Abstract
The H19 gene, which localizes within a chromosomal region on human chromosome 11p15 that is commonly lost in Wilms tumor (WT), encodes an imprinted untranslated RNA. However, the biological significance of the H19 noncoding transcript remains unresolved because replacement of the RNA transcript with a neocassette has no obvious phenotypic effect. Here we show that the human H19 locus also encodes a maternally expressed, translated gene, antisense to the known H19 transcript, which is conserved in primates. This gene, termed HOTS for H19 opposite tumor suppressor, encodes a protein that localizes to the nucleus and nucleolus and that interacts with the human enhancer of rudimentary homolog (ERH) protein. WTs that show loss of heterozygosity of 11p15 or loss of imprinting of IGF2 also silence HOTS (7/7 and 10/10, respectively). Overexpression of HOTS inhibits Wilms, rhabdoid, rhabdomyosarcoma, and choriocarcinoma tumor cell growth, and silencing HOTS by RNAi increases in vitro colony formation and in vivo tumor growth. These results demonstrate that the human H19 locus harbors an imprinted gene encoding a tumor suppressor protein within the long-sought WT2 locus.
Keywords: antisense RNA, DNA methylation, epigenetics, genomic, imprinting
Genomic imprinting is an epigenetic modification leading to parent-of-origin–dependent differential expression of the two alleles of a gene. Much of our knowledge of this phenomenon comes from studies of the maternally expressed H19 and the nearby imprinted Igf2 gene, which is reciprocally expressed from the paternal allele (1). In the past decade, several deletion mutants and transgenic mouse models targeting the H19-Igf2 locus have been developed (1). These studies show that regulation of Igf2 and H19 imprinting is dependent on a differentially methylated region (DMR) located −4 to −2 kb upstream from the H19 transcriptional start site (2–4). Only when the DMR is unmethylated does the methylation-sensitive zinc-finger protein CTCF bind, thus insulating Igf2 from its enhancer creating a silenced maternal Igf2 allele (5, 6). Knockout of the mouse H19 transcript has provided mixed results; in one case, no function was inferred (7), and in another case, overgrowth was reported due to activation of the normally silent paternal allele of Igf2, an important autocrine growth factor (4, 8). In humans, loss of imprinting (LOI) of IGF2 is an important epigenetic mechanism first found in WT (9), the most common childhood kidney cancer; in Beckwith–Wiedemann syndrome (BWS) (10), which predisposes to WT; and in many other childhood and adult malignancies (11). LOI can also be caused, at least in BWS, by microdeletions within the H19 DMR (12), although most loss of heterozygosity (LOH) in WT includes IGF2 itself (13) and so presumably acts through a mechanism other than LOI. In addition to LOI of IGF2, about 30% of WTs and other embryonal tumors show LOH of 11p15, which has been narrowed down to a 1-Mb region including the H19 gene (13–15). LOH of 11p15 is also commonly found in rhabdomyosarcoma, rhabdoid, cervical, ovarian, lung, bladder, breast, and hepatocellular cancers (13, 15). Preferential LOH of the maternal allele in WT and embryonal tumors implies that the undiscovered WT2 gene is imprinted and expressed from the maternal allele (16). However, no imprinted tumor suppressor gene in this region has yet been identified. The maternally expressed imprinted gene CDKN1C is not a likely candidate gene because it is expressed even in LOH-positive WT (17). There are also contradictory data on the role of H19 in cancer, as the sense RNA has been described by some as an oncogene (18) and others have demonstrated that the transfected H19 gene suppresses cellular proliferation, clonogenicity, and tumorigenicity in certain tumor cell lines (19). Moreover, recent studies on the H19 locus have identified the H19-derived miR-675 and a H19 antisense RNA named 91H (20, 21). The miR-675 targets and down-regulates the RB gene and is thought to contribute to colorectal cancer development when overexpressed (20), and 91H is a single 120-kb transcript that is stabilized in breast cancer cells and overexpressed in human breast tumors (21). Because the H19 loci is inactive in WT due to LOH or imprinting, neither of these recent studies demonstrate a tumor suppressor role for the H19 locus. Other large intervening noncoding RNAs (lincRNAs) like HOTAIR show increased expression in primary breast tumors and metastases and are thought to induce genome-wide retargeting of Polycomb repressive complex 2 (PRC2) to an occupancy pattern more resembling embryonic fibroblasts, leading to altered histone H3 lysine 27 methylation, gene expression, and increased cancer invasiveness and metastasis in a manner dependent on PRC2 (22). The potential of antisense transcripts to modulate the cancer epigenome in trans highlights a direction in elucidating the function of such transcripts and may help to understand the role of 91H. The absence of conservation at the protein level and the evolutionary conservation of structure at the RNA level has led to the proposal that the functional product of the H19 gene is a structured RNA or a miRNA (23, 24).
Results and Discussion
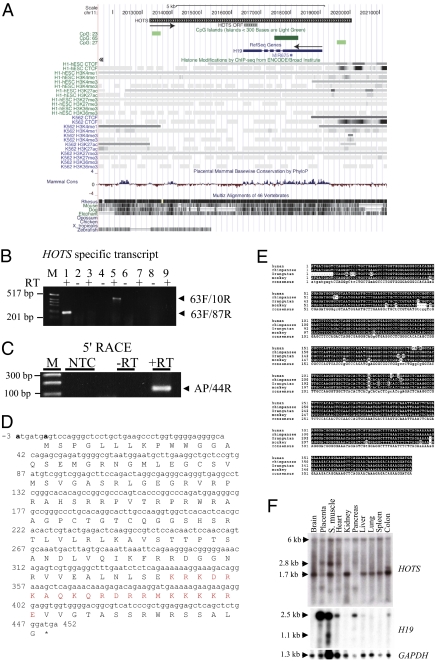
The human genome is thought to contain far more transcripts than were previously appreciated, and up to 14% of the genome may be transcribed (25). To identify previously unappreciated transcripts in the human 11p15.5 imprinted region, we designed 278 reverse transcription (RT) and PCR primers, with a mean amplicon length of 2 kb, identifying 10 previously unappreciated transcripts in human kidney and placenta spread over 200 kb including the H19 gene locus. To characterize the H19 antisense transcript, we performed RT-PCR with 13 additional gene-specific RT (GS-RT) primers spanning a 7.6-kb genomic region, positioned at 0.1- to 1-kb intervals and in sense orientation to H19 (Fig. 1A). Following amplification of the cDNAs using 10 paired nested PCR primers in addition to performing 5′ and 3′ RACE, we found that the longest transcript was 6 kb, polyadenylated, contained a CpG island promoter, and extended 1 kb upstream and 2.8 kb downstream of H19 (Fig. 1 A–C and SI Appendix, Fig. S1). To confirm specificity of HOTS transcription, we included two negative controls, GS-RT primers without RT enzyme, and GS-RT primers with RT enzyme. The two negative control reactions failed to yield any RT-PCR amplification products (Fig. 1B). Strand-specific RT-PCR analyses of the 6-kb genomic locus failed to reveal any evidence of splicing, including the use of strand-specific RT-PCR primers initiated within the H19 intron (RT primer: 1119R; PCR primers: 10R and 689R) or the H19 exon (RT primer: 1R; PCR primers: 1144R and 115R) (Fig. 1A). Recently, a H19 antisense transcript named 91H RNA with the potential to produce a 120-kb transcript was reported for the human and the mouse H19/IGF2 loci (21). We cannot exclude the possibility that the H19 antisense transcript that we report here could be part of 91H RNA owing to the technical limitations in both synthesis and Northern blotting of high-molecular-weight RNA. We named this smaller transcript H19 opposite tumor suppressor (HOTS). Earlier studies have shown that the human H19 locus can suppress tumor cell growth (19). We therefore focused our studies on the protein-coding potential of the ORFs contained in the H19 antisense segment and found entirely within the H19 transcriptional unit. The longest ORF that met this selection criteria within HOTS encodes a predicted polypeptide of 150 amino acids (Fig. 1D), and the +4 (purine) and −3 (purine) bases fit the Kozak consensus sequence for translation initiation (Fig. 1D). Protein database searches did not reveal any homology or functional motifs; however, we identified nuclear localization signals at residues 131 (KKKK) and 132 (KKKR) and nuclear localization bipartite signals at 116 (KRKDRKAQKQRDRRMKK) and at 117 and 120 in HOTS by using the PSORT II program (http://psort.hgc.jp/form2.html) software (Fig. 1D).
Fig. 1.
HOTS is a primate-conserved, ubiquitous transcript antisense to H19. (A) Genomic organization of HOTS and H19 genes on human chromosome 11p15.5. The direction of HOTS and H19 transcriptional orientation is shown by black arrows. HOTS ORF is shown. The display is populated with CpG island (green) CTCF-binding sites, ENCODE histone modification marks, and mammalian DNA sequence conservation for the region [University of California at Santa Cruz (UCSC) on Human Genome Sequence: Feb. 2009 (GRCh37/hg19) Assembly, http://genome.ucsc.edu/]. (B) RT-PCR amplification of HOTS using strand-specific RT primer 1119R located within the first intron of H19, followed by amplification with intronic PCR primer pairs 63F/87R (lanes 1 and 2) and 63F/10R (lanes 5 and 6). RT-PCR amplification of H19 using strand-specific RT primer 1F, followed by intronic PCR primer pair 63F/87R (lanes 3 and 4) or 63F/10R (lanes 7 and 8). (Lane 9) No RT primer included but RT enzyme and 63F/87R primer pair. (C) 5′ RACE using primer 4534R for RT and primers 44R and PCR anchor primer. M, 1-kb DNA ladder; NTC, no template control. (D) Sequence for the HOTS ORF, nuclear localization signal shown in red type and the putative Kozak consensus sequences at −3 (A) and +4 (A) in boldface type. (E) Multiple sequence alignment of predicted HOTS primates sequences from human, chimpanzee, orangutan, and monkey. Sequence areas depicted against a black background represent identity, and small letters in the consensus sequence at the bottom show areas of sequence variability. (F) HOTS and H19 human multiple tissues Northern blots. (Top) HOTS. (Middle) H19. (Bottom) GAPDH.
Multiple sequence alignment revealed that HOTS was conserved in primates with 96% nucleotide identity in the ORF (Fig. 1E). HOTS was not well conserved in mouse, showing 43% identity and no ORF (SI Appendix, Fig. S2). HOTS was expressed ubiquitously in all tissues tested, as shown by RT-PCR and by Northern blot hybridization with a HOTS strand-specific antisense RNA probe (Fig. 1F and SI Appendix, Fig. S3). The HOTS Northern blot shows a 6-, 2.8-, and 1.7-kb transcript, suggesting tissue-specific alternative splicing (Fig. 1F). H19 was expressed only in placenta, skeletal muscle, heart, pancreas, liver, and colon (Fig. 1F), suggesting that the regulation of HOTS and H19 might not be coordinated.
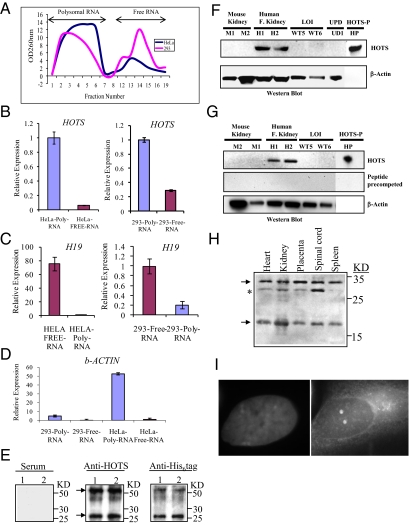
Because H19 is uniformly described as a noncoding RNA, we took three approaches to confirm or disprove the existence of a HOTS protein. First, we assayed for association of the HOTS transcript with polysomes, the ribosomal protein complexes that recruit mRNAs for protein translation, an indicator that a transcript is translated. To this end, we purified RNA-associated polysomes by sucrose gradient differential centrifugation (Methods) from the cervical carcinoma cell lines HeLa and SiHa, human embryonic kidney 293 (HEK293) cells, and the rhabdomyosarcoma cell line RD (Fig. 2A and SI Appendix, Fig. S4). Strand-specific quantitative real-time PCR revealed preferential association of the HOTS transcript with polysomes, suggesting that it is translated, whereas H19 was associated with free RNA, consistent with its being untranslated (Fig. 2 B and C and SI Appendix, Fig. S5). As a positive control, β-actin also was preferentially associated with polysomes (Fig. 2D). Second, we raised a polyclonal antibody against His6-tagged HOTS protein. We were able to detect His-tagged HOTS with HOTS antibody and anti-His6 antibody, as a 25-kDa protein, the additional weight beyond the expected 17 kDa is due to 9 kDa of His6 plus trailing vector amino acid sequences, whereas no signal was obtained with preimmune serum (Fig. 2E). Because purified His-tagged HOTS displayed an additional signal that was twice the size of the HOTS protein's molecular weight (Fig. 2E), we performed immunoprecipitation to confirm if the higher band was indeed a dimer. Only the monomer band of His-tagged HOTS was detected with HOTS antibody after immunoprecipitation with anti-His6 antibodies from HEK293 transfected cells, thus confirming the prediction that HOTS exists as a dimer in vivo (Fig. 2E).To determine the specificity of the HOTS antibody, we performed a Western blot on protein samples from human kidney as a positive control and on protein extracts from mouse kidney, Wilms tumor, and a BWS UPD as negative controls (Fig. 2F). As expected, we saw a strong positive signal only on the human kidney (Fig. 2F), which was specific, because it was eliminated by precompetition with purified HOTS protein (Fig. 2F), and there was no detectable signal in the mouse kidney (Fig. 2F), consistent with the sequencing data. We additionally detected on a denaturing gel a 17-kDa polypeptide using purified HOTS antibody (Fig. 2G). Western blot using the anti-HOTS antibody of a non-denatured gel containing fetal tissues revealed the expected 17 kDa HOTS monomer, 34 kDa HOTS dimer, and a differentially expressed 29 kDa isoform (Fig. 2H). Third, we investigated the subcellular localization of HOTS by transfecting Cos7 and SiHa cell lines with HOTS cDNA tagged with green fluorescence protein (GFP) in a pEGFP-N3 vector (Clontech). HOTS localized to the nucleus and was sequestered in a subnuclear organelle (SI Appendix, Fig. S6), confirming our earlier prediction that HOTS was a nuclear protein. Colocalization of HOTS with the nucleolar protein nucleophosmin (B23) showed that HOTS protein predominantly colocalized with nucleophosmin (SI Appendix, Fig. S7). In addition, immunohistochemical studies with the HOTS antibodies revealed nuclear and nucleolar localization (Fig. 2I).
Fig. 2.
HOTS is a polysome-associated RNA encoding a nucleolar protein. (A) Polysomal and free total cellular RNA fractionated by sucrose gradient centrifugation from homogenates of HeLa (blue) and HEK293 (pink) human cells. (B) Strand-specific quantitative real-time PCR amplification of HOTS transcripts shows enrichment in polysomes from HeLa and HEK293 cells, in contrast to (C) H19, which shows enrichment in the free RNA fraction, with (D) β-actin a positive control for polysome enrichment. (B–D) Analyses were performed in triplicate (n = 6). (E) Western blots of purified His6-tag HOTS protein loaded in duplicate lanes using preimmune serum (Left), HOTS antibody (Center), and His6-tag antibody (Right). Both the HOTS and His6-tag antibodies detect the expected recombinant protein (arrows) of 26 kDa (17 kDa of predicted HOTS polypeptide sequence and 9 kDa of His6 plus trailing vector amino acid sequences) and a dimer of 52 kDa. (F) Western blot with anti-HOTS antibody on protein extracts from mouse kidney (M1 and M2), human fetal kidney (H1 and H2), WTs with LOI (WT5 and WT6), a BWS sample with chromosome 11p UPD (UD1), and purified HOTS protein (HP). The LOI and UD samples are negative controls with loss of expression of the maternally expressed HOTS. β-Actin antibody was used on the same blot as a loading control. (G) Western blot similar to the previous but precompeted with HOTS purified protein (1 μg/mL). (H) Western blots of a nondenaturing gel using anti-HOTS antibody on human fetal tissues. Arrows indicate the HOTS monomer of 17 kDa and a dimer of 34 kDa; asterisk indicates a 29-kDa band that might represent an isoform or posttranslationally modified protein abundant in the spinal cord. (I) Subcellular localization of native HOTS protein to the nucleolus using anti-HOTS antibody on human SiHa cells. (Left) DAPI stained nucleus. (Right) HOTS antibody image superimposed on the DAPI stain.
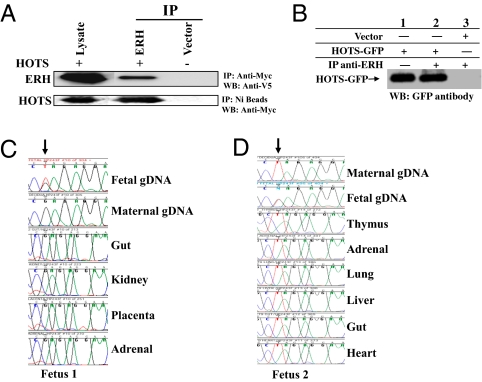
To further validate the protein-coding potential of HOTS and to begin to understand the cellular and molecular pathways important in HOTS function, we searched for HOTS interacting proteins. Immunoprecipitation of myc-tagged HOTS from HEK293 cells revealed eight potential HOTS interacting proteins (SI Appendix, Fig. S8 and Table S1), which were then gel-purified and identified by mass spectrometry (Proteomic Research Systems). To corroborate the interaction of HOTS with each of the candidate interacting proteins, we performed immunoprecipitations from HEK293 cells cotransfected with HOTS-myc and with each potentially interacting protein tagged with His6-V5. We confirmed by Western blot the interaction of HOTS with all eight proteins by using anti-myc antibody directed against HOTS-myc to immunoprecipitate each of the proteins detected by anti-V5 antibody. However, only one of these, human enhancer of rudimentary homolog (ERH), also showed interaction with HOTS when we performed the reciprocal pulldown using Ni-beads to bind His6-tagged ERH, and we detected HOTS-myc by Western blot (Fig. 3A). We could not immunoprecipitate HOTS directly with anti-HOTS antibody from HEK293 cells transfected with ERH, perhaps due to low expression levels of native HOTS. However, we immunoprecipitated HOTS-GFP using ERH antibodies (Abcam) from HEK293 cells transfected with HOTS-GFP, demonstrating that native ERH interacts with HOTS protein (Fig. 3B).
Fig. 3.
HOTS interacts with ERH in vivo and is imprinted. (A) (Upper) Immunoprecipitation (IP) with anti–myc-tag antibodies specific to myc-tagged HOTS from HEK293 cells cotransfected with myc-tagged HOTS and His6/V5-tagged ERH. V5 antibody was used to detect ERH on the Western blot (WB). (Lower) IP with nickel beads specific for His-tagged ERH from HEK293 cells transfected with myc-tagged HOTS and His6/V5-tagged ERH; myc-tagged antibody was used to detect HOTS by WB. (B) Immunoprecipitation of HOTS from protein extracts of HEK293 cells transfected with HOTS tagged with GFP and immunoprecipitated using ERH antibody. Ten percent (30 μg) of HEK293 input extract was loaded in lane 1. Anti-GFP antibody was used for Western blot. (C) Strand-specific cDNA synthesis was carried out to study HOTS expression in the mother and in fetal tissues of fetus 1. The polymorphism is shown by an arrow. The maternal decidua and fetal genomic DNA sequence is included to distinguish the origin of the expressed allele. Note the exclusive expression the maternal G allele in all fetal tissues. (D) Maternal T allele expression in all fetus 2 tissues (indicated by an arrow). Note that the fetus genomic DNA is heterozygous and therefore informative for allele-specific gene expression. All cDNA synthesis was performed with gene-specific primers that produced only HOTS transcripts.
Several imprinted gene loci transcribe both an imprinted sense and an antisense transcript, including IGF2R and its antisense Air (26); Xist and its antisense Tsix (27); and Kvlqt1 and its antisense LIT1 (28). We therefore suspected that HOTS might also be imprinted, which we tested by exploiting a polymorphism at nucleotide +588 in the 3′ UTR. Synthesis of HOTS cDNA using RT primers specific for HOT transcript from the kidneys of five individuals revealed exclusive expression from only one of the two alleles found in the genomic DNA (Fig. 3 C and D, and SI Appendix, Fig. S9). Genomic and fetal cDNA sequence from two informative paired maternal and fetal samples revealed the expressed allele to be of maternal origin (Fig. 3 C and D).
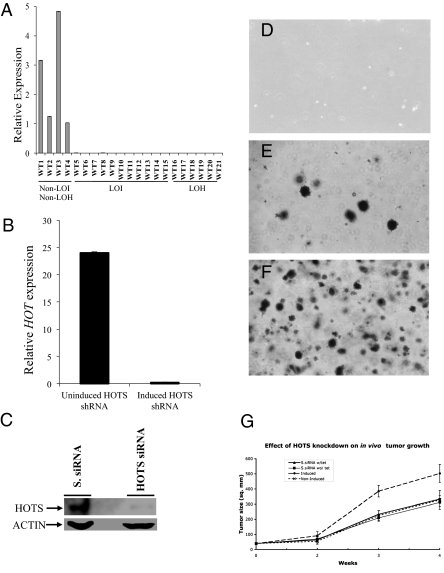
WTs and rhabdomyosarcoma showed preferential loss of a specific parental allele in LOH, suggesting the existence of an imprinted tumor suppressor gene on 11p15 (16). Because alleles that carry an imprint are presumed to be inactive on one allele as a result of the imprinting process, tumorigenesis arising from isodisomy of the imprinted allele would have no requirement that such an allele be further mutated. Likewise, LOH affecting the normally active 11p15.5 maternal allele would be sufficient to exhibit a null (tumorigenic) phenotype. These predictions fit Sapienza's modified Knudson's “two hit’’ model for tumorigenesis, which incorporates genomic imprinting (16). Consistent with this model, real-time quantitative RT-PCR analysis of HOTS in seven WTs with LOH showed loss of expression in all cases (Fig. 4A). Furthermore, LOI of IGF2, which silences the maternal H19 allele as well as activates the maternal IGF2 allele, was also associated with HOTS silencing in all 10 cases tested (Fig. 4A). To rule out RNA degradation in the LOH and LOI WT samples, DNase I-treated RNA samples were separated on an agarose gel before cDNA synthesis. All but one showed a high 28S/18S ratio, and all of the samples showed abundant expression of GAPDH (SI Appendix, Fig. S10 and Fig. S11).
Fig. 4.
HOTS is lost in LOH and LOI WT and suppresses tumor cell growth in vitro and in vivo. (A) Quantitative real-time expression analysis of HOTS in WT samples shows loss of expression in LOI (WT5–14) and LOH (WT15–21) cases, but not in non-LOH, non-LOI cases (WT1–4). Error bars (SD) are plotted but are too small to see. (B) Tetracycline-inducible knockdown of HOTS by HOTS shRNA in HeLa cells. Induction with tetracycline activates the HOTS shRNA-expressing plasmid, leading to silencing of HOTS. (C) Western blot showing siRNA knockdown of HOTS with HOTS siRNA but not with scrambled siRNA (S. siRNA). β-Actin was used as a loading control. (D–F) HeLa tumor cell growth assayed on soft agar, showing that growth is inhibited by HOTS expressed from a tetracycline-inducible vector after Zeocin selection. (D) Nontransfected HeLa cells, selected with 0.5 mg/mL Zeocin. No visible colonies were observed due to cell death from Zeocin drug selection. (E) HeLa cell colonies from a culture that was not induced to express anti-HOTS RNAi. (F) Fifteen-fold more HeLa cell colonies occur when cells are induced by tetracycline to express anti-HOTS RNAi. (Scale bar, D–F: 0.3 mm.) n = 3. (G) Increased tumor cell growth in nude mice upon RNAi knockdown of HOTS. n = 6. Mean tumor volume was plotted against time. Statistically significant difference in tumor area between the HOTS knockdown animals (Induced, •) and the HOTS-expressing animals (Non-Induced, X-dotted line) was scored (P < 0.01 at 3 wk and P < 0.02 at 4 wk, Student's t-test). As controls, we included scrambled siRNA (S.siRNA) tetracycline-induced (▲, w/tet) and Non-Induced S.siRNA (■, wo/tet) HeLa transfected cells.
These data suggest that HOTS is a tumor suppressor gene. However, we did not identify mutations within the coding sequence of 30 WTs, even though mutations might not be expected for an imprinted tumor suppressor gene and are not found, for example, in the imprinted tumor suppressor gene ARHI in breast cancer (29). Nevertheless, to confirm the tumor suppressor activity of HOTS, we expressed a HOTS-GFP fusion construct and a H19 cDNA in WT (SK-NEP-1), rhabdoid tumor (G401), rhabdomyosarcoma (RD), choriocarcinoma (JEG-3), and cervical carcinoma (SiHa) cell lines. Transfection efficiency was comparable between the native HOTS-GFP (20.2%) and mutated HOT-GFP (21.4%) transfected cells (SI Appendix, Fig. S18). We observed potent growth inhibition in cells transfected with HOTS-GFP (SI Appendix, Table S2), whereas cell lines transfected with GFP-expressing control vector, HOTS-GFP carrying truncating mutations in HOTS, and H19 cDNA grew to confluence with no growth inhibition (SI Appendix, Tables S2 and S3; Fig. S12).
We also considered the possibility that an overexpressed gene might show nonspecific growth inhibition or influence the expression of the counterpart sense or antisense transcript. To exclude this possibility, we performed the reverse experiment, where we knocked down HOTS instead of overexpressing HOTS and looked at reduced HOTS expression influence on tumor growth. We therefore generated three HeLa tumor cell lines containing tetracycline-inducible RNAi constructs for HOTS using the shRNA system and also directly transfected anti-HOTS siRNA and scrambled anti-HOTS siRNA. We verified HOTS RNA and protein knockdown by quantitative RT-PCR and Western blot (Fig. 4 B and C). HeLa cells in which HOTS expression was knocked down using a tetracycline-inducible HOTS-specific shRNA formed >15-fold more colonies in soft agar than did cells expressing HOTS or a scrambled shRNA for HOTS; representative data from three experiments are shown in Fig. 4 D–F and SI Appendix, Fig. S13. HOTS shRNA was specific for HOTS and did not significantly reduce H19 expression (SI Appendix, Fig. S14). To test for the effect of expression of HOTS on H19 and vice versa, we transfected each gene into HEK293 cells. Transfection of either gene did not result in significant change in the expression of the other gene, HOT (0.96-fold expression) and H19 (1.0-fold expression, not statistically significant; n = 3).
Similar to the in vitro tumor suppressor activity of HOTS, in vivo HOTS knockdown in HeLa cells resulted in decreased HOTS expression and in increased in vivo tumor growth in nude mice (Fig. 4G and SI Appendix, Fig. S15). The mean tumor size at 3 and 4 wk was 385 (P < 0.01) and 503 mm2 (P < 0.02), respectively, for nude mice (n = 6) with HOTS knockdown and was smaller in nude mice (n = 6) retaining HOTS expression: 224 and 327 mm2, respectively (Fig. 4G). Control cell lines transfected with scrambled HOTS siRNA (S.siRNA) induced with tetracycline and noninduced were included and did not show increased tumor growth, discounting any tetracycline influence on tumor growth (Fig. 4G). Thus, HOTS encodes a tumor suppressor gene.
To investigate the possibility that altered methylation of the HOTS region may play a role in WT, we bisulfite-sequenced a CpG island that lies within the HOTS transcript near the 5′ end (Fig. 1A) in 10 LOI and 10 non-LOI WT cases. Analysis of all 26 CpG dinucleotides in the island showed full methylation in every sample (SI Appendix, Fig. S16). These data suggest that methylation of this island has no role in HOTS regulation. However, as the HOTS CpG island is not sequence conserved (http://genome.ucsc.edu; SI Appendix, Figs. S16 and S17), we postulated that it may not represent the true HOTS regulatory region. We therefore searched upstream of the HOTS CpG island and identified a 330-bp GC-rich sequence, 768 bp 5′ to HOTS and highly conserved across mammals. Bisulfite sequence analysis of this sequence revealed two invariant sites of DNA methylation across all samples and a striking difference in methylation at the remaining 10 sites: The non-LOI samples showed methylation at 0–1 of these 10 sites, whereas the LOI samples showed methylation at 5–8 of these 10 sites, which was statistically significant (P < 0.001, t test). These data are consistent with an epigenetic change at this locus related to LOI, but presumably affecting both alleles because the methylation in LOI was complete. It thus seems unlikely that HOTS itself regulates imprinting, but it appears to be regulated in a complex way, with silencing epigenetically in LOI and genetically in LOH.
In summary, we have shown that the human H19 locus includes a maternally expressed, translated transcript that is transcribed in antisense orientation to the H19 gene, and whose gene product is targeted to the nucleus and sequestered in the nucleolus. WTs with LOI of H19 or LOH of human chromosomal band 11p15.5 lose HOTS expression. Expression of HOTS in cancer cell lines inhibits tumor cell growth, and silencing of HOTS promotes in vitro anchorage-independent growth and in vivo tumorigenicity. Because HOTS is imprinted and its expression is lost in WTs with both LOH and LOI, it fits the predictions for an imprinted tumor suppressor gene (16). As such, it is at least one member of the WT2 gene locus on 11p15.
We also identified at least one protein that interacts with HOTS, namely ERH. ERH has been shown to be important in pyrimidine biosynthesis, cell cycle regulation, and transcriptional repression (30). ERH is known to interact with the zinc-finger protein CIZ1, a promoter of DNA replication that interacts with p21 (Cip1), a CDK2 inhibitor critical for cell cycle regulation (31). Moreover, ERH interacts with SKAR, a cell growth regulator in the S6K1-signaling pathway (32). We propose from these protein interactions that HOTS may mediate its tumor suppressor activity by targeting DNA replication and cell cycle regulation.
Finally, we note that HOTS joins a relatively small group of primate-specific proteins, including the POTE (expressed in prostate, ovary, testis, and placenta) gene family (33) and MGC8902, a human lineage-specific protein thought to be involved in higher cortical function (34).
Methods
HOTS cloning was performed by RT-PCR, and a full-length ORF was cloned into pEGFP.N3 and p3XFLAG-Myc-CMV-24 plasmid vectors. To generate riboprobes, partial HOTS/H19 sequences were cloned into the pCRII TOPO T7/Sp6 vector. Polysome association of HOTS transcript was investigated by purification of free and polysome-associated RNA by using sucrose gradient differential centrifugation, and transcript levels were measured by quantitative real time PCR using internal controls. To generate HOTS antibody, we cloned HOTS into the bacterial expression vector pDEST17 and purified His6-tagged proteins. For subcellular localization, we used HOTS antibodies and also transfected Cos-7 cells with HOTS cloned in pEGFP.N3 by lipofection and stained organelles using specific antibodies. Tumor growth assays were performed with wild-type and mutated HOTS cloned in pEGFP.N3 and transfected into SiHa, Cos7, RD, JEG-3, SK-NEP-1, and G401 cells. To study HOTS knockdown on tumor cell growth, we cloned shRNA targeting HOTS into the tetracycline-inducible pENTR H1/TO vector and transfected HeLa-TREx cells by lipofection, generating stable cell lines. Tumor growth effects were studied by both soft agar assay and in vivo in athymic nude mice. To identify HOTS-interacting proteins, we transfected HEK293 cells with HOTS in p3XFLAG-Myc-CMV-24 and used anti-cmyc antibodies to immunoprecipitate interacting proteins that were subjected to mass spectrometry for identification. Candidate HOTS-interacting proteins were isolated by RT-PCR and cloned into the pEF6/V5-His TOPO TA vector and were validated by coimmunoprecipitation and Western blot analysis. See SI Appendix for full description of methods.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant CA65435 (to A.P.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1110904108/-/DCSupplemental.
References
- 1.Arney KL. H19 and Igf2: Enhancing the confusion? Trends Genet. 2003;19:17–23. doi: 10.1016/s0168-9525(02)00004-5. [DOI] [PubMed] [Google Scholar]
- 2.Tremblay KD, Duran KL, Bartolomei MS. A 5′ 2-kilobase-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol Cell Biol. 1997;17:4322–4329. doi: 10.1128/mcb.17.8.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thorvaldsen JL, Duran KL, Bartolomei MS. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 1998;12:3693–3702. doi: 10.1101/gad.12.23.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ripoche MA, Kress C, Poirier F, Dandolo L. Deletion of the H19 transcription unit reveals the existence of a putative imprinting control element. Genes Dev. 1997;11:1596–1604. doi: 10.1101/gad.11.12.1596. [DOI] [PubMed] [Google Scholar]
- 5.Hark AT, et al. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 6.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 7.Jones BK, Levorse JM, Tilghman SM. Igf2 imprinting does not require its own DNA methylation or H19 RNA. Genes Dev. 1998;12:2200–2207. doi: 10.1101/gad.12.14.2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leighton PA, Ingram RS, Eggenschwiler J, Efstratiadis A, Tilghman SM. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature. 1995;375:34–39. doi: 10.1038/375034a0. [DOI] [PubMed] [Google Scholar]
- 9.Rainier S, et al. Relaxation of imprinted genes in human cancer. Nature. 1993;362:747–749. doi: 10.1038/362747a0. [DOI] [PubMed] [Google Scholar]
- 10.Weksberg R, et al. Tumor development in the Beckwith-Wiedemann syndrome is associated with a variety of constitutional molecular 11p15 alterations including imprinting defects of KCNQ1OT1. Hum Mol Genet. 2001;10:2989–3000. doi: 10.1093/hmg/10.26.2989. [DOI] [PubMed] [Google Scholar]
- 11.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 12.Sparago A, et al. Microdeletions in the human H19 DMR result in loss of IGF2 imprinting and Beckwith-Wiedemann syndrome. Nat Genet. 2004;36:958–960. doi: 10.1038/ng1410. [DOI] [PubMed] [Google Scholar]
- 13.Feinberg AP, Williams BR. Wilms tumor as a model for cancer biology. In: El-Deiry WS, editor. Tumor Suppressor Genes. Vol 1. Totowa, NJ: Humana; 2003. pp. 239–248. [DOI] [PubMed] [Google Scholar]
- 14.Dao D, et al. Multipoint analysis of human chromosome 11p15/mouse distal chromosome 7: Inclusion of H19/IGF2 in the minimal WT2 region, gene specificity of H19 silencing in Wilms’ tumorigenesis and methylation hyper-dependence of H19 imprinting. Hum Mol Genet. 1999;8:1337–1352. doi: 10.1093/hmg/8.7.1337. [DOI] [PubMed] [Google Scholar]
- 15.Karnik P, Chen P, Paris M, Yeger H, Williams BR. Loss of heterozygosity at chromosome 11p15 in Wilms tumors: Identification of two independent regions. Oncogene. 1998;17:237–240. doi: 10.1038/sj.onc.1201959. [DOI] [PubMed] [Google Scholar]
- 16.Scrable H, et al. A model for embryonal rhabdomyosarcoma tumorigenesis that involves genome imprinting. Proc Natl Acad Sci USA. 1989;86:7480–7484. doi: 10.1073/pnas.86.19.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Overall ML, Spencer J, Bakker M, Dziadek M, Smith PJ. p57K1P2 is expressed in Wilms’ tumor with LOH of 11p15.5. Genes Chromosomes Cancer. 1996;17:56–59. doi: 10.1002/(SICI)1098-2264(199609)17:1<56::AID-GCC8>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 18.Rachmilewitz J, et al. H19 expression and tumorigenicity of choriocarcinoma derived cell lines. Oncogene. 1995;11:863–870. [PubMed] [Google Scholar]
- 19.Hao Y, Crenshaw T, Moulton T, Newcomb E, Tycko B. Tumour-suppressor activity of H19 RNA. Nature. 1993;365:764–767. doi: 10.1038/365764a0. [DOI] [PubMed] [Google Scholar]
- 20.Tsang WP, et al. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis. 2010;31:350–358. doi: 10.1093/carcin/bgp181. [DOI] [PubMed] [Google Scholar]
- 21.Berteaux N, et al. A novel H19 antisense RNA overexpressed in breast cancer contributes to paternal IGF2 expression. Mol Cell Biol. 2008;28:6731–6745. doi: 10.1128/MCB.02103-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Juan V, Crain C, Wilson C. Evidence for evolutionarily conserved secondary structure in the H19 tumor suppressor RNA. Nucleic Acids Res. 2000;28:1221–1227. doi: 10.1093/nar/28.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai X, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA. 2007;13:313–316. doi: 10.1261/rna.351707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.ENCODE Project Consortium. Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lyle R, et al. The imprinted antisense RNA at the Igf2r locus overlaps but does not imprint Mas1. Nat Genet. 2000;25:19–21. doi: 10.1038/75546. [DOI] [PubMed] [Google Scholar]
- 27.Lee JT, Davidow LS, Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet. 1999;21:400–404. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- 28.Lee MP, et al. Loss of imprinting of a paternally expressed transcript, with antisense orientation to KVLQT1, occurs frequently in Beckwith-Wiedemann syndrome and is independent of insulin-like growth factor II imprinting. Proc Natl Acad Sci USA. 1999;96:5203–5208. doi: 10.1073/pnas.96.9.5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu Y, et al. NOEY2 (ARHI), an imprinted putative tumor suppressor gene in ovarian and breast carcinomas. Proc Natl Acad Sci USA. 1999;96:214–219. doi: 10.1073/pnas.96.1.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin T, Guo F, Serebriiskii IG, Howard A, Zhang YZ. A 1.55 A resolution X-ray crystal structure of HEF2/ERH and insights into its transcriptional and cell-cycle interaction networks. Proteins. 2007;68:427–437. doi: 10.1002/prot.21343. [DOI] [PubMed] [Google Scholar]
- 31.Coverley D, Marr J, Ainscough J. Ciz1 promotes mammalian DNA replication. J Cell Sci. 2005;118:101–112. doi: 10.1242/jcs.01599. [DOI] [PubMed] [Google Scholar]
- 32.Richardson CJ, et al. SKAR is a specific target of S6 kinase 1 in cell growth control. Curr Biol. 2004;14:1540–1549. doi: 10.1016/j.cub.2004.08.061. [DOI] [PubMed] [Google Scholar]
- 33.Bera TK, et al. POTE, a highly homologous gene family located on numerous chromosomes and expressed in prostate, ovary, testis, placenta, and prostate cancer. Proc Natl Acad Sci USA. 2002;99:16975–16980. doi: 10.1073/pnas.262655399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Popesco MC, et al. Human lineage-specific amplification, selection, and neuronal expression of DUF1220 domains. Science. 2006;313:1304–1307. doi: 10.1126/science.1127980. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.