Abstract
The emerging amphibian disease chytridiomycosis is caused by the fungal pathogen Batrachochytrium dendrobatidis (Bd). Amphibian populations and species differ in susceptibility to Bd, yet we know surprisingly little about the genetic basis of this natural variation. MHC loci encode peptides that initiate acquired immunity in vertebrates, making them likely candidates for determining disease susceptibility. However, MHC genes have never been characterized in the context of chytridiomycosis. Here, we performed experimental Bd infections in laboratory-reared frogs collected from five populations that show natural variation in Bd susceptibility. We found that alleles of an expressed MHC class IIB locus associate with survival following Bd infection. Across populations, MHC heterozygosity was a significant predictor of survival. Within populations, MHC heterozygotes and individuals bearing MHC allele Q had a significantly reduced risk of death, and we detected a significant signal of positive selection along the evolutionary lineage leading to allele Q. Our findings demonstrate that immunogenetic variation affects chytridiomycosis survival under controlled experimental conditions, confirming that host genetic polymorphisms contribute to chytridiomycosis resistance.
Keywords: emerging infectious disease, experimental infection, major histocompatibility complex, molecular evolution, Ranidae
Amphibians face a biodiversity crisis (1) brought on in large part by the emerging infectious disease chytridiomycosis (2–5). The fungal pathogen Batrachochytrium dendrobatidis (Bd) causes chytridiomycosis in amphibian species worldwide, but the extent of morbidity and mortality varies within and among populations (6, 7), species (7), and assemblages (7, 8). Some of this variation is attributable to regional and local ecological factors such as climate (9), elevation (2), host life history traits (10), and different Bd strains (11). However, host immunity likely also contributes to disease outcomes. In the model frog Silurana tropicalis, experimental Bd inoculations significantly change host gene expression profiles (12), and innate (13) and acquired (14) immune responses are activated at host-optimal temperatures. In addition, experimental infections of the toad Alytes obstetricans show that Bd survival varies among individuals and populations under constant environmental conditions (15), and upon reinfection, previously Bd-infected Leiopelma archeyi clear the pathogen significantly faster than control animals (16), suggesting a secondary acquired immune response. Identifying heritable determinants of Bd resistance has the potential to enhance the success of amphibian conservation efforts (17). However, to date, immunogenetic correlates of survival remain underexplored (18).
Vertebrate MHC genes are good candidates for influencing host Bd dynamics because they encode cell-surface glycoproteins that regulate the vertebrate acquired immune response and affect the development and progression of wildlife disease (19). To initiate acquired immunity, MHC proteins bind pathogen molecules on their peptide-binding regions (PBRs) and present them to T cells (20). This central function in pathogen defense creates strong diversifying selection within host populations for numerous MHC proteins that can collectively bind a wide array of pathogens (21). MHC genes are divided into three classes (I, II, and III) that are genomically linked across tetrapods (22). Class I loci are expressed on all nucleated somatic cells and present peptides derived from intracellular pathogens, whereas class II genes are expressed in epithelial cells in the thymus and antigen-presenting cells in the periphery, and primarily present peptides derived from extracellular pathogens (23, 24). The class III region encodes a group of structurally unrelated proteins that are not involved in antigen presentation (but have immune function) (24). Class II MHC loci are the most likely candidate immunity genes for chytridiomycosis for two reasons: (i) they are the primary presenters of extracellular fungal pathogens (23) and (ii) class II-expressing dendritic and Langerhans lymphocytes are present in amphibian skin (25, 26), the primary location of Bd infections. Despite these data, class II genes have only been described in a few amphibians (24, 27–32) and never, to our awareness, in the context of Bd susceptibility.
Associations between disease susceptibility and MHC polymorphisms are widespread in natural wildlife populations (19). Most studies focus on variation in the PBR of class I and class IIB loci because PBR amino acid variability determines the repertoire of pathogen peptides to which T cells can respond (20). Across diverse vertebrate taxa, survival after disease exposure has been associated with particular PBR alleles (33), elevated PBR heterozygosity (34), and intermediate levels of PBR diversity (35). These multiple mechanisms of disease resistance indicate that MHC-associated immunity is context-dependent and varies with each combination of host, pathogen, and environment. Chytridiomycosis is caused by a single pathogen affecting numerous amphibian hosts that occupy diverse habitats, thereby providing a unique epidemiological combination for immunogenetic studies of independent hosts evolving under varied environmental regimes.
The lowland leopard frog (Lithobates yavapaiensis) is a North American amphibian that began experiencing mass chytridiomycosis die-offs in the early 1990s (6). Currently, populations persist with Bd infections that vary in prevalence and intensity across space and time (36, 37). In some populations, a proportion of individuals die from chytridiomycosis during winter months when temperatures are pathogen-optimal (36). In other populations, Bd infection prevalence and intensity increase during winter months, but host frogs never develop signs of disease (36). This pattern suggests two nonexclusive hypotheses: (i) genetic resistance to chytridiomycosis occurs in some populations or (ii) environmental variation explains differences in disease dynamics among populations. However, the spatial and seasonal variation in pathogen and disease occurrence across populations precludes distinguishing genetic from environmental determinants of Bd-associated mortality in the field.
Here, we determined the role of host genetic variation in Bd-associated mortality by experimentally infecting laboratory-reared L. yavapaiensis collected from five geographically distinct populations that naturally differ in disease dynamics. We exposed individuals to Bd and induced chytridiomycosis under constant, pathogen-optimal environmental conditions. We then tested the following four hypotheses: (i) laboratory survival varies across individuals and/or populations in patterns that correspond to observed chytridiomycosis mortality in the field; (ii) pathogen burdens are lower in surviving individuals compared with individuals that die; (iii) genetic variation at MHC loci associates with survival following infection; and (iv) MHC alleles associated with survival show a signature of positive selection. Combined, these hypotheses address the overarching question of whether heritable host immune factors explain differences in Bd susceptibility in natural populations.
Results
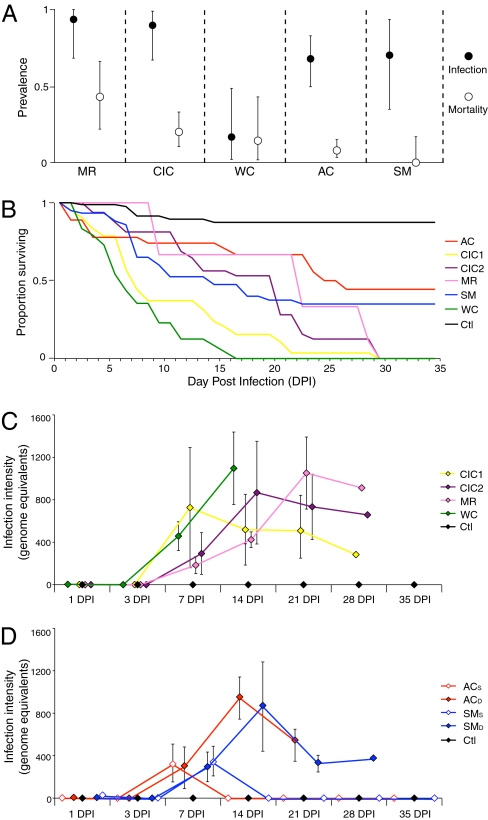
We characterized natural chytridiomycosis in five L. yavapaiensis populations during 2007 through 2010 (Fig. 1A and Fig. S1) (36) to generate a priori expectations for survival patterns in our experimental Bd challenges. We collected egg masses in 2008 and reared them through metamorphosis in a light-, temperature-, and humidity-controlled growth chamber. Before the experiment, we tested for, but did not detect, native Bd infections among our experimental animals. Metamorphosed frogs were randomly assigned to two replicated treatments: (i) infected with Arizona Bd strain PsTr2004 via exposure to 1 × 105 zoospores in liquid media or (ii) kept as uninfected controls exposed to liquid media only. Among the 99 frogs experimentally infected with Bd, 14 individuals survived the experiment (Fig. 1B and Table S1); survivors were from Aravaipa Canyon (AC) and the Santa Maria River (SM). These two populations showed 41% and 27% survival in the laboratory, respectively, and are also the two populations with the lowest proportion of mortalities in the field (Fig. 1A). No experimentally infected frogs from Cienega Creek (CIC), Muleshoe Ranch (MR), or Willow Creek (WC) survived, corroborating the hypothesis that laboratory survival varies across populations according to chytridiomycosis mortality in the field. Mean infection intensity, defined as the number of Bd genome equivalents recovered per swab, was not significantly different from zero at 1 d post infection (DPI; P = 0.74) or 3 DPI (P = 0.39), but zoospore counts increased significantly in all populations at 7 DPI (P = 0.0005; Fig. 1 C and D). At 14 DPI, mean infection intensity was no longer significantly different from zero for individuals from AC and SM that survived (P = 0.5; Fig. 1D, open symbols) but remained significantly higher than zero for all individuals that eventually died (P < 0.0001; Fig. 1D, closed symbols). Thus, among Bd-infected individuals, those that died maintained Bd infection intensities higher than zero for the duration of the experiment, whereas surviving individuals cleared Bd infections by 14 DPI (Fig. 1 C and D).
Fig. 1.
Bd infection and mortality vary among populations of L. yavapaiensis in nature and in the laboratory. (A) Field estimates of winter Bd infection prevalence (proportion of frogs testing positive for Bd; filled symbols) and mortality (proportion of Bd-positive frogs that died; open symbols) in five natural populations; 95% Clopper–Pearson binomial confidence intervals are indicated (modified from ref. 37). (B) Survival following Bd infection varies among individuals from the same five populations when reared in a growth chamber. Survival was tracked for 35 DPI with 1 × 105 zoospores of Bd (colored lines) or sham infection (black line). CIC1 and CIC2 denote genetically distinct clutches from population CIC. Numbers of individuals per population (Bd-infected/uninfected) are as follows: AC, 17/11; CIC, 34/22; MR, 5/2; WC, 17/11; SM, 26/15; all, 99/59. (C) Mean Bd infection intensity (zoospore equivalents ± SD) measured at various DPI intervals in populations with no survival (CIC, MR, and WC). (D) Mean Bd infection intensity over time for survivors (open symbols; ACS and SMS) and nonsurvivors (filled symbols; ACD and SMD) from populations with survivors (AC and SM). Control frogs (black symbols) remained uninfected for the duration of the experiment.
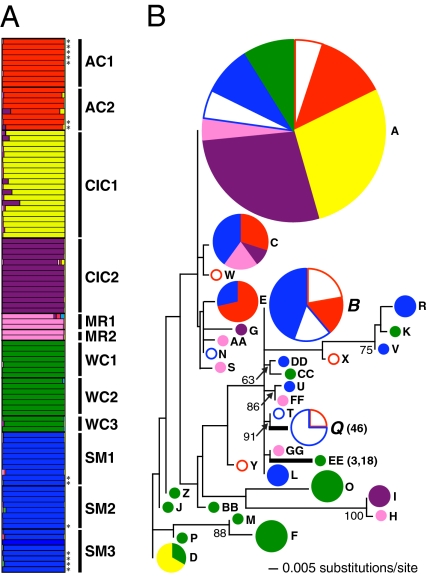
Population genetic background can bias disease association analyses (38); therefore, we genotyped all 99 Bd-infected frogs at 14 unlinked microsatellite loci (39) to characterize population structure for comparison with immunogenetic variation. We implemented a Bayesian assignment test (40) that confirmed the independent evolutionary history and current genetic isolation of each sampled population: frogs were assigned to six genetic demes (lnL, −2,984.1) corresponding to their five natal populations, with further differentiation between clutches from population CIC (Fig. 2A). We next characterized the PBR of an expressed MHC class IIB gene (41) and sequenced PBR genotypes for all Bd-infected individuals to test for associations between allelic polymorphisms and responses to Bd infection. We recovered 33 unique PBR alleles (GenBank accession nos. JN638850--JN638882; Fig. S2), of which only six occurred in multiple genetic demes. Ten of the 12 sampled clutches showed evidence of multiple paternity based on microsatellite and PBR genotypes (Fig. S3 and Table S1); thus, the majority of clutches included half-sibships, rather than full sibships.
Fig. 2.
(A) Clutch genetic structure inferred by Bayesian assignment of 99 Bd-infected L. yavapaiensis individuals. Thin bars separate individuals, thick bars separate clutches, and asterisks indicate individuals that survived experimental infection. Horizontal bars show the coefficient of membership across genetic demes for each Bd-infected frog, with each color representing a unique genetic deme. (B) ML genealogy of 33 MHC class IIB PBR alleles. Circle size is proportional to frequency, colors correspond to the genetic demes in A, alleles recovered from survivors are shown with open symbols, and alleles recovered from nonsurvivors are shown with filled symbols. Bold denotes terminal branches with significantly elevated nonsynonymous substitution rates (i.e., ω > 1) followed by the codon positions under selection in parentheses. Alleles showing significant associations with survival are enlarged and italicized. Bootstrap values greater than 50% are indicated.
To identify parameters with significant effects on survival, we used the Cox proportional-hazards model including treatment, genetic deme, clutch, replicate, number of frogs per replicate, initial mass, change in mass, PBR heterozygosity, maximum infection intensity (MII), and interactions between MII and all other parameters (complete model, ΔlnL = 36.2, df = 22, P < 0.0001). As expected, significantly more uninfected individuals survived compared with Bd-infected individuals (P = 0.001). Among infected individuals, PBR heterozygosity (P = 0.0002), MII (P = 0.005), and the interaction between MII and PBR heterozygosity (P = 0.0005) were significant explanatory variables predicting survival. In contrast, density-dependent dynamics within replicates did not influence survival, as the number of frogs per replicate and the interaction between number of frogs per replicate and MII were not significant.
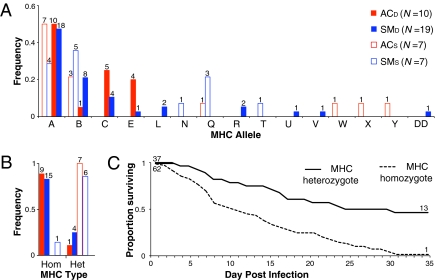
We generated an unrooted genealogy of unique PBR sequences to examine the distribution of alleles in relation to genetic demes and disease outcomes (Fig. 2B). PBR alleles did not cluster by genetic deme. Of the alleles recovered from multiple populations (A–E and Q), frogs with alleles B and Q showed a significantly reduced risk of mortality (Table 1). This pattern could arise because alleles B and Q occur only within AC and SM, the two genetic demes in which some individuals survived. Thus, we tested for allelic associations within each of those demes, and found a significant reduction in risk associated with allele Q in SM but not AC (Table 1). Similarly, SM (signed-rank test, P = 0.03), but not AC (P = 0.09), showed significantly different PBR allele frequency distributions between survivors and nonsurvivors (Fig. 3A). Across populations, PBR heterozygotes had a significantly reduced risk of death (Table 1 and Fig. 3C). Within populations, risk of death remained significantly reduced for heterozygotes in demes AC and SM (Table 1 and Fig. 3B), demonstrating that heterozygote advantage was not an artifact of higher heterozygote frequency in demes with survivors.
Table 1.
RR for survival-associated MHC risk factors among Bd-infected frogs
| Risk factor |
||||||
| Heterozygote |
Allele B |
Allele Q |
||||
| Genetic deme | RR | P value | RR | P value | RR | P value |
| AC (n = 17) | 0.13 (0.13–0.43) | 0.001* | 0.46 (0.14–1.2) | 0.16 | 0.0 (0.0–1.3) | 0.41 |
| SM (n = 26) | 0.43 (0.32–0.77) | 0.005* | 0.64 (0.26–1.2) | 0.31 | 0.0 (0.0–0.71) | 0.017* |
| All (N = 99) | 0.58 (0.54–0.78) | <0.00001* | 0.47 (0.27–0.74) | 0.001* | 0.0 (0.0–0.55) | <0.00001* |
Values in parentheses are 95% confidence intervals, followed by two-tailed Fisher exact test P values.
*Significant RR value after sequential Bonferroni correction.
Fig. 3.
MHC class IIB PBR allele and heterozygote frequencies for Bd-infected survivors and nonsurvivors. PBR allele distribution (A) and PBR heterozygote and homozygote frequency (B) among survivors (open bars) and nonsurvivors (filled bars) in AC (red bars) and SM (blue bars), the two genetic demes with survivors. Allele counts and numbers of heterozygous and homozygous individuals are indicated above frequency bars. (C) Survival curves for PBR heterozygotes (solid line) and homozygotes (dotted line) across Bd-infected individuals from all five genetic demes.
To distinguish between selection and neutral evolution, we estimated ω, the ratio of nonsynonymous (i.e., amino acid-changing) to synonymous (i.e., silent) nucleotide substitutions. Values of ω equal to 1 indicate neutral evolution, whereas values greater than 1 indicate positive selection and values less than 1 indicate purifying selection (42). Along the 82 PBR codons, models including positive selection (allowing ω > 1 as a rate class) fit the alignment significantly better than models excluding positive selection (Table 2). We detected significant positive selection in codons 3, 18, and 46, which correspond to three of 14 codon positions that determine peptide binding in the human class II PBR (43). These substitution rate analyses detect selection over the entire history of allelic lineages (44) and do not distinguish among ancient versus recent selective events. Thus, to identify PBR alleles that may have recently fixed nonsynonymous substitutions in response to chytridiomycosis, we tested for positive selection (ω > 1) along terminal branches of the genealogy, which represent the most recent molecular changes. We detected a ω greater than 1 in two terminal branches (Fig. 2B and Table S2). Codons 3 and 18 showed a ω value greater than 1 along the branch leading to an allele present in an individual that died; thus, we conclude this signal of selection is unrelated to Bd. In contrast, codon position 46 showed a ω value greater than 1 along the branch leading to allele Q, the only allele recovered exclusively from surviving individuals (Fig. 2B). Codon 46 may therefore be particularly important in determining Bd peptide binding, and allele Q is a candidate allele for recent immunogenetic adaptation in response to disease pressure.
Table 2.
Evidence of positive selection among 82 amino acid residues of L. yavapaiensis MHC class IIB PBR sequences
| Method | LRT for model allowing ω > 1 rate classes* | Percentage of sites in ω > 1 rate classes | Mean ω per ω > 1 rate class |
| Rate class | |||
| PARRIS | 9.16 (0.01) | 15% | 4.3 |
| Evolutionary fingerprint | 26.1 (0.006) | 27% | 4.4 |
| Codon-specific | Codon 3 dN–dS† | Codon 18dN–dS† | Codon 46 dN–dS† |
| SLAC | 18.1 (0.09) | — | — |
| FEL | 1.1 (0.03) | 0.93 (0.03) | 0.74 (0.04) |
| REL | 3.2 (0.99) | 3.3 (0.99) | — |
dN–dS, normalized difference between nonsynonymous substitutions per nonsynonymous sites (dN) and synonymous substitutions per synonymous sites (dS); FEL, fixed-effects likelihood; LRT, likelihood ratio test; PARRIS, partitioning approach for robust inference of selection; PP, posterior probability; REL, random-effects likelihood; SLAC, single likelihood ancestral counting; ω, dN/dS ratio.
*Values in parentheses are P values.
†Values in parentheses are P values/posterior probability.
Discussion
We have shown that MHC class IIB-associated genetic factors explain differences in Bd susceptibility in L. yavapaiensis by confirming our four hypotheses: (i) Bd survival was significantly different among individuals and populations, and was concordant with observed variation in field mortality (Fig. 1 A and B); (ii) maximum pathogen burdens were significantly lower in surviving individuals (Fig. 1 C and D); (iii) MHC heterozygotes and MHC allele Q showed significant associations with survival (Table 1 and Fig. 2B); and (iv) allele Q had a signal of recent positive selection acting on codon 46 (Fig. 2B). Thus, MHC class IIB genotypes are significantly associated with differences in Bd survival in L. yavapaiensis populations. Our results should be interpreted with the caveat that the MHC contains numerous immune-related loci that are in strong linkage disequilibrium (22), and MHC loci experience frequent and diverse forms of selection for pathogen resistance, self and nonself discrimination, and kin selection (19, 45). These varied evolutionary processes present a challenge for identifying specific effects of individual MHC genes. Indeed, we cannot exclude the possibility that the class IIB locus examined here shows significant associations with survival as a result of linkage with another locus, because at least two class II genes are expressed in L. yavapaiensis (41). However, the signal of positive selection we detected along the PBR lineage leading to the only allele significantly associated with Bd survival challenges linkage as the sole explanation for our results. To the contrary, recent positive selection acting in the PBR of a resistance-associated allele found in natural populations provides compelling evidence that this locus encodes a molecule with specific involvement in the immunological response to Bd.
Our results underscore the evolutionary advantage of diverse genetic resistance pathways in an ever-changing environment. Lowland leopard frogs show spatial and temporal variation in demography and Bd exposure (36), which likely creates population-specific selective regimes for the evolution of chytridiomycosis resistance. Furthermore, host Bd-resistance alleles may carry some fitness cost with regard to other Bd strains or pathogens (45, 46). Because of the specificity of MHC-peptide binding, individual alleles often provide a fitness advantage against some pathogen epitopes while simultaneously incurring a fitness cost against others (19, 47). Thus, the strength and targets of selection on the MHC are likely to differ across populations, depending on the genetic background of the host and the full complement of pathogens at each site. In this context, our finding that PBR heterozygosity was a fitness advantage across all populations underscores that the broader peptide binding capabilities inherent to having two MHC alleles can be more advantageous than having one allele with particular efficacy against one pathogen epitope (46). Heterozygote advantage may thus be the general mechanism for enhancing chytridiomycosis resistance in natural populations. However, we also detected a resistance allele in population SM, highlighting that individual alleles can be selectively advantageous for specific genetic backgrounds and environmental regimes. Numerous studies of wildlife populations demonstrate that genetic disease resistance is a host-, strain-, and context-dependent process (19, 45), and our results confirm that lowland leopard frogs have reduced chytridiomycosis susceptibility via multiple evolutionary mechanisms.
The ability of host populations to evolve Bd resistance could be limited by the recent timescale of chytridiomycosis emergence. Lowland leopard frogs have suffered from Bd-induced declines for only 20 generations (6), and genetic theory predicts that the shorter generation time of pathogens should provide them an advantage in the coevolutionary arms race (47). However, hosts can benefit from high standing genetic diversity, because the rate of pathogen evolution is negatively correlated with host genetic variability (48–50). Empirical studies also demonstrate that relative generation times of hosts and pathogens do not influence local adaptation (51). In fact, when pathogen genetic variability is low, as is the case for Bd in the New World (52), short generations increase the strength of selection for host resistance and decrease the capacity of pathogens to adapt to an evolving host (53). Chytridiomycosis in L. yavapaiensis thus fits a scenario with strong potential for the evolution of heritable disease resistance in host populations. In our study, SM and AC were the only populations in which individuals varied in Bd survival and the only populations with the PBR allele (i.e., Q) showing a signal of recent positive selection. Importantly, these populations also had the lowest proportions of Bd mortalities in our field surveys, indicating that our experimental results have biological relevance to Bd dynamics in natural populations. These populations are therefore the best candidates for future studies of the evolutionary dynamics of Bd resistance in natural populations.
Animal hosts evolve two mechanisms to combat infections: resistance, defined as the ability to limit pathogen burden, and tolerance, the ability to limit the damage caused by a given pathogen burden (54). The distinction between tolerance and resistance is critical for understanding the evolutionary consequences of chytridiomycosis disease dynamics because resistance has a negative effect on pathogen fitness and tolerance does not (54). Under our experimental conditions, surviving frogs cleared Bd infections, demonstrating disease resistance. However, the same lowland leopard frog populations (AC and SM) show evidence of disease tolerance in the field, where individuals carry high pathogen burdens with no disease signs (36). These differences underscore the need to consider experimental infection studies and disease dynamics in wild populations. Controlled experiments allow us to pinpoint the effects of immunogenetic factors in the absence of environmental variation. However, the critical next step will be studies of natural populations incorporating environmental factors that mediate host resistance and tolerance, pathogen virulence, and population MHC variability.
Given the scale of the amphibian biodiversity crisis (4), understanding host–pathogen chytridiomycosis dynamics is essential for amphibian conservation. Our study shows that genetically distinct lowland leopard frog populations experimentally infected with Bd have significant differences in survival that correlate with immunogenetic heterozygosity and polymorphism, and thus that host genetic variability determines Bd infection outcomes under controlled environmental conditions. To date, studies of natural amphibian populations have compared chytridiomycosis susceptibility across sympatric species (2, 8, 9), among intraspecific populations (55), and between individuals (56). Our findings emphasize the need for studies that quantify host population genetics and immunogenetics at these same hierarchical levels. For highly susceptible species, captive breeding is one of the few remaining strategies for maintaining natural populations (57). Species reintroductions with assisted selection could be a viable chytridiomycosis mitigation strategy, but will require a better understanding of amphibian immune responses and immunity genes targeted by natural selection (19). Although allele Q may be unique to L. yavapaiensis, we also detected significant heterozygote advantage, and selection for MHC heterozygosity is a strategy that could be implemented for any Bd-susceptible amphibian species. Here we have identified genetic determinants of chytridiomycosis resistance in natural populations of a declining amphibian, a critical step in developing genetically informed breeding programs for species recovery.
Methods
Experimental Bd Exposures.
Frogs were infected with Bd strain PsTr2004, an Arizona strain isolated from a Pseudacris triseriata individual collected from Coconino County, AZ (Universal Mobile Telecommunications System coordinates: 446716E, 3871380N), in 2003. This locality is geographically distinct from our five populations, minimizing the potential that they were locally adapted to this Bd strain. Frogs from each replicate group were split into two cages; one cage was seeded with 1 × 105 PsTr2004 Bd zoospores in 1 mL of tryptone/hydrolyzed gelatin/lactose (TGhL) broth and the other cage was sham-infected with 1 mL of TGhL broth only. The film of water/TGhL/Bd or water/TGhL covering the floor of each cage was left undisturbed for 24 h, and a new film of dechlorinated laboratory water was added to each cage daily. Cages were rotated daily to avoid the potential for microclimatic effects. Frogs were weighed immediately before infection and at death or the end of the experiment, whichever came first.
Infection Intensity Over Time.
To quantify Bd infections, we used sterile swabs and followed standard amphibian swabbing protocols (58). Bd DNA was extracted using PrepMan Ultra, and we used quantitative PCR (59) to determine Bd infection intensity. Before experiments began, we tested individuals for preexisting Bd infection at all three developmental stages (eggs, larvae, and metamorphs). One day before experimental Bd infections, we assayed all metamorphs for Bd infection. Frogs were handled with unused latex gloves to prevent Bd transmission. Following Bd exposures, we quantified infection intensity for each surviving replicate group on 1, 3, 7, 14, 21, 28, and 35 DPI.
Statistical Analysis of Survival.
We analyzed survival data by using the Cox proportional-hazards model (60), which assumes an underlying function describing how hazard changes over time and fits effect parameters using Cox likelihood. Survival was censored, meaning that individuals may die after the end of the study period. To identify potential predictors, we first performed a univariate analysis for each variable and each interaction among variables. We performed log-rank tests of equality across strata for categorical variables (replicate, clutch, source population, number of cage mates, and MHC heterozygosity), and univariate Cox proportional-hazards regression for continuous variables (initial mass, change in mass, and MII). Predictors with a P value of 0.1 or less in the univariate analyses were included as potential effect parameters in the final model. Significant P values were determined by using sequential Bonferroni correction (61). We tested whether infection intensities were significantly different from zero for survivors and nonsurvivors by using Wilcoxon signed-rank tests. Tests were performed in JMP software, version 9.0 (SAS).
Screening of Microsatellite Loci.
We genotyped all Bd-infected individuals at 14 previously characterized microsatellite loci (39). DNA was extracted by using a 5% Chelex 100 solution with 0.5 μg proteinase K per sample. Chelex extractions were incubated at 55 °C for 120 min and 99 °C for 10 min. PCR amplification was performed under the following conditions: 5 min initial denaturation at 94 °C; 35 cycles of 1 min denaturing at 94 °C, 1 min annealing at primer-specific annealing temperatures, 1 min extension at 72 °C, and a final extension of 75 °C for 5 min. Amplified products with different labels or nonoverlapping size ranges were multiplexed and run on a 3730 Genetic Analyzer (Applied Biosystems). Fragment sizes were determined by comparison with a LIZ-500 standard by using GeneMapper, version 3.5 (Applied Biosystems).
Population Structure.
Genetic structuring among populations was inferred for all Bd-infected individuals. We used the program Structure, version 2.3 (40), to identify the most likely number of genetic clusters (K) present and the proportion of membership in those clusters for each Bd-infected clutch. This method uses Bayesian assignment techniques to identify clusters of genetically similar individuals from multilocus genotypes without prior knowledge of their population affinities. Twenty independent runs were performed for each value of K ranging from 1 to 12, with a burn-in of 1 million Markov chain Monte Carlo iterations and a data collection period of 3 million iterations.
MHC Amplification, Cloning, and Sequencing.
We extracted genomic DNA from toe-clips of Bd-infected individuals by using Qiagen DNeasy kits. We used a degenerate MHC class IIB forward primer (MHC-F) (31) and an intron-specific reverse primer (B1intron2_R) (41) to amplify 246 bp of exon 2 and 189 bp of the adjacent 3′ flanking intron. This primer pair was designed to amplify alleles from a single locus and ensure that only orthologues were compared, because at least two MHC class IIB loci are expressed in L. yavapaiensis (41). Subsequent analyses included the exon 2 coding region only. Amplifications were performed for 35 cycles at 95 °C for 50 s, 60 °C for 45 s, and 72 °C for 1 min. We cloned PCR products into the Promega pGEM T vector and transformed recombinant DNA into Invitrogen TOP-10 Escherichia coli. Cells were grown on Luria agar plates for 18 to 22 h at 37 °C. We used blue/white screening to choose eight to 32 clones from each transformation and amplified them by using M13 primers with standard reaction conditions. PCR products were purified using an alkaline phosphatase-exonuclease reaction and sequenced on an ABI 3730 sequencer using Big Dye (version 3.1) chemistry. We discarded MHC sequences recovered singly with two or fewer nucleotide polymorphisms to other sequences, attributing these small differences to PCR or cloning error. After discarding these sequences, no more than two alleles were recovered from any individual. We used translated amino acid queries in GenBank to confirm MHC class IIB homology and aligned sequences in Sequencher, version 4.10 (Gene Codes).
Genealogy Reconstruction.
We tested the alignment of MHC sequences for recombination by using Genetic Algorithm for Recombination Detection (62), and detected no recombination; therefore, we performed an unrooted maximum-likelihood (ML) analysis. Model parameters were determined using Akaike and Bayesian information criteria in jModeltest, version 0.1.1 (63). The best-fit model (HKY85) was used in a ML analysis in PAUP* (Sinauer) using the heuristic search option, TBR branch swapping, MulTrees option in effect, and a single neighbor-joining tree as a starting topology. Node support was estimated from 1,000 bootstrap replicates.
Tests of Selection.
We ran tests of selection using the HyPhy software package (64) with the ML genealogy as our input tree. We used the partitioning approach for robust inference of selection method to test for positive selection across the alignment, and the evolutionary fingerprinting method to infer positive selection rate classes and selection intensity. We tested for residue-specific selection in PBR lineages using three ML methods: single likelihood ancestral counting (SLAC), fixed-effects likelihood (FEL), and random-effects likelihood (REL) (64).
Supplementary Material
Acknowledgments
We thank S. Bogdanowicz, M. Lenker, K. Kiemnec, J. Q. Richmond, M. Schlaepfer, E. Davidson, V. Miera, R. Retallick, and E. Retallick for laboratory and field support, and A. Longo, G. Becker, and J. Cavatorta for constructive comments on the manuscript. This work was funded by Cornell University, Population and Evolutionary Process National Science Foundation (NSF) Grant DEB-0815315 (to K.R.Z.), and NSF Doctoral Dissertation Improvement Grant DEB-0909013 (to A.E.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. JN638850–JN638882).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1106893108/-/DCSupplemental.
References
- 1.Stuart SN, et al. Status and trends of amphibian declines and extinctions worldwide. Science. 2004;306:1783–1786. doi: 10.1126/science.1103538. [DOI] [PubMed] [Google Scholar]
- 2.Lips KR, et al. Emerging infectious disease and the loss of biodiversity in a Neotropical amphibian community. Proc Natl Acad Sci USA. 2006;103:3165–3170. doi: 10.1073/pnas.0506889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skerratt LF, et al. Spread of chytridiomycosis has caused the rapid global decline and extinction of frogs. EcoHealth. 2007;4:125–134. [Google Scholar]
- 4.Wake DB, Vredenburg VT. Colloquium paper: are we in the midst of the sixth mass extinction? A view from the world of amphibians. Proc Natl Acad Sci USA. 2008;105(suppl 1):11466–11473. doi: 10.1073/pnas.0801921105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rohr JR, Raffel TR, Romansic JM, McCallum H, Hudson PJ. Evaluating the links between climate, disease spread, and amphibian declines. Proc Natl Acad Sci USA. 2008;105:17436–17441. doi: 10.1073/pnas.0806368105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradley GA, Rosen PC, Sredl MJ, Jones TR, Longcore JE. Chytridiomycosis in native Arizona frogs. J Wildl Dis. 2002;38:206–212. doi: 10.7589/0090-3558-38.1.206. [DOI] [PubMed] [Google Scholar]
- 7.Lips KR. Decline of a tropical montane amphibian fauna. Conserv Biol. 1998;12:106–117. [Google Scholar]
- 8.Crawford AJ, Lips KR, Bermingham E. Epidemic disease decimates amphibian abundance, species diversity, and evolutionary history in the highlands of central Panama. Proc Natl Acad Sci USA. 2010;107:13777–13782. doi: 10.1073/pnas.0914115107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pounds JA, et al. Widespread amphibian extinctions from epidemic disease driven by global warming. Nature. 2006;439:161–167. doi: 10.1038/nature04246. [DOI] [PubMed] [Google Scholar]
- 10.Lips KR, Reeve JD, Witters LR. Ecological traits predicting amphibian population declines in Central America. Conserv Biol. 2003;17:1078–1088. [Google Scholar]
- 11.Retallick RWR, Miera V. Strain differences in the amphibian chytrid Batrachochytrium dendrobatidis and non-permanent, sub-lethal effects of infection. Dis Aquat Organ. 2007;75:201–207. doi: 10.3354/dao075201. [DOI] [PubMed] [Google Scholar]
- 12.Rosenblum EB, et al. Genome-wide transcriptional response of Silurana (Xenopus) tropicalis to infection with the deadly chytrid fungus. PLoS ONE. 2009;4:e6494. doi: 10.1371/journal.pone.0006494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ribas L, et al. Expression profiling the temperature-dependent amphibian response to infection by Batrachochytrium dendrobatidis. PLoS ONE. 2009;4:e8408. doi: 10.1371/journal.pone.0008408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramsey JP, Reinert LK, Harper LK, Woodhams DC, Rollins-Smith LA. Immune defenses against Batrachochytrium dendrobatidis, a fungus linked to global amphibian declines, in the South African clawed frog, Xenopus laevis. Infect Immun. 2010;78:3981–3992. doi: 10.1128/IAI.00402-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tobler U, Schmidt BR. Within- and among-population variation in chytridiomycosis-induced mortality in the toad Alytes obstetricans. PLoS ONE. 2010;5:e10927. doi: 10.1371/journal.pone.0010927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shaw SD, et al. Experimental infection of self-cured Leiopelma archeyi with the amphibian chytrid Batrachochytrium dendrobatidis. Dis Aquat Organ. 2010;92:159–163. doi: 10.3354/dao02227. [DOI] [PubMed] [Google Scholar]
- 17.Woodhams DC, et al. Mitigating amphibian disease: strategies to maintain wild populations and control chytridiomycosis. Front Zool. 2011;8:8–31. doi: 10.1186/1742-9994-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richmond JQ, Savage AE, Rosenblum EB, Zamudio KR. Towards immunogenetic studies of amphibian chytridiomycosis: Linking innate and acquired immunity. Bioscience. 2009;59:311–320. [Google Scholar]
- 19.Bernatchez L, Landry C. MHC studies in nonmodel vertebrates: what have we learned about natural selection in 15 years? J Evol Biol. 2003;16:363–377. doi: 10.1046/j.1420-9101.2003.00531.x. [DOI] [PubMed] [Google Scholar]
- 20.Jones EY, Fugger L, Strominger JL, Siebold C. MHC class II proteins and disease: A structural perspective. Nat Rev Immunol. 2006;6:271–282. doi: 10.1038/nri1805. [DOI] [PubMed] [Google Scholar]
- 21.Hughes AL, Yeager M. Natural selection at major histocompatibility complex loci of vertebrates. Annu Rev Genet. 1998;32:415–435. doi: 10.1146/annurev.genet.32.1.415. [DOI] [PubMed] [Google Scholar]
- 22.Ohta Y, Goetz W, Hossain MZ, Nonaka M, Flajnik MF. Ancestral organization of the MHC revealed in the amphibian Xenopus. J Immunol. 2006;176:3674–3685. doi: 10.4049/jimmunol.176.6.3674. [DOI] [PubMed] [Google Scholar]
- 23.Braciale TJ, et al. Antigen presentation pathways to class I and class II MHC-restricted T lymphocytes. Immunol Rev. 1987;98:95–114. doi: 10.1111/j.1600-065x.1987.tb00521.x. [DOI] [PubMed] [Google Scholar]
- 24.Kaufman JF, Flajnik MF, Du Pasquier L, Riegert P. Xenopus MHC class II molecules. I. Identification and structural characterization. J Immunol. 1985;134:3248–3257. [PubMed] [Google Scholar]
- 25.Du Pasquier L, Flajnik MF. Expression of MHC class II antigens during Xenopus development. Dev Immunol. 1990;1:85–95. doi: 10.1155/1990/67913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carrillo-Farga J, Castell A, Pérez A, Rondán A. Langerhans-like cells in amphibian epidermis. J Anat. 1990;172:39–45. [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y, Kasahara M, Rumfelt LL, Flajnik MF. Xenopus class II A genes: Studies of genetics, polymorphism, and expression. Dev Comp Immunol. 2002;26:735–750. doi: 10.1016/s0145-305x(02)00034-4. [DOI] [PubMed] [Google Scholar]
- 28.Sato K, Flajnik MF, Du Pasquier L, Katagiri M, Kasahara M. Evolution of the MHC: Isolation of class II beta-chain cDNA clones from the amphibian Xenopus laevis. J Immunol. 1993;150:2831–2843. [PubMed] [Google Scholar]
- 29.Sammut B, et al. Axolotl MHC architecture and polymorphism. Eur J Immunol. 1999;29:2897–2907. doi: 10.1002/(SICI)1521-4141(199909)29:09<2897::AID-IMMU2897>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 30.Bos DH, DeWoody JA. Molecular characterization of major histocompatibility complex class II alleles in wild tiger salamanders (Ambystoma tigrinum) Immunogenetics. 2005;57:775–781. doi: 10.1007/s00251-005-0038-5. [DOI] [PubMed] [Google Scholar]
- 31.Hauswaldt JS, Stuckas H, Pfautsch S, Tiedemann R. Molecular characterization of MHC class II in a nonmodel anuran species, the fire-bellied toad Bombina bombina. Immunogenetics. 2007;59:479–491. doi: 10.1007/s00251-007-0210-1. [DOI] [PubMed] [Google Scholar]
- 32.May S, Zeisset I, Beebee TJC. Larval fitness and immunogenetic diversity in chytrid-infected and uninfected natterjack toad (Bufo calamita) populations. Conserv Genet. 2011;12:805–811. [Google Scholar]
- 33.Langefors A, Lohm J, Grahn M, Andersen O, von Schantz T. Association between major histocompatibility complex class IIB alleles and resistance to Aeromonas salmonicida in Atlantic salmon. Proc Biol Sci. 2001;268:479–485. doi: 10.1098/rspb.2000.1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Penn DJ, Damjanovich K, Potts WK. MHC heterozygosity confers a selective advantage against multiple-strain infections. Proc Natl Acad Sci USA. 2002;99:11260–11264. doi: 10.1073/pnas.162006499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wegner KM, Kalbe M, Kurtz J, Reusch TB, Milinski M. Parasite selection for immunogenetic optimality. Science. 2003;301:1343. doi: 10.1126/science.1088293. [DOI] [PubMed] [Google Scholar]
- 36.Savage AE, Sredl MJ, Zamudio KR. Disease dynamics vary spatially and temporally in a North American amphibian. Biol Conserv. 2011;144:1910–1915. [Google Scholar]
- 37.Schlaepfer MA, Sredl MJ, Rosen PC, Ryan MJ. High prevalence of Batrachochytrium dendrobatidis in wild populations of Lowland Leopard Frogs Rana yavapaiensis in Arizona. EcoHealth. 2007;4:421–427. [Google Scholar]
- 38.Pritchard JK, Stephens M, Rosenberg NA, Donnelly P. Association mapping in structured populations. Am J Hum Genet. 2000;67:170–181. doi: 10.1086/302959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Savage AE, Jaeger JR. Isolation and characterization of microsatellite markers in the lowland leopard frog (Rana yavapaiensis) and the relict leopard frog (R. onca), two declining frogs of the North American desert southwest. Mol Ecol Resour. 2009;9:199–202. doi: 10.1111/j.1755-0998.2008.02343.x. [DOI] [PubMed] [Google Scholar]
- 40.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kiemnec-Tyburczy KM, Richmond JQ, Savage AE, Zamudio KR. Selection, trans-species polymorphism, and locus identification of major histocompatibility complex class IIβ alleles of New World ranid frogs. Immunogenetics. 2010;62:741–751. doi: 10.1007/s00251-010-0476-6. [DOI] [PubMed] [Google Scholar]
- 42.Kimura M. Preponderance of synonymous changes as evidence for the neutral theory of molecular evolution. Nature. 1977;267:275–276. doi: 10.1038/267275a0. [DOI] [PubMed] [Google Scholar]
- 43.Tong JC, et al. Modeling the bound conformation of Pemphigus vulgaris-associated peptides to MHC Class II DR and DQ alleles. Immunome Res. 2006;2:1–10. doi: 10.1186/1745-7580-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li WH, Wu CI, Luo CC. A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Mol Biol Evol. 1985;2:150–174. doi: 10.1093/oxfordjournals.molbev.a040343. [DOI] [PubMed] [Google Scholar]
- 45.Apanius V, Penn D, Slev PR, Ruff LR, Potts WK. The nature of selection on the major histocompatibility complex. Crit Rev Immunol. 1997;17:179–224. doi: 10.1615/critrevimmunol.v17.i2.40. [DOI] [PubMed] [Google Scholar]
- 46.Zuk M, Stoehr AM. Immune defense and host life history. Am Nat. 2002;160(suppl 4):S9–S22. doi: 10.1086/342131. [DOI] [PubMed] [Google Scholar]
- 47.Gandon S. Local adaptation and the geometry of host–parasite coevolution. Ecol Lett. 2002;5:246–256. [Google Scholar]
- 48.Lipsitch M, Herre EA, Nowak MA. Host population structure and the evolution of virulence – a law-of-diminishing-returns. Evolution. 1995;49:743–748. doi: 10.1111/j.1558-5646.1995.tb02310.x. [DOI] [PubMed] [Google Scholar]
- 49.Regoes RR, Nowak MA, Bonhoeffer S. Evolution of virulence in a heterogeneous host population. Evolution. 2000;54:64–71. doi: 10.1111/j.0014-3820.2000.tb00008.x. [DOI] [PubMed] [Google Scholar]
- 50.Zhan J, Mundt CC, Hoffer ME, McDonald BA. Local adaptation and effect of host genotype on the rate of pathogen evolution: An experimental test in a plant pathosystem. J Evol Biol. 2002;15:634–647. [Google Scholar]
- 51.Morgan AD, Buckling A. Relative number of generations of hosts and parasites does not influence parasite local adaptation in coevolving populations of bacteria and phages. J Evol Biol. 2006;19:1956–1963. doi: 10.1111/j.1420-9101.2006.01148.x. [DOI] [PubMed] [Google Scholar]
- 52.Morgan JAT, et al. Population genetics of the frog-killing fungus Batrachochytrium dendrobatidis. Proc Natl Acad Sci USA. 2007;104:13845–13850. doi: 10.1073/pnas.0701838104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gandon S, Michalakis Y. Local adaptation, evolutionary potential and host-parasite coevolution: interactions between migration, mutation, population size and generation time. J Evol Biol. 2002;15:451–462. [Google Scholar]
- 54.Råberg L, Sim D, Read AF. Disentangling genetic variation for resistance and tolerance to infectious diseases in animals. Science. 2007;318:812–814. doi: 10.1126/science.1148526. [DOI] [PubMed] [Google Scholar]
- 55.Rachowicz LJ, et al. Emerging infectious disease as a proximate cause of amphibian mass mortality. Ecology. 2006;87:1671–1683. doi: 10.1890/0012-9658(2006)87[1671:eidaap]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 56.Briggs CJ, Vredenburg VT, Knapp RA, Rachowicz LJ. Investigating the population-level effects of chytridiomycosis, an emerging infectious disease of amphibians. Ecology. 2005;86:3149–3159. [Google Scholar]
- 57.Mendelson JR, 3rd, et al. Biodiversity. Confronting amphibian declines and extinctions. Science. 2006;313:48–51. doi: 10.1126/science.1128396. [DOI] [PubMed] [Google Scholar]
- 58.Hyatt AD, et al. Diagnostic assays and sampling protocols for the detection of Batrachochytrium dendrobatidis. Dis Aquat Organ. 2007;73:175–192. doi: 10.3354/dao073175. [DOI] [PubMed] [Google Scholar]
- 59.Boyle DG, Boyle DB, Olsen V, Morgan JAT, Hyatt AD. Rapid quantitative detection of chytridiomycosis (Batrachochytrium dendrobatidis) in amphibian samples using real-time Taqman PCR assay. Dis Aquat Organ. 2004;60:141–148. doi: 10.3354/dao060141. [DOI] [PubMed] [Google Scholar]
- 60.Cox DR. Regression models and life tables. J R Stat Soc B. 1972;34:187–220. [Google Scholar]
- 61.Rice WR. Analyzing tables of statistical tests. Evolution. 1989;43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- 62.Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SDW. Automated phylogenetic detection of recombination using a genetic algorithm. Mol Biol Evol. 2006;23:1891–1901. doi: 10.1093/molbev/msl051. [DOI] [PubMed] [Google Scholar]
- 63.Posada D. jModelTest: Phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 64.Pond SL, Frost SDW, Muse SV. HyPhy: Hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.