Abstract
BDNF is produced from many transcripts that display distinct subcellular localization, suggesting that spatially restricted effects occur as a function of genetic and physiological regulation. Different BDNF 5′ splice variants give a restricted localization in the cell body or the proximal and distal compartments of dendrites; however, the functional consequences are not known. Silencing individual endogenous transcripts or overexpressing BDNF-GFP transcripts in cultured neurons demonstrated that whereas some transcripts (1 and 4) selectively affected proximal dendrites, others (2C and 6) affected distal dendrites. Moreover, segregation of BDNF transcripts resulted in a highly selective activation of the BDNF TrkB receptor. These studies indicate that spatial segregation of BDNF transcripts enables BDNF to differentially shape distinct dendritic compartments.
Keywords: development, neurotrophins, plasticity
Brain-derived neurotrophic factor (BDNF) is a neurotrophin with multifaceted functions such as survival, neurite outgrowth, synaptogenesis, and synaptic plasticity (1–3). In addition, BDNF induces dendritic sprouting in the presence of synaptic activity (4, 5), causes local instability in dendrites (6, 7), and increases spine density and dimension (8). Several levels of regulation of BDNF, including proteolytic processing (9, 10) and the use of distinct receptors and signaling cascades (11, 12), may explain how this neurotrophin exerts so many different functions. Another level of BDNF regulation is represented by the production of multiple transcripts. In the rat bdnf gene (Fig. 1A), 22 different transcripts are generated by 11 different 5′ untranslated regions (UTRs) that are encoded by 9 exons. Each 5′ exon is alternatively spliced to a downstream exon that contains the coding region of BDNF with a 3′UTR containing two potential polyadenylation signals (13). However, the functional consequences of multiple transcripts that encode the same protein are not understood.
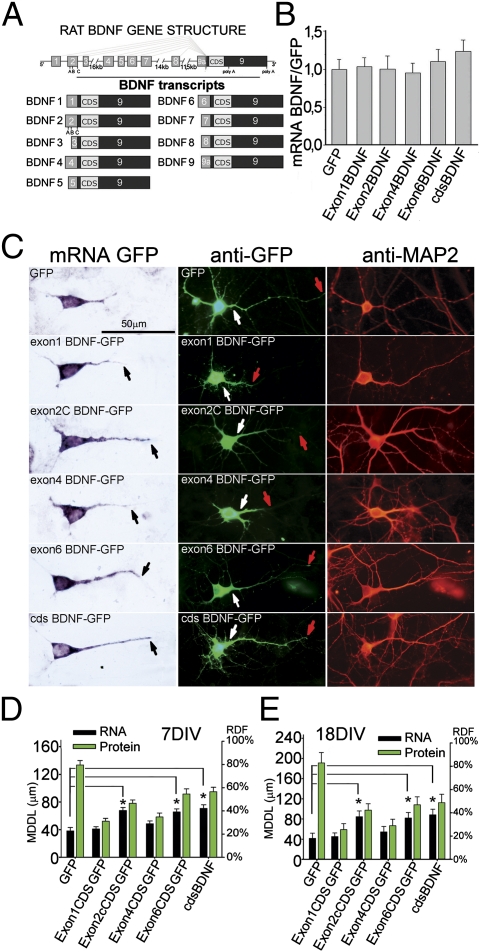
Fig. 1.
BDNF transcripts and protein cosegregate. (A) Structure of rat bdnf gene and transcripts. (B) Densitometric analysis of chimeric constructs in the somata of transfected hippocampal neurons (n = 100 neurons per bar). (C) (Left) In situ hybridization on 7DIV cultures of hippocampal neurons transfected with 5′UTR-cdsBDNF-GFP constructs. (Center) Fluorescence of BDNF-GFP proteins. (Right) MAP2 staining. Black arrows show the end point of MDDL; white and red arrows show respectively the starting and ending points of dendritic BDNF-GFP protein labeling. (D and E) Quantification of maximal distance reached by BDNF-GFP mRNAs (black bars) and BDNF-GFP protein (green bars) in dendrites (n = 40 neurons per bar; error whiskers indicate SE) expressed both in MDDL and RDF in 7- (D) and 18-d-old (E) neurons. (*P < 0.05; ANOVA.)
We recently carried out a densitometric analysis of the subcellular distribution of the most abundant BDNF transcripts in the hippocampus and cortex during epilepsy. We found that exons 2 and 6 localized into distal dendrites following 3 h of status epilepticus, whereas exons 1 and 4 were restricted to the soma even after this strong neuronal activation (14, 15). On this basis, we proposed a hypothesis, the “spatial code hypothesis of BDNF transcripts,” in which different BDNF splice variants, through the spatial segregation of their mRNA, represent a code to direct the protein to either the soma or proximal or distal dendrites (14, 16). In this study, we have directly tested this hypothesis by measuring the morphology of dendrites after silencing or overexpressing specific BDNF splice variants to change the local availability of BDNF.
Results
BDNF Transcripts and Protein Cosegregate.
Because all BDNF transcripts encode for the same BDNF protein, it is not possible to assess by a simple immunohistochemical analysis whether the protein generated by different transcripts displays different localization in vivo. To address this question, we analyzed the subcellular distribution of chimeric BDNF-GFP mRNAs and that of the corresponding proteins in cultured hippocampal neurons using previously described constructs with the coding sequence (cds) of BDNF alone or preceded by one of the BDNF 5′UTR exons, 1, 2C, 4, or 6, without 3′UTR (17). Constructs with exons 2C or 6 were previously shown to have a constitutive distal dendritic localization, whereas those with exons 1 or 4 remain restricted in the cell soma and proximal dendrites (17). To detect the localization of chimeric transcripts and proteins, in situ hybridization and immunocytochemistry, respectively, were carried out to label the common GFP domain in day in vitro (DIV)7 neurons transfected at DIV4 or DIV18 neurons transfected at DIV15 (Fig. 1 C–E). The different constructs with exons 1, 2C, 4, or 6 showed equal levels of mRNA expression as determined by densitometric analysis of the signals on cell somata (Fig. 1B), and their subcellular localization matched that of the corresponding GFP protein (Fig. 1C). To confirm these results, the dendritic distribution of chimeric BDNF-GFP transcripts and protein was quantified in two ways. First, we measured the distance (in microns) from the soma at which the in situ or immunofluorescence signal reached and remained below background levels for the remainder of the dendrite (maximal distance of dendritic labeling; MDDL). Second, the relative dendritic filling index (RDF) was determined by dividing the MDDL by the mean length of the dendrites identified by MAP2 immunostaining (Fig. 1C and Fig. S1A). The mean length of the apical dendrites at DIV7 or DIV18 was not significantly affected by transfection with any of the different BDNF-GFP chimera (Fig. S1B). At both DIV7 and DIV18, transcripts with exons 1 and 4 segregated within the first 40–50 μm from the soma (20–30% RDF), whereas exon 2C or 6 or total BDNF mRNAs were detected up to 70 μm from the soma at DIV7 (45% RDF) or to 95 μm (40% RDF) at DIV18 (Fig. 1 D and E). The chimeric BDNF-GFP proteins showed a strikingly similar distribution to that of their corresponding mRNAs, although the proteins were slightly more distal than the mRNAs (Fig. 1 D and E). This effect was general and may be due to a higher mobility of the secretory vesicles containing chimeric BDNF-GFP versus their mRNAs. Indeed, since BDNF contains very strong signal peptides, it is entirely processed into the secretory pathway.
To evaluate whether the 3′UTR sequence is relevant to the expression of BDNF constructs, we determined the dendritic distribution of selected constructs bearing a short or long BDNF 3′UTR sequence (18) (Fig. S1C). In DIV7 neurons under basal conditions, GFP-3′UTR short and GFP-3′UTR long mRNAs were located in the soma and proximal dendrites (46, 42 μm from the soma) and, in response to KCl, were both targeted to distal dendrites (92, 86 μm). Chimeric exon 1 and exon 6 full-length transcripts (with either the short or long 3′UTR) and proteins were localized in proximal dendrites (Fig. S1C), but after stimulation with 10 mM KCl for 3 h, exon 6, but not exon 1, mRNA and protein were able to reach the distal dendrites (Fig. S1C). These results indicate that (i) both the short and the long 3′UTR could mediate activity-dependent dendritic localization of the mRNA, and (ii) the ultimate destination of BDNF mRNAs is dictated by the presence of 5′UTR exons, with either dendritic permissive (e.g., exon 6) or soma retention (e.g., exon 1) properties. The behavior of full-length constructs was identical to that observed at both DIV7 and DIV18 (Fig. S1E) for the endogenous BDNF mRNAs analyzed using transcript-specific probes used previously in cortex (15) and hippocampus (14). Indeed, endogenous BDNF transcripts with exon 1, 2, 4, or 6 were located in the soma and proximal dendrites in unstimulated cultures, and KCl depolarization for 3 h induced targeting in distal dendrites of only exons 2 and 6 or total BDNF (Fig. S1 D and E). BDNF mRNA distribution in unstimulated cultures was similar between DIV7 and DIV18 but, in younger neurons, all BDNF transcripts were found in a more proximal location than in older neurons, showing an MDDL range of 36–38 μm at DIV7 and 44–78 μm at DIV18 (Fig. S1E). Of note, the chimeric protein derived from the exon 6 construct without a 3′UTR had a distal dendritic localization under basal conditions (Fig. 1 D and E), similar to the protein derived from full-length exon 6 (with either 3′UTR short or long) after stimulation (Fig. S1C). We conclude that truncated chimeric transcripts missing the 3′UTR are constitutively segregated in different cellular domains and therefore can be used to study the effects of different BDNF transcripts on dendrite shape.
Demonstration of Local Translation of BDNF mRNAs in Dendrites.
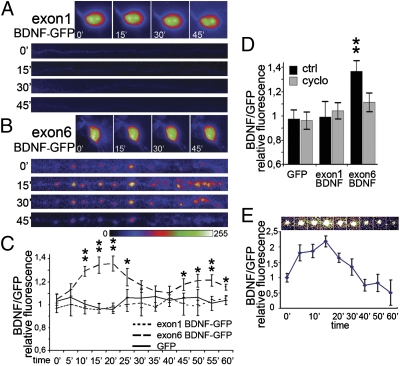
The cosegregation of BDNF-GFP mRNAs and proteins suggests that these proteins may be translated locally. However, a formal demonstration that BDNF mRNA targeted to dendrites is locally translated has been lacking. Therefore, we tested whether different BDNF-GFP constructs could be translated in dendrites separated from the cell body in response to depolarization with 10 mM KCl. For exon 1 BDNF-GFP, fluorescence puncta were clearly visible in the cell soma at time 0 min (Fig. 2A). Exon 1 BDNF-GFP fluorescence was nearly absent in severed dendrites during 60 min of KCl treatment (Fig. 2A). In contrast, in severed dendrites, exon 6 BDNF-GFP fluorescence intensity displayed a significant biphasic increase at 15–20 and 45–60 min of KCl treatment (P < 0.01; Fig. 2 B and C). The increase of exon 6 BDNF-GFP in dendrites was abolished by cycloheximide (Fig. 3D), indicating that local translation was affected, rather than spatial redistribution of preexisting protein. Similar results were observed for exon 4 and 2C constructs (Fig. S2). When single puncta of exon 6 BDNF-GFP were tracked in video time-lapse experiments, we found little lateral displacement (Fig. 2E), in agreement with previous studies on cdsBDNF-GFP (19). Individual puncta showed more than a twofold increase in fluorescence within 15 min of stimulation with KCl, followed by a downturn, suggesting a release by secretion (Fig. 2E). These results demonstrate that BDNF transcripts can be locally translated in dendrites.
Fig. 2.
Local translation of BDNF transcripts. Video time-lapse tracking of fluorescence in the soma and dendrites severed from the cell body of hippocampal neurons transfected with (A) exon1 BDNF-GFP and (B) exon6 BDNF-GFP and depolarized with 10 mM KCl. Images of dendrites show a straightened 150 μm-long segment. Proximal stump on the left. (C) Quantification of fluorescence in isolated dendrites depolarized with 10 mM KCl for the indicated times (0–60 min). (D) Protein synthesis inhibitor cycloheximide abolishes the fluorescence increase for exon6 BDNF-GFP in isolated dendrites. (E) Video time-lapse tracking of a single spot of exon6 BDNF-GFP fluorescence during KCl depolarization in a dendrite isolated from the soma, and quantification of exon6 BDNF-GFP fluorescence of 10 different puncta during KCl depolarization at the indicated time points (0–60 min). Quantitative data are expressed as the mean of 10 dendrites or somata for each condition. Error bars indicate SE. (*P < 0.05; **P < 0.01; Kruskal–Wallis one-way ANOVA.)
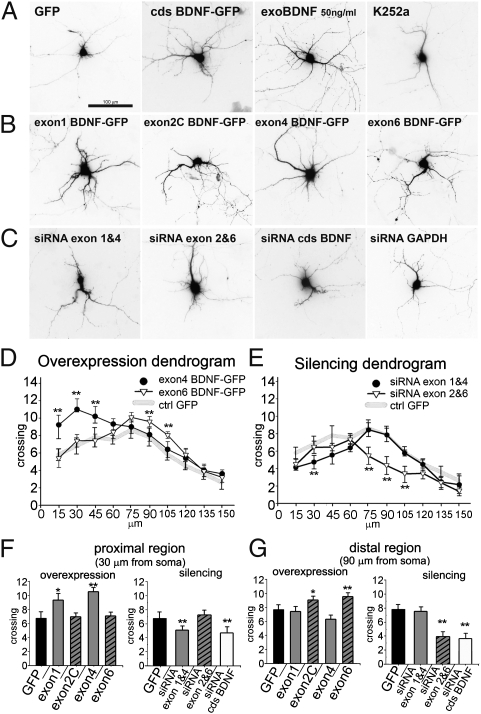
Fig. 3.
BDNF transcripts produce local effects on the number of dendritic branchings. Examples of 7-d-old neurons, visualized by GFP fluorescence 3 d after transfection with (A) GFP or cdsBDNF-GFP, treatment with 50 ng/mL BDNF or K252a, (B) the indicated BNDF-GFP transcripts, (C) siRNA against endogenous BDNF transcripts, or GAPDH. (D) Expression dendrograms displaying the complete Sholl analysis for exon4 BDNF-GFP (somatic) and exon6 BDNF-GFP (dendritic) constructs. (**P < 0.01 vs. control GFP; ANOVA; n = 50 neurons.) (E) A dendrogram showing the effects on dendritic crossings after silencing of endogenous transcripts. (**P < 0.01 vs. control GFP; n = 50 neurons.) (F and G) Detail of crossing dendrites at 30 μm (F) or 90 μm (G) from the soma in neurons treated as indicated. (*P < 0.05; **P < 0.01 vs. control GFP; n = 50 neurons.)
Segregated BDNF Variants Regulate Dendrite Complexity Locally.
To test the impact of endogenous, full-length BDNF mRNA variants on dendritic morphology, we analyzed the effects of downregulating BDNF splice variants by making use of transcript-specific small interfering RNAs (siRNAs). The efficiency of siRNAs directed to endogenous exons 1, 2C, 4, and 6 splice variants ranged between 60 and 80% in DIV7 rat hippocampal neurons that were transfected at DIV4 (Fig. S3 A and B). Neurons transfected with GFP only displayed a basal number of 6.81 ± 1.15 crossing dendrites at 30 μm from the soma and 7.74 ± 0.78 at 90 μm (Fig. 3 A, F, and G). Silencing of all endogenous BDNF transcripts with siRNA against the cds reduced the number and length of all dendrites (Fig. 3C), whereas silencing of the housekeeping gene GAPDH had no detectable effect (Fig. 3C and Fig. S3 A and C). Knockdown of endogenous exon 6 variant resulted in 30% reduction in the number of dendrites (P < 0.01) 75–105 μm from the soma, but had no discernible effect within 60 μm of the soma.
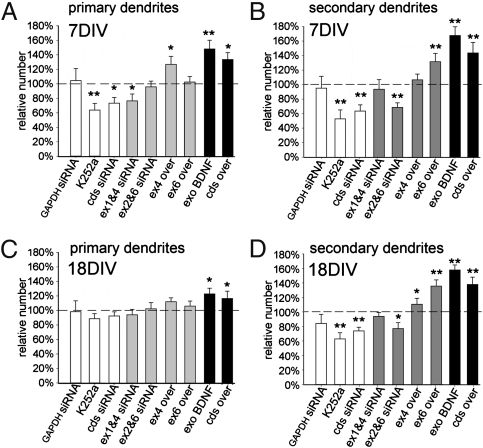
To test for the possibility of compensation due to cosegregation of different endogenous splice variants with similar subcellular distribution, we simultaneously silenced the two somatic variants 1 and 4, or the dendritic variants 2C and 6 (Fig. 3C). Co-silencing of endogenous somatic exons 1 and 4 resulted in a significant decrease (28% reduction) of dendrite crossings at 30 μm, but not at 90 μm, from the soma (Fig. 3 E–G), whereas co-silencing of endogenous dendritic exons 2C and 6 showed the opposite, that is, no effect at 30 μm and a 35% reduction of crossings at 90 μm (Fig. 3 E–G). To extend the findings obtained with silencing of endogenous BDNF variants, we carried out overexpression experiments using truncated BDNF-GFP chimeric constructs. Transfection with cdsBDNF-GFP led, 72 h later, to a 75% increase in the number of dendrite crossings at 30 μm (11.81 ± 1.7) and a 55% increase at 90 μm (11.97 ± 1.2). Similarly, treatment for 72 h with purified BDNF produced a marked growth of the whole dendritic tree, whereas treatment with the K252a Trk inhibitor produced a reduction (Fig. 3A). In neurons expressing somatic exon 1 and 4 BDNF-GFP constructs, we found a higher number of primary, poorly ramified dendrites (Fig. 3B) with a significant increase in the number of crossings at 30 μm from the soma (29% increase for exon 1, P < 0.05; 37% for exon 4, P < 0.01; Fig. 3 D and F), but no effect beyond 60 μm from soma (Fig. 3 D and G). In contrast, transfection of dendritic exon 2C or 6 produced no effects up to 75 μm (Fig. 3 D and F), but gave a significant increase of dendritic crossings at 90 μm (20% increase for exon 2C, P < 0.05; 30% increase for exon 6, P < 0.01; Fig. 3 D and G). Taken together, these results support the conclusion that the dendritic BDNF splice variants regulate dendritic complexity in the periphery, whereas somatic transcripts do so in proximity to the soma. Sholl analysis provides information about which region of the dendritic tree is affected, but does not necessarily identify whether primary or secondary branches are selectively involved. To assess the impact of single BDNF variants on primary and secondary branching, neurons were treated at DIV4 and analyzed after DIV7. In these cultures, we found that the number of primary dendrites was influenced only by somatic variants 1 and 4, but not by dendritic variants 2C and 6 (Fig. 4A). Indeed, siRNA against endogenous somatic exons 1 and 4 induced a slight (−21%) but significant reduction (P < 0.05) in the number of primary dendrites, whereas co-silencing of dendritic transcripts 2 and 6 had no effect (Fig. 4A). Accordingly, the number of primary dendrites increased by 30% in neurons transfected with somatic variant exon 4, but not with the dendritic variant 6 (Fig. 4A). By contrast, secondary dendrites were unaffected by co-silencing of somatic exons 1 and 4 or up-regulation of exon 4, but were clearly regulated by dendritic exons 2C and 6 (Fig. 4B). GAPDH siRNA had no effect. Other controls with K252a or cdsBDNF siRNA treatment produced a significant reduction of both dendrite orders. As positive controls, treatment with extracellular BDNF or transfection with a plasmid encoding cdsBDNF produced a strong increase of both dendrite orders. Thus, at DIV7, somatic BDNF variants affect the number of primary dendrites, whereas dendritic BDNF variants affect the number of secondary dendrites. Because, during development in vitro, maturation of the dendritic arborization occurs between 14 and 21 DIV (20), the effects observed at DIV7 may not reflect plasticity in adult neurons. Therefore, these experiments were repeated at DIV18. We found that the number of primary dendrites was unaffected by silencing or overexpression of any BDNF splice variants tested (Fig. 4C). Conversely, the number of secondary dendrites was slightly increased by up-regulation of somatic exon 4. Strikingly, expression of the exon 6 dendritic splice variant produced a prominent increase in the number of secondary dendrites (+38%, P < 0.01), and co-silencing of endogenous exons 2C and 6 produced a 27% decrease (Fig. 4D). These results provide evidence that the number of primary dendrites in more mature neurons is not modified by changes in BDNF expression. However, secondary dendrites remain highly plastic in response to dendritic BDNF exon 6 transcript, with smaller effects produced by exon 4.
Fig. 4.
Primary and secondary dendrite modulation by BDNF variants in young and mature hippocampal neurons. Quantification of primary (A) and secondary (B) dendrites in young (7 d) hippocampal neurons in relation to control GFP-transfected neurons (100%). White bars represent control experiments using siRNA against GAPDH, or siRNA against cdsBDNF or treatment with K252a. Gray bars represent experiments using silencing or overexpression of BDNF variants. Black bars represent positive control experiments using exogenous BDNF (50 ng/mL) or cdsBDNF overexpression. Quantification of the number of primary (C) and secondary dendrites (D) in mature neurons (18 d). (*P < 0.05; **P < 0.01 vs. control GFP; ANOVA.)
Local Up-Regulation of BDNF Variants Results in Local Activation of TrkB.
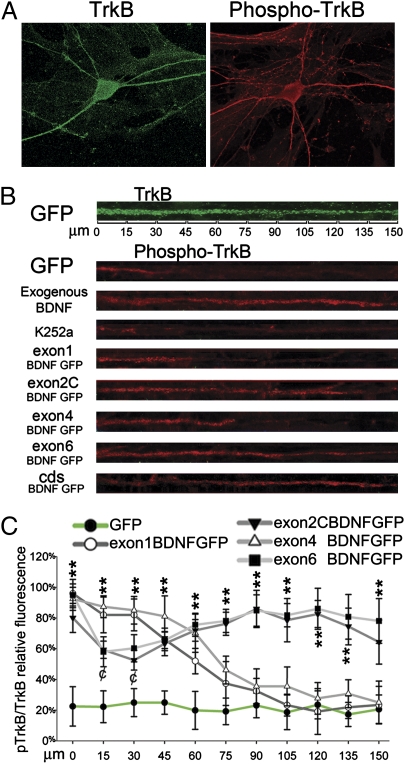
Since BDNF is known to be secreted from dendrites (21), we hypothesized that the selective effects of somatic/dendritic splicing variants may be the result of a spatially restricted activation of the TrkB receptor in the same cellular domain. In our cultures, TrkB is expressed in all neuronal compartments (Fig. 5A, Left). Using an antibody that selectively recognizes phosphorylated TrkB (pTrkB-Y816; Fig. 5A, Right) (22), we first confirmed that K252a, a potent inhibitor of Trk phosphorylation, abolished pTrkB staining, whereas addition of 50 ng/mL of purified BDNF in the culture media led to increased pTrkB immunoreactivity over the entire dendritic tree (Fig. 5 A and B). Neurons transfected with a plasmid encoding only GFP showed basal pTrkB immunoreactivity located mainly in the first 45 μm from the soma, which can be accounted for by the higher abundance of TrkB receptors in this domain (Fig. 5 A and B). Quantitative analysis of TrkB and pTrkB staining showed a constant amount (ca. 20%) of TrkB activation throughout the first 150 μm from the soma of neurons regardless of the local abundance of TrkB receptors (Fig. 5C). Overexpression of exon 1 or 4 transcript led to phosphorylation of 80–100% of TrkB receptors within the first 45 μm from the soma, 50% at 60 μm, and close to background levels (20%) at 90 μm from the soma (Fig. 5C). By contrast, in neurons overexpressing exon 2C or 6 BDNF variants, TrkB phosphorylation was 50% at 30 μm from the soma, reached 80% at 80 μm from the soma, and persisted at high levels beyond 100 μm from the soma (Fig. 5C). Thus, up-regulation of individual BDNF splice variants resulted in a striking, spatially restricted activation of TrkB receptors that correlates with the site of morphological changes observed in the dendritic tree.
Fig. 5.
Local up-regulation of BDNF variants results in local phosphorylation of TrkB. (A) Immunofluorescence for full-length TrkB (green) and phosphorylated TrkB (red) in two similar hippocampal neurons treated for 3 h with 50 ng/mL BDNF. (B) Straightened dendrites of neurons labeled with an anti-TrkB (green) or transfected with the indicated GFP constructs and labeled with anti-pTrkB (red). Treatment with the TrkB inhibitor K252a abolishes staining for pTrkB, whereas addition of 50 ng/mL BDNF led to labeling for pTrkB throughout the entire dendrite length. (C) Semiquantitative densitometric analysis of pTrkB fluorescence intensity in neurons transfected with the different constructs. (**P < 0.01; ANOVA.)
Discussion
Our findings indicate that the differential localization of BDNF transcripts, followed by their local translation and secretion, leads to a spatially restricted increase in TrkB receptor phosphorylation in dendrites as well as spatially restricted effects upon dendritic complexity. Accordingly, we propose that BDNF splice variants represent a spatial code used by neurons to selectively target the effects of BDNF to distinct dendritic compartments. Generation of multiple transcripts with different 5′ and 3′UTR segments, but sharing a common coding region, appears to be a general feature of neurotrophins (23, 24). Thus, this strategy might represent a general mechanism used to differentially regulate the levels of expression of a protein in different subcellular compartments.
In hippocampal pyramidal neurons, the spatial code for BDNF variants shows two main features: (i) the results define three principal regions, namely the cell soma (which includes the dendritic hillock, up to 20-μm distance from the soma), the proximal dendrites (i.e., primary dendrites up to 60 μm away from the soma), and the distal dendrites (i.e., primary and higher-order dendrites beyond 60 μm); and (ii) there is partial redundancy in that splice variants 1 and 4 control the architecture of the proximal compartment and exons 2 and 6 of the distal one. A similar segregation of BDNF mRNA variants has also been observed in hypothalamic (25) and cortical neurons (15), suggesting that our model can be applied to several neuronal types. A slight discrepancy in the actual distances or RDF values between this and our previous studies (15, 17) does not affect the general conclusions on the differential localization of BDNF transcripts, and is accounted for by different culture conditions and by the fact that only apical dendrites were measured here, whereas previous studies also included basal dendrites, which are generally shorter and tend to have a higher RDF. Our findings also indicate that the differential segregation of BDNF transcripts is able to generate localized changes in BDNF protein. This inference is consistent with a growing consensus that BDNF protein trafficking in dendrites is limited (19, 26), as opposed to the situation in axons where a fast anterograde transport of BDNF has been observed (19, 27). In this way, local translation of BDNF transcripts in dendrites results in local BDNF protein accumulation and secretion. This conclusion is corroborated by our finding that overexpression of localized BDNF splice variants results in local phosphorylation of TrkB. In this study, we further demonstrate that the mRNA containing the coding region alone is transported in dendrites and the protein localizes accordingly. This is important, because many previously published studies (more than 50) have used a construct only with the coding region of BDNF fused with GFP. One further aspect that must be considered is that the translational regulation of each transcript is expected to be different due to the intrinsic differences in the translatability of the different 5′UTRs. This issue, which is beyond the purpose of the present study, warrants further investigation.
BDNF Variants Regulate Local Dendritic Architecture.
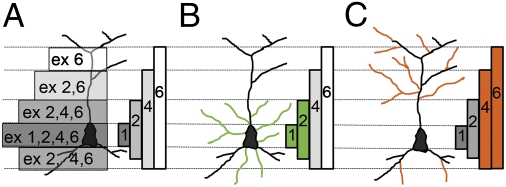
A large body of evidence supports the view that BDNF is a major extracellular cue involved in dendritic specification (28–30). However, it is unclear how a molecule that is expressed in all neuronal compartments may promote selective effects in different dendritic regions. Here we show that, in immature neurons, spatial segregation of BDNF transcripts mediates differential regulation of the number of primary and secondary dendrites in proximal and distal dendritic compartments (Fig. 6). In more mature neurons, which show limited changes in dendritic branching (31), we found that distal dendrites of secondary order retain sensitivity to local levels of BDNF determined by dendritic isoform exons 2 and 6 and, in part, by the exon 4 transcript. In contrast, proximal dendrites become insensitive to changes in local BDNF levels. It is worth noting that transfection for 24 h with the different transcripts was not sufficient to induce any significant change in the mean length of the primary dendrites or the dendrogram (17). Here we show that overexpression or silencing for 72 h of the different alternatively spliced BDNF transcripts affects the relative number of primary/secondary dendrites or of their branching but not their length. The time course of this phenomenon is consistent with the molecular processes of dendritic rearrangements, which require several days (30). In addition, this finding clearly indicates that up-regulation of a single BDNF isoform preferentially affects the relative number of dendrites, rather than their length. During development, BDNF 1, 2, 4, and 6 splice variants increase rapidly from postnatal day (P)0 to P7 in the rat hippocampus and reach a plateau at P14–P35, after which exon 4 and 6 transcripts decrease (32). Significantly, exon 4 and 6 variants showed a prominent effect on secondary dendrites in mature cultures (Fig. 4D), suggesting that limiting amounts of these transcripts might regulate plasticity of distal dendrites into adulthood. A study of conditional bdnf KO adult mice found a significant decrease in dendritic complexity and number of mushroom spines on distal apical dendrites of hippocampal neurons (33). This finding can be reconciled by our data showing that only distal, secondary dendrites, but not proximal dendrites, on mature neurons (Fig. 4) are sensitive to changes in BDNF levels.
Fig. 6.
Model of the BDNF spatial code in hippocampal pyramidal neurons. Localization of morphological effects induced by up-regulation of segregated BDNF transcripts. (A) Gradient of expression of the BDNF protein generated by the different transcripts. (B) In young but not mature neurons, proximal BDNF transcripts containing exons 1 and 4 affect dendrite architecture close to the soma. In green are shown dendrites induced by expression of BDNF transcripts containing exon 1 or 4. (C) In both young and mature neurons, distal dendritic transcripts regulate the morphology of the periphery of the dendritic tree. In orange are shown dendrites induced by expression of exon 2C or 6 BDNF transcripts.
Model of BDNF mRNA Trafficking.
It is becoming increasingly clear that mRNA transport in glial and neuronal processes depends upon multiple dendritic targeting elements (34, 35). These targeting elements were mainly found in the 3′UTR of the transported mRNAs but are also present in the coding region of some dendritic messenger RNAs (36–38). We previously showed that the coding region of BDNF contains a constitutively active dendritic targeting signal mediated by the RNA-binding protein translin (17). Of note, a cis-recognition sequence for translin was also found in the coding region of CaMKII-α. Moreover, translin resulted to be directly involved in the dendritic targeting process of CamKII-α mRNAs (37). In our previous studies, we provided evidence that the 3′UTR long can be targeted to distal dendrites in response to electrical activity (17). Here we further show that even the 3′UTR short displays dendritic targeting in response to KCl depolarization in vitro. However, we also demonstrate that the conjunction of either the 3′UTR short or long with a nonpermissive 5′UTR sequence such as, for instance, exon 1 results in a complete suppression of dendritic targeting. These results suggest a need to revise a previously proposed hypothesis that all BDNF mRNAs containing the 3′UTR short are segregated to the soma, whereas those with the 3′UTR long are targeted to dendrites (18). This conclusion did not fully consider the effects of neuronal activity or the role of the 5′UTR sequences. Hence, the emerging unified model that takes into account both 5′ and 3′UTR elements in a spatial code can represent an activity-dependent mechanism for dendritic targeting. This unified mechanism could include the role of constitutively active dendritic targeting signals in the cds and the use of different 5′UTRs acting as “selectors” for the final subcellular destination.
Implications of BDNF Spatial Code for Neuropsychiatric Diseases.
Antidepressants, antipsychotic drugs, and physical exercise regulate transcription of selected BDNF splice variants in restricted brain areas (39–41). The spatial code hypothesis predicts that these treatments may specifically up-regulate BDNF transcripts in one region of the dendritic tree. Since GABAergic, monoaminergic, and glutamatergic terminations form spatially segregated synaptic contacts on the soma or proximal or distal dendritic compartments, respectively (42, 43), selective production of specific BDNF splice variants may result in activation and reinforcement of specific neural networks. Thus, defining the BDNF splice variants that are critical for mediating the therapeutic effects of pharmacological or behavioral treatments may provide key information about the neuronal circuits involved in this response.
Materials and Methods
Hippocampal Cell Cultures.
Dissociated primary cultures of hippocampal neurons from embryonic day 18 (E18) rats were prepared from Sprague–Dawley rats as described previously (44) and maintained in Neurobasal media (Invitrogen) with B27 supplement (Invitrogen) and l-glutamine (0.5 mM; Euroclone) (details in SI Materials and Methods).
Cell Transfections and Treatments.
Chimeric BDNF-GFP construct preparation and transfections were described previously (17) (details in SI Materials and Methods).
RNA Interference.
RNAi “pools” against each transcript of BDNF were generated using Silencer siRNA Mixture Kit (Ambion) (details in SI Materials and Methods). Primers and PCR program details are reported in Tables S1–S3.
Riboprobe Preparation.
GFP riboprobe preparation was performed as previously described (17) (details in SI Materials and Methods).
In Situ Hybridization and Immunocytofluorescence.
In situ hybridization on cultures was performed as described (17). See SI Materials and Methods for details.
Quantitative Imaging Analysis and Statistics.
Images of labeled cultures were acquired with a CCD camera (Nikon; ADX-1200) on a Nikon E800 microscope and analyzed with Image-ProPlus (Media Cybernetics) or with a 63× oil objective on a Bio-Rad confocal system (MRC-1024) on a BX50WI Olympus microscope. See SI Materials and Methods for details.
Supplementary Material
Acknowledgments
We thank Jay Baraban for critically reading the manuscript. This work was supported by Telethon-GGP08258 and Compagnia San Paolo (E.T.), and by National Institutes of Health Grants NS21072 and HD23315 (to M.V.C.). G.B. was supported by Sanofi-Aventis and SIBioC.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. S.R.J. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1014168108/-/DCSupplemental.
References
- 1.Casaccia-Bonnefil P, Gu C, Chao MV. Neurotrophins in cell survival/death decisions. Adv Exp Med Biol. 1999;468:275–282. doi: 10.1007/978-1-4615-4685-6_22. [DOI] [PubMed] [Google Scholar]
- 2.McAllister AK, Katz LC, Lo DC. Neurotrophins and synaptic plasticity. Annu Rev Neurosci. 1999;22:295–318. doi: 10.1146/annurev.neuro.22.1.295. [DOI] [PubMed] [Google Scholar]
- 3.Huang EJ, Reichardt LF. Neurotrophins: Roles in neuronal development and function. Annu Rev Neurosci. 2001;24:677–736. doi: 10.1146/annurev.neuro.24.1.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McAllister AK, Lo DC, Katz LC. Neurotrophins regulate dendritic growth in developing visual cortex. Neuron. 1995;15:791–803. doi: 10.1016/0896-6273(95)90171-x. [DOI] [PubMed] [Google Scholar]
- 5.McAllister AK, Katz LC, Lo DC. Neurotrophin regulation of cortical dendritic growth requires activity. Neuron. 1996;17:1057–1064. doi: 10.1016/s0896-6273(00)80239-1. [DOI] [PubMed] [Google Scholar]
- 6.Horch HW, Krüttgen A, Portbury SD, Katz LC. Destabilization of cortical dendrites and spines by BDNF. Neuron. 1999;23:353–364. doi: 10.1016/s0896-6273(00)80785-0. [DOI] [PubMed] [Google Scholar]
- 7.Horch HW, Katz LC. BDNF release from single cells elicits local dendritic growth in nearby neurons. Nat Neurosci. 2002;5:1177–1184. doi: 10.1038/nn927. [DOI] [PubMed] [Google Scholar]
- 8.Tanaka J, et al. Protein synthesis and neurotrophin-dependent structural plasticity of single dendritic spines. Science. 2008;319:1683–1687. doi: 10.1126/science.1152864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu B, Pang PT, Woo NH. The yin and yang of neurotrophin action. Nat Rev Neurosci. 2005;6:603–614. doi: 10.1038/nrn1726. [DOI] [PubMed] [Google Scholar]
- 10.Hempstead BL. Dissecting the diverse actions of pro- and mature neurotrophins. Curr Alzheimer Res. 2006;3:19–24. doi: 10.2174/156720506775697061. [DOI] [PubMed] [Google Scholar]
- 11.Chao MV. Neurotrophins and their receptors: A convergence point for many signalling pathways. Nat Rev Neurosci. 2003;4:299–309. doi: 10.1038/nrn1078. [DOI] [PubMed] [Google Scholar]
- 12.Reichardt LF. Neurotrophin-regulated signalling pathways. Philos Trans R Soc Lond B Biol Sci. 2006;361:1545–1564. doi: 10.1098/rstb.2006.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aid T, Kazantseva A, Piirsoo M, Palm K, Timmusk T. Mouse and rat BDNF gene structure and expression revisited. J Neurosci Res. 2007;85:525–535. doi: 10.1002/jnr.21139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiaruttini C, Sonego M, Baj G, Simonato M, Tongiorgi E. BDNF mRNA splice variants display activity-dependent targeting to distinct hippocampal laminae. Mol Cell Neurosci. 2008;37:11–19. doi: 10.1016/j.mcn.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 15.Pattabiraman PP, et al. Neuronal activity regulates the developmental expression and subcellular localization of cortical BDNF mRNA isoforms in vivo. Mol Cell Neurosci. 2005;28:556–570. doi: 10.1016/j.mcn.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 16.Tongiorgi E. Activity-dependent expression of brain-derived neurotrophic factor in dendrites: Facts and open questions. Neurosci Res. 2008;61:335–346. doi: 10.1016/j.neures.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 17.Chiaruttini C, et al. Dendritic trafficking of BDNF mRNA is mediated by translin and blocked by the G196A (Val66Met) mutation. Proc Natl Acad Sci USA. 2009;106:16481–16486. doi: 10.1073/pnas.0902833106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.An JJ, et al. Distinct role of long 3′ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell. 2008;134:175–187. doi: 10.1016/j.cell.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adachi N, Kohara K, Tsumoto T. Difference in trafficking of brain-derived neurotrophic factor between axons and dendrites of cortical neurons, revealed by live-cell imaging. BMC Neurosci. 2005;6:42. doi: 10.1186/1471-2202-6-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaech S, Banker G. Culturing hippocampal neurons. Nat Protoc. 2006;1:2406–2415. doi: 10.1038/nprot.2006.356. [DOI] [PubMed] [Google Scholar]
- 21.Matsuda N, et al. Differential activity-dependent secretion of brain-derived neurotrophic factor from axon and dendrite. J Neurosci. 2009;29:14185–14198. doi: 10.1523/JNEUROSCI.1863-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arévalo JC, et al. Cell survival through Trk neurotrophin receptors is differentially regulated by ubiquitination. Neuron. 2006;50:549–559. doi: 10.1016/j.neuron.2006.03.044. [DOI] [PubMed] [Google Scholar]
- 23.Kendall S, Yeo M, Henttu P, Tomlinson DR. Alternative splicing of the neurotrophin-3 gene gives rise to different transcripts in a number of human and rat tissues. J Neurochem. 2000;75:41–47. doi: 10.1046/j.1471-4159.2000.0750041.x. [DOI] [PubMed] [Google Scholar]
- 24.Salin T, Timmusk T, Lendahl U, Metsis M. Structural and functional characterization of the rat neurotrophin-4 gene. Mol Cell Neurosci. 1997;9:264–275. doi: 10.1006/mcne.1997.0625. [DOI] [PubMed] [Google Scholar]
- 25.Aliaga EE, Mendoza I, Tapia-Arancibia L. Distinct subcellular localization of BDNF transcripts in cultured hypothalamic neurons and modification by neuronal activation. J Neural Transm. 2009;116:23–32. doi: 10.1007/s00702-008-0159-8. [DOI] [PubMed] [Google Scholar]
- 26.Horton AC, Ehlers MD. Neuronal polarity and trafficking. Neuron. 2003;40:277–295. doi: 10.1016/s0896-6273(03)00629-9. [DOI] [PubMed] [Google Scholar]
- 27.Altar CA, Criden MR, Lindsay RM, DiStefano PS. Characterization and topography of high-affinity 125I-neurotrophin-3 binding to mammalian brain. J Neurosci. 1993;13:733–743. doi: 10.1523/JNEUROSCI.13-02-00733.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McAllister AK. Cellular and molecular mechanisms of dendrite growth. Cereb Cortex. 2000;10:963–973. doi: 10.1093/cercor/10.10.963. [DOI] [PubMed] [Google Scholar]
- 29.Chen Y, Ghosh A. Regulation of dendritic development by neuronal activity. J Neurobiol. 2005;64:4–10. doi: 10.1002/neu.20150. [DOI] [PubMed] [Google Scholar]
- 30.Urbanska M, Blazejczyk M, Jaworski J. Molecular basis of dendritic arborization. Acta Neurobiol Exp (Warsz) 2008;68:264–288. doi: 10.55782/ane-2008-1695. [DOI] [PubMed] [Google Scholar]
- 31.Wu GY, Zou DJ, Rajan I, Cline H. Dendritic dynamics in vivo change during neuronal maturation. J Neurosci. 1999;19:4472–4483. doi: 10.1523/JNEUROSCI.19-11-04472.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Timmusk T, Belluardo N, Persson H, Metsis M. Developmental regulation of brain-derived neurotrophic factor messenger RNAs transcribed from different promoters in the rat brain. Neuroscience. 1994;60:287–291. doi: 10.1016/0306-4522(94)90242-9. [DOI] [PubMed] [Google Scholar]
- 33.Rauskolb S, et al. Global deprivation of brain-derived neurotrophic factor in the CNS reveals an area-specific requirement for dendritic growth. J Neurosci. 2010;30:1739–1749. doi: 10.1523/JNEUROSCI.5100-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kindler S, Wang H, Richter D, Tiedge H. RNA transport and local control of translation. Annu Rev Cell Dev Biol. 2005;21:223–245. doi: 10.1146/annurev.cellbio.21.122303.120653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carson JH, et al. Multiplexed RNA trafficking in oligodendrocytes and neurons. Biochim Biophys Acta. 2008;1779:453–458. doi: 10.1016/j.bbagrm.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mori Y, Imaizumi K, Katayama T, Yoneda T, Tohyama M. Two cis-acting elements in the 3′ untranslated region of α-CaMKII regulate its dendritic targeting. Nat Neurosci. 2000;3:1079–1084. doi: 10.1038/80591. [DOI] [PubMed] [Google Scholar]
- 37.Severt WL, et al. The suppression of testis-brain RNA binding protein and kinesin heavy chain disrupts mRNA sorting in dendrites. J Cell Sci. 1999;112:3691–3702. doi: 10.1242/jcs.112.21.3691. [DOI] [PubMed] [Google Scholar]
- 38.Shan J, Munro TP, Barbarese E, Carson JH, Smith R. A molecular mechanism for mRNA trafficking in neuronal dendrites. J Neurosci. 2003;23:8859–8866. doi: 10.1523/JNEUROSCI.23-26-08859.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dias BG, Banerjee SB, Duman RS, Vaidya VA. Differential regulation of brain derived neurotrophic factor transcripts by antidepressant treatments in the adult rat brain. Neuropharmacology. 2003;45:553–563. doi: 10.1016/s0028-3908(03)00198-9. [DOI] [PubMed] [Google Scholar]
- 40.Garza AA, Ha TG, Garcia C, Chen MJ, Russo-Neustadt AA. Exercise, antidepressant treatment, and BDNF mRNA expression in the aging brain. Pharmacol Biochem Behav. 2004;77:209–220. doi: 10.1016/j.pbb.2003.10.020. [DOI] [PubMed] [Google Scholar]
- 41.Khundakar AA, Zetterström TS. Biphasic change in BDNF gene expression following antidepressant drug treatment explained by differential transcript regulation. Brain Res. 2006;1106:12–20. doi: 10.1016/j.brainres.2006.05.063. [DOI] [PubMed] [Google Scholar]
- 42.Gulyás AI, Megías M, Emri Z, Freund TF. Total number and ratio of excitatory and inhibitory synapses converging onto single interneurons of different types in the CA1 area of the rat hippocampus. J Neurosci. 1999;19:10082–10097. doi: 10.1523/JNEUROSCI.19-22-10082.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Megías M, Emri Z, Freund TF, Gulyás AI. Total number and distribution of inhibitory and excitatory synapses on hippocampal CA1 pyramidal cells. Neuroscience. 2001;102:527–540. doi: 10.1016/s0306-4522(00)00496-6. [DOI] [PubMed] [Google Scholar]
- 44.Aibel L, Martin-Zanca D, Perez P, Chao MV. Functional expression of TrkA receptors in hippocampal neurons. J Neurosci Res. 1998;54:424–431. doi: 10.1002/(SICI)1097-4547(19981101)54:3<424::AID-JNR13>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.