #21857). BSA was dissolved at 22 mg/mL in water, and 50 μL 10× PBS pH 7.2 (Invitrogen) was added. SPDP was dissolved in DMSO (100 mg/mL) and added to the BSA, shaking for 3 h at room temperature. The protein was dialyzed overnight using a Slide-a-Lyzer with a molecular-weight cutoff of 20 kDa, against 0.1× PBS pH 7.4, containing 0.02% NaN3 with three changes of the buffer. The volume increased during the dialysis because of the change in salt conditions. The solution was recovered and passed through a 0.22-μm syringe filter. Final concentration of the protein was determined by UV-visible spectroscopy to be 6.5 mg/mL with a content of approximately 5 SPDP per BSA, found through pyridyl reduction (extinction at 343 nm of 8,080 M-1 cm-1).
BSA modified by the previous reaction of the sulfo-NHS group on SPDP with lysines interacts with the silver surface of the colloid. The BSA binds strongly through the SPDP molecule with the pyridine group appearing in the SERS spectrum prior to Raman reporter being added. Each protein carries several free hanging SPDP groups that can interact with the cys-peptides, thus holding peptides on the outside of the SBT coating for interacting with cell membranes.
Ag SBTs Synthesis.
The silver colloid was synthesized according to the Lee and Meisel protocol (31): 500 mL of deionized water (DI, resistivity 18 MΩ) with 1 mM silver nitrate were brought to a boil. Then, 10 mL of 1% sodium citrate was added. The mixture was kept at boiling temperature for about 90 min until the color turned dark green/gray. Aliquots of the colloid were taken and centrifuged at 0.8 × g to remove the smallest particles. The yellow supernatant was discarded, and the pellet was resuspended in DI. The pellet was diluted until the absorbance of the band at 406 nm was 0.3 at 0.1-mm path length. The resulting colloid was called Ag03. NRP and PC-SBT were then prepared by adding to every 100 μL of Ag03 3.5 μL phosphate buffer (250 mM, pH 7.5), 4 μL HMD (0.4 mg/mL in DI, pH 4.0), waiting for 2 min, then adding 4 μL 1% PVP 40 kDa in DI and 100 μL DI, and finally waiting for 5 min before proceeding with further functionalization. The particles resulting from this controlled aggregation stage were called AgPHP and were then used to prepare NRP and PC-SBTs: For PC-SBT, to every 200 μL AgPHP we added 2 μL BSA-SPDP (6.5 mg/mL in 0.1× PBS), waited 15 min, then added 4 μL MB (600 μM in DI) and 2 μL KCl (500 μM) and incubated at room temperature for 30 min; then, we added 2 μL FAM-Cys-TAT (500 μM in DI), incubated for 15 min, and backfilled with 1 μL BSA 5% (0.1× PBS). For the NRP-SBT, to every 200 μL of AgPHP, we added 2 μL BSA-SPDP (6.5 mg/mL in 0.1× PBS), waited 15 min, added 4 μL of thionin (0.2 mg/mL in DI), incubated for 30 min at room temperature, added 2 μL BSA-SPDP (6.5 mg/mL in 0.1× PBS), waited for 15 min, then added 1 μL BSA 5% (in 0.1× PBS) and 5 μL FAM-C-X-GRPARPAR-OH (200 μM in DI). After adding 0.005% final concentration of Tween-20 (T20), the SBTs were washed by centrifugation (10 min at 0.8 × g), the supernatant was discarded (to remove most of the non-SERS bright single silver nanoparticle biotags), and the pellet resuspended in 1/20th the initial volume in 0.1× PBS/0.1 % BSA/0.005% T20.
Cell Culture.
PPC-1 cells were a generous gift from Erkki Rouslahti’s group (Sanford–Burnham Medical Research Institute, University of California, Santa Barbara, CA). They were grown in DMEM/high glucose (HyClone) supplemented with 10% FBS. RWPE-1 cells (ATCC) were grown in Keratinocyte serum free medium (Invitrogen) supplemented with bovine pituitary extract (0.05 ng/mL) and recombinant EGF (5 ng/mL). Both were incubated at 37 °C in a 5% CO2 atmosphere. For SERRS mapping experiments, cells were plated in multiwell plates and after either 24 or 48 h harvested using a nonenzymatic cell dissociation buffer (Invitrogen) that does not disrupt the membrane receptors. Cells were then washed by centrifugation for 2 min, and the pellet was resuspended in the appropriate volume of DMEM + 10% FBS in order to obtain a concentration of 1.2–1.3 × 105 cells/100 μL. SBTs were then added to cell suspension (with PC versus NRP ratio of 1∶2 or 1∶7 vol/vol) and incubated for 60 min at room temperature; then, a 10-μL aliquot was placed on a microscopy glass slide, covered with a coverslip, sealed with nail polish, and mapped by SERS. For microscopy characterization, cells were plated in multiwall plates and after either 24 or 48 h incubated with SBT for 120 min at 37 °C in a 5% CO2 atmosphere, then imaged.
SERS Measurements.
All SERS measurements (live cell mapping and composition calibration) were carried out using a Horiba Jobin–Yvon LabARAMIS instrument equipped with a 633-nm laser, confocal microscope with 10×, 50×, and 100× objectives. Measurements for concentration calibration on SBT ensembles were carried out using a 10× objective (100 μW power at sample), 633 laser line, hole 400 μm, slit 400 μm, exposure time 1 s, averaged three times.
Live cell mapping was performed in point scan mode, with hole 600 μm, slit 300 μm, using a 100× objective and 633-nm laser (power at sample 5 μW); exposure time 1 s, step size 1.5 μm.
Data Analysis.
Initial data analysis (mapping, whole cell averaging of signal, baseline correction, normalization) was performed with the help of the onboard program LabSpec. The spectral deconvolution and fitting of single spectra for the composition calibration was done using a weighted least-squares algorithm in IgorPro (WaveMetrics Inc.), whereas the deconvolution and NRP/PC ratio calculations for point-to-point maps were done using Mathematica’s FindFit, a nonlinear least-squares-fitting algorithm, and the SBT pure spectra, taken from Fig. 2, were processed by a customized notebook. The statistics on the whole cell average data were performed using OriginPro (Version 8.1). Student’s t test was performed, and p < 0.001 was considered highly significant.
Supplementary Material
Acknowledgments.
We are grateful to Professor Erkki Ruoslahti for providing us with advice and samples of the FAM-C-X-GRPARPAR-OH peptide. We thank Professor Norbert Reich at University of California, Santa Barbara, and Dr. Tambet Teesalu in the Ruoslahti lab for many helpful discussions. This work was supported by the Institute for Collaborative Biotechnologies through Grant DAAD19-03-D-0004 from the US Army Research Office and made extensive use of the Materials Research Laboratory Central Facilities at University of California, Santa Barbara, supported by the National Science Foundation under Awards DMR-0080034 and DMR-0216466 for the high-resolution transmission electron microscopy/scanning transmission electron microscopy. G.B.B. was also supported in part by a fellowship from the Santa Barbara Cancer Center.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1109490108/-/DCSupplemental.
References
- 1.Rouprêt M, et al. Molecular detection of localized prostate cancer using quantitative methylation-specific PCR on urinary cells obtained following prostate massage. Clin Cancer Res. 2007;13:1720–1725. doi: 10.1158/1078-0432.CCR-06-2467. [DOI] [PubMed] [Google Scholar]
- 2.Eschwege P, et al. Prognostic value of prostate circulating cells detection in prostate cancer patients: A prospective study. Br J Cancer. 2009;100:608–610. doi: 10.1038/sj.bjc.6604912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scher HI, et al. Circulating tumour cells as prognostic markers in progressive, castration-resistant prostate cancer: A reanalysis of IMMC38 trial data. Lancet Oncol. 2009;10:233–239. doi: 10.1016/S1470-2045(08)70340-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pantel K, Brakenhoff RH, Brandt B. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nat Rev Cancer. 2008;8:329–340. doi: 10.1038/nrc2375. [DOI] [PubMed] [Google Scholar]
- 5.Nagrath S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harvey TJ, et al. Spectral discrimination of live prostate and bladder cancer cell lines using Raman optical tweezers. J Biomed Opt. 2008;13:064004–064012. doi: 10.1117/1.2999609. [DOI] [PubMed] [Google Scholar]
- 7.Crow P, et al. The use of Raman spectroscopy to differentiate between different prostatic adenocarcinoma cell lines. Br J Cancer. 2005;92:2166–2170. doi: 10.1038/sj.bjc.6602638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taleb A, et al. Raman microscopy for the chemometric analysis of tumor cells. J Phys Chem B. 2006;110:19625–19631. doi: 10.1021/jp061981q. [DOI] [PubMed] [Google Scholar]
- 9.Harris A, et al. Raman spectroscopy and advanced mathematical modelling in the discrimination of human thyroid cell lines. Head Neck Oncol. 2009;1:1–6. doi: 10.1186/1758-3284-1-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun L, et al. Composite organic-inorganic nanoparticles as Raman labels for tissue analysis. Nano Lett. 2007;7:351–356. doi: 10.1021/nl062453t. [DOI] [PubMed] [Google Scholar]
- 11.Lutz B, et al. Raman nanoparticle probes for antibody-based protein detection in tissues. J Histochem Cytochem. 2008;56:371–379. doi: 10.1369/jhc.7A7313.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuen JM, Shah NC, Walsh JT, Glucksberg MR, Van Duyne RP. Transcutaneous glucose sensing by surface-enhanced spatially offset Raman spectroscopy in a rat model. Anal Chem. 2010;82:8382–8385. doi: 10.1021/ac101951j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng SY, et al. Nasopharyngeal cancer detection based on blood plasma surface-enhanced Raman spectroscopy and multivariate analysis. Biosens Bioelectron. 2010;25:2414–2419. doi: 10.1016/j.bios.2010.03.033. [DOI] [PubMed] [Google Scholar]
- 14.Braun GB, et al. Generalized approach to SERS-active nanomaterials via controlled nanoparticle linking, polymer encapsulation, and small-molecule infusion. J Phys Chem C. 2009;113:13622–13629. [Google Scholar]
- 15.Sabatté G, et al. Comparison of surface-enhanced resonance Raman scattering and fluorescence for detection of a labeled antibody. Anal Chem. 2008;80:2351–2356. doi: 10.1021/ac071343j. [DOI] [PubMed] [Google Scholar]
- 16.Pallaoro A, Braun GB, Reich NO, Moskovits M. Mapping local pH in live cells using encapsulated fluorescent SERS nanotags. Small. 2010;6:618–622. doi: 10.1002/smll.200901893. [DOI] [PubMed] [Google Scholar]
- 17.Green M, Loewenstein PM. Autonomous functional domains of chemically synthesized human immunodeficiency virus tat trans-activator protein. Cell. 1988;55:1179–1188. doi: 10.1016/0092-8674(88)90262-0. [DOI] [PubMed] [Google Scholar]
- 18.Frankel AD, Pabo CO. Cellular uptake of the tat protein from human immunodeficiency virus. Cell. 1988;55:1189–1193. doi: 10.1016/0092-8674(88)90263-2. [DOI] [PubMed] [Google Scholar]
- 19.Zorko M, Langel Ü. Cell-penetrating peptides: Mechanism and kinetics of cargo delivery. Adv Drug Deliv Rev. 2005;57:529–545. doi: 10.1016/j.addr.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 20.Teesalu T, Sugahara KN, Kotamraju VR, Ruoslahti E. C-end rule peptides mediate neuropilin-1-dependent cell, vascular, and tissue penetration. Proc Natl Acad Sci USA. 2009;106:16157–16162. doi: 10.1073/pnas.0908201106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jia H, et al. Neuropilin-1 antagonism in human carcinoma cells inhibits migration and enhances chemosensitivity. Br J Cancer. 2010;102:541–552. doi: 10.1038/sj.bjc.6605539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen B, Liu Q, Zhang Y, Xu L, Fang X. Transmembrane delivery of the cell-penetrating peptide conjugated semiconductor quantum dots. Langmuir. 2008;24:11866–11871. doi: 10.1021/la802048s. [DOI] [PubMed] [Google Scholar]
- 23.Braun GB, et al. Laser-activated gene silencing via gold nanoshell-siRNA conjugates. ACS Nano. 2009;3:2007–2015. doi: 10.1021/nn900469q. [DOI] [PubMed] [Google Scholar]
- 24.Vajna B, Farkas I, Szabó A, Zsigmond Z, Marosi G. Raman microscopic evaluation of technology dependent structural differences in tablets containing imipramine model drug. J Pharm Biomed Anal. 2010;51:30–38. doi: 10.1016/j.jpba.2009.07.030. [DOI] [PubMed] [Google Scholar]
- 25.Zhang L, Henson MJ, Sekulic SS. Multivariate data analysis for Raman imaging of a model pharmaceutical tablet. Anal Chim Acta. 2005;545:262–278. [Google Scholar]
- 26.Hutchinson K, Hester RE, Albery WJ, Hillman AR. Raman spectroscopic studies of a thionine-modified electrode. J Chem Soc Faraday Trans 1. 1984;80:2053–2071. [Google Scholar]
- 27.Naujok RR, Duevel RV, Corn RM. Fluorescence and Fourier transform surface-enhanced Raman scattering measurements of methylene blue adsorbed onto a sulfur-modified gold electrode. Langmuir. 1993;9:1771–1774. [Google Scholar]
- 28.Guarrotxena N, Ren Y, Mikhailovsky A. Raman response of dithiolated nanoparticle linkers. Langmuir. 2010;27:347–351. doi: 10.1021/la103114b. [DOI] [PubMed] [Google Scholar]
- 29.Bello D, Webber MM, Kleinman HK, Wartinger DD, Rhim JS. Androgen responsive adult human prostatic epithelial cell lines immortalized by human papillomavirus 18. Carcinogenesis. 1997;18:1215–1223. doi: 10.1093/carcin/18.6.1215. [DOI] [PubMed] [Google Scholar]
- 30.Webber MME, Rhim JS. 5,824,488. US Patent. 2001
- 31.Lee PC, Meisel D. Adsorption and surface-enhanced Raman of dyes on silver and gold sols. J Phys Chem. 1982;86:3391–3395. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.