Abstract
Whereas the neurodegeneration associated with various polyglutamine (polyQ) diseases has prompted extensive studies of polyQ-induced cell death, the neuronal loss that typically appears during late stages of the diseases may not account for the preceding movement and mental disorders. The cellular basis for polyQ-induced neuronal dysfunction preceding neuronal cell death remains largely unknown. Here we report defective dendrite morphogenesis within a specific subset of neurons due to polyQ toxicity that can be dissociated from caspase-dependent cell death. Expressing pathogenic spinocerebellar ataxia type 1 (SCA1) or type 3 (SCA3) proteins in Drosophila larval dendritic arborization neurons caused neuronal type-specific dendrite phenotypes primarily affecting terminal branches. We further show that expression of pathogenic polyQ proteins in adult flies after the formation of neuronal dendrites also greatly reduced dendritic complexity. These defects are associated with disruption of dendritic F-actin structures that can be partially mitigated by increasing Rac-PAK signaling. Together, these findings suggest that specific actin cytoskeletal alterations that alter dendrite morphology and function may contribute to the pathogenesis of at least a subset of polyQ disorders, including SCA3 and SCA1.
Keywords: disease model, genetic study
Polyglutamine (polyQ) diseases are inherited neurodegenerative diseases caused by expansion of a trinucleotide CAG repeat within the protein coding region of the disease gene (1, 2). To date, nine neurological disorders, including several types of spinocerebellar ataxia (SCA) and Huntington disease (HD), have been identified as polyQ diseases (2, 3). Characteristic of expanded (pathogenic) polyQ proteins is their tendency to form aggregates together with many target molecules in afflicted neurons (1, 2, 4). Recent studies suggest that the different protein context in various polyQ proteins may affect the kinetics of aggregate formation and the severity of toxicity as well as possible interactions with specific, but not all, targets (1, 5). Notably, among the many identified molecules that interact with various pathogenic polyQ proteins accumulating within the nucleus, several nuclear proteins, such as CREB binding protein (CBP), interact with multiple different polyQ proteins (4), indicative of the possible presence of common targets/pathogenic mechanisms shared by at least a subset of the polyQ proteins.
Because polyQ diseases are usually accompanied by substantial neuronal loss (1, 2), it has long been assumed that the accumulated polyQ proteins trapping multiple target proteins may be toxic to the affected neuronal cells, causing massive neurodegeneration and neurological symptoms in patients. Besides neuronal cell death, neuronal abnormalities, including changes in dendrite morphology, have been reported in several studies of transgenic animal models or human patients for certain types of polyQ diseases, such as SCA1, SCA3, and HD (6–10). These neuronal abnormalities warrant experimental scrutiny, because neuronal loss tends to be associated with the late stages of polyQ diseases, following more than 10 y of manifestation of motor symptoms as well as mental disorders (2). Indeed, behavioral symptoms are observed without massive neuronal loss in several HD and SCA1 transgenic mouse models (8, 11) and in a recent HD fly model (12). Thus, a more extensive understanding of the changes in a neuron's cellular properties before cell death may uncover novel pathways implicated in the pathogenesis of polyQ diseases.
It is of interest that the aforementioned studies regarding polyQ-induced neuronal abnormalities (6–10) describe a range of dendritic changes with variable severity in several different types of neurons. Dendrite morphogenesis is a complicated process affected by many factors (13), and it is unclear whether different polyQ proteins affect dendrite morphogenesis in similar or different ways. Thus, the cellular and molecular basis of the polyQ-induced dendrite defects remains largely unknown despite their potential importance in the disease pathogenesis. To better understand polyQ diseases, it would be desirable to establish a model system to dissect the diverse and potentially complicated dendrite defects caused by various polyQ proteins.
Drosophila is a useful model system for polyQ disease studies. For example, expressing pathogenic SCA3, SCA1, or other polyQ proteins specifically in Drosophila eyes causes progressive eye degeneration (14–16), which is well suited for genetic modifier screens. These genetic approaches have identified a number of molecules, including HSP70 chaperone (17) as well as novel pathogenic mechanisms such as polyQ RNA toxicity (18), to have significant roles in polyQ-induced cell death. However, given that Drosophila photoreceptor cells do not extend dendrites, additional assays are needed for the study of morphological defects of neuronal dendrites.
Recently, a group of Drosophila peripheral nervous system (PNS) sensory neurons, the dendritic arborization (da) neurons, have emerged as a suitable system for studying the cellular and molecular basis of dendritic abnormalities associated with neurological disorders. Da neurons have stereotyped dendrite branching patterns (19), which can be labeled with genetically coded GFP for visualizing dendrite morphogenesis within live, intact animals. The da neurons are grouped into four classes (I–IV) based on their stereotyped dendrite arborization patterns (19) that are controlled in a neuronal type-specific manner by multiple factors, such as signaling molecules and transcription factors as well as specific intracellular organelles and cytoskeletons (13). Here, we report our use of Drosophila da neurons as a unique model system to gain insight into the neuronal type-specific dendrite phenotypes caused by a subset of polyQ proteins and the underlying cellular and molecular mechanisms.
Results
Pathogenic SCA3 and SCA1 Proteins Induce Dendrite Phenotypes in da Neurons That Are Distinct from Dendrite Degeneration.
To determine whether the Drosophila PNS can be used as a model system to uncover the cellular basis of polyQ toxicity affecting neuronal dendrites, we expressed several pathogenic polyQ proteins in da neurons. Because recent studies indicate that the normal protein activity in addition to the polyQ expansion in polyQ-containing proteins can contribute to the polyQ diseases (5, 20, 21), we tested both truncated and full-length (FL) forms of several polyQ proteins (MJD-78Q, MJDFL-78Q, and SCA1FL-82Q). The MJD-78Q and MJDFL-78Q transgenes correspond to the truncated and the full-length mammalian MJD1/ataxin3/SCA3 proteins containing expanded glutamine (Q) repeat, respectively (14, 22), and the SCA1FL-82Q transgene corresponds to the pathogenic form of full-length mammalian ataxin1/SCA1 protein (15). For comparison, we also examined the effect of expressing truncated or full-length polyQ transgenes with a much shorter Q tract (MJD-27Q, MJDFL-27Q, and SCA1FL-30Q) on da neuron dendrite morphogenesis. It has been previously shown that expression of long polyQ-containing MJD-78Q, MJDFL-78Q, or SCA1FL-82Q but not short polyQ-containing MJD-27Q or MJDFL-27Q transgene in Drosophila eyes causes severe eye degeneration phenotypes (14, 15, 22). Interestingly, expression of SCA1FL-30Q can have effects in some tissues in the flies that are relatively milder than the effects caused by SCA1FL-82Q (15).
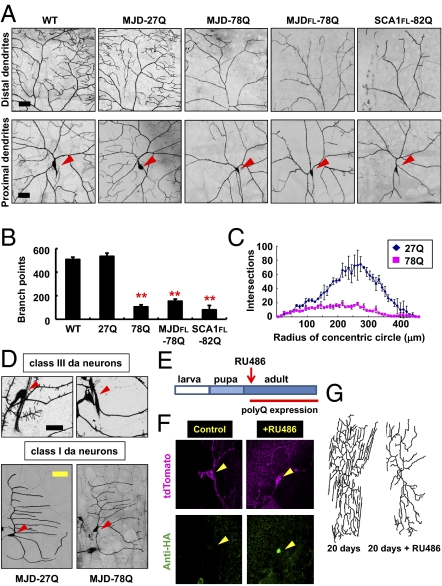
Live imaging of dendrite morphologies of da neurons at 120 h after egg-laying (AEL), a time when the dendritic arbors are fully formed in normal larvae, revealed that expression of the MJD-78Q transgene in the class IV da neurons at either high or low levels caused striking dendritic abnormalities (Fig. S1 and Fig. 1A). Similar dendrite defects were observed in class IV da neurons expressing other pathogenic polyQ proteins (MJDFL-78Q and SCA1FL-82Q; Fig. 1A and Fig. S2). Because all control polyQ proteins containing a shorter Q tract (MJD-27Q, MJDFL-27Q, and SCA1FL-30Q) did not induce any detectable dendrite phenotypes in larval da neurons (Fig. S2), we used one of them (MJD-27Q) as a control hereafter. These striking dendrite phenotypes became partially suppressed by coexpression of chaperone proteins (HSP-A1L), a well-known genetic suppressor of the polyQ-induced neuronal cell death (17), with pathogenic polyQ proteins (Fig. S2), suggesting that polyQ toxicities associated with multiple different neuronal pathologies may share certain common features/mechanisms. Nonetheless, the observed dendritic changes in polyQ-expressing class IV da neurons—greatly reduced dendritic branching (Fig. 1B) and complexity (Fig. 1C)—were distinct from dendrite blebbing and breakages characteristic of dead or dying da neurons (23). Moreover, coexpression of MJD-78Q with p35, a caspase inhibitor, failed to suppress the dendrite phenotypes (Fig. S2). Therefore, the dendrite phenotypes of neurons expressing these pathogenic polyQ proteins are distinct from degenerative cellular changes associated with caspase-dependent cell death.
Fig. 1.
Pathogenic SCA3 and SCA1 proteins induce dendrite defects in da neurons primarily involving terminal branches. (A) PolyQ-induced dendrite defects in class IV da neurons. ppk-gal4–driven expression of mCD8-GFP enabled imaging of dendrites of class IV da neurons. Red arrowheads indicate cell bodies (Lower). WT, w1118. (Scale bars: 30 μm.) (B and C) PolyQ expression reduced the number of total dendritic branch points (B) and the complexity of dendrites via Sholl analysis (C) in class IV da neurons. Bars indicate mean ± SD; n = 4 (B), n = 3 (C). **P < 0.01 (Student's unpaired t test) for the reduction of branch points of pathogenic polyQ-expressing neurons relative to MJD-27Q-expressing control neurons (B). (D) Images of dendrites of class III (Upper) or class I (Lower) da neurons expressing the denoted transgenes. (Scale bars: black, 25 μm; yellow, 50 μm.) Red arrowheads indicate cell bodies. (E) Schematic diagram showing conditional induction of polyQ expression at the adult stage. (F) Immunostaining of polyQ proteins (HA-MJD-78Q, green) in adult class IV da neurons at 30 d after eclosion from puparium. Yellow arrowheads indicate cell bodies. (G) Images of dendrites of adult class IV da neurons with (+RU486) or without conditionally induced MJD-78Q expression.
To examine the neuronal type-specific effects of polyQ-induced dendrite defects, we expressed MJD-78Q within class III da neurons, which are characterized by extensive dendritic spikes/filopodia (19), and class I da neurons, which are characterized by simple dendritic structures with few side branches (19). Expression of MJD-78Q reduced filopodia formation of class III da neurons at 120 h AEL (Fig. 1D), but had no obvious effects on the dendrites of class I da neurons (Fig. 1D and Fig. S1). These results demonstrate that polyQ toxicity arising from the subset of pathogenic polyQ proteins examined in our study can induce neuronal type-specific dendrite phenotypes in da neurons. A plausible explanation for this neuronal type-dependent susceptibility of dendrite morphology to polyQ toxicity will be discussed elsewhere in this article.
PolyQ Toxicity from Pathogenic SCA3 or SCA1 Proteins Primarily Affects Terminal Dendrite Branches in da Neurons.
To determine how the dendrite morphology was altered, we monitored the establishment and maintenance of dendrites in polyQ-expressing da neurons. We found that the reduced dendritic complexity in class IV da neurons expressing pathogenic SCA3 or SCA1 proteins results from a reduction in the terminal branches but not the major dendrite branches (Fig. 1A). Similarly, pathogenic SCA3 polyQ proteins could inhibit filopodia (dendrite terminal structures) formation in class III da neurons without affecting major dendrite branches (Fig. 1D).
A reduction in terminal dendritic branches could have arisen from a reduction in branch initiation or an increase in branch loss, or both. To better understand this effect of polyQ toxicity affecting terminal dendrites, we examined the dynamic changes of dendrite terminals of class IV da neurons expressing MJD-78Q by time-lapse live imaging. Dendrite terminal dynamics are readily detected within 30-min intervals in control (w1118) larvae and can be categorized as branch initiation, growth, retraction, and branch loss (Fig. S3). At 96 h AEL, MJD-78Q but not MJD-27Q almost completely blocked growth and branch initiation of terminal dendrites (Fig. S3). In contrast, though the total number of terminal dendrites showing retraction and branch loss was also highly reduced by MJD-78Q expression, after normalization to the smaller total number of terminal dendrites of these polyQ-expressing neurons, the fraction of terminal dendrites exhibiting branch retraction or loss in MJD-78Q-expressing neurons was comparable to that in control neurons expressing MJD-27Q (Fig. S3). These data suggest that the observed dendrite defects in larval da neurons result from impaired growth of dendrite terminals.
To examine the effect of polyQ toxicity on dendrite maintenance after establishment of dendritic field, we induced polyQ protein expression in da neurons during the adult stage after those neurons have developed (Fig. 1E) by using the RU486-inducible gene-switch GAL4 system (24). First, we confirmed the strong induction of MJD-78Q expression following RU486 administration but not in control without RU486 (Fig. 1F). This late induction of polyQ proteins caused progressive dendrite defects that primarily involve dendritic terminals (Fig. 1G) similar to what we observed in developing da neurons during larval stage (Fig. 1A), indicating that certain polyQ proteins not only affect dendrite terminal growth when expressed during development, but also that adult-specific expression of these proteins can compromise the maintenance of dendrite terminals in da neurons. In addition, we also examined whether these dendrite phenotypes can be reversed after short-term RU486 administration. Given the reported slow decay of gene-switch activity after RU486 withdrawal (25), we minimized the duration of RU486 administration and found that with only a 3-d administration after eclosion, adult da neurons displayed comparable defects as neurons experiencing constant drug exposure when both groups were examined 20 d after eclosion (Fig. S4). These dendrite phenotypes were not noticeably reversed with aging when examined 40 d after eclosion (Fig. S4), suggestive of the potential difficulty in efficient reversal of polyQ-induced neuronal changes.
PolyQ-Induced Dendrite Phenotypes Are Associated with Specific Alterations of F-Actin Structures in Dendrites.
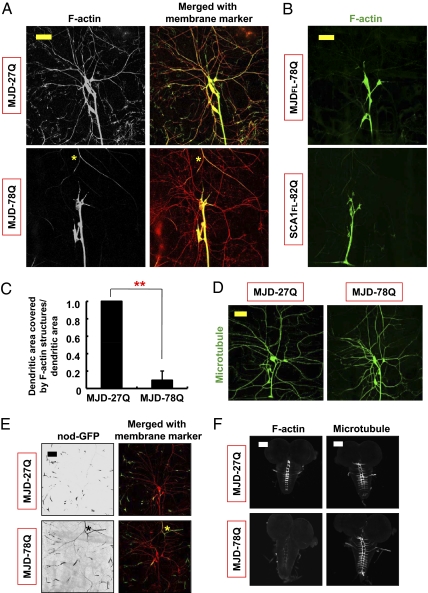
The specific effects of pathogenic SCA3 and SCA1 polyQ proteins on class III and IV da neurons but not class I da neurons prompted us to examine the dendritic F-actin and microtubule cytoskeletons, which contribute differently to the dendrite morphology of different classes of da neurons (26–28). We specifically labeled cytoskeletal structures in da neurons of living animals, using transgenic markers for F-actin (GMA, an F-actin–binding GFP fusion protein) (29) and for microtubules (tau-GFP) (24). In wild-type class IV da neurons, F-actin structures extended throughout the entire dendritic arbor and were enriched at many dendrite terminals (Fig. S5). In contrast, microtubular structures were present only in the major dendrite branches (Fig. S5). Remarkably, these pathogenic polyQ proteins affected primarily the actin-rich class IV da neuron dendrite terminals and the class III da neuron filopodia containing only F-actin (26) but not the class I da neuron dendrites composed of mainly microtubular structures (26, 27) (Fig. 1D). For this reason we explored the possibility that these pathogenic polyQ proteins preferentially disrupt F-actin structures in dendrites.
We found that the expression of pathogenic SCA3 or SCA1 proteins severely affected the distribution of F-actin structures in distal dendrites of da neurons (Fig. 2 A–C). In many da neurons expressing polyQ proteins, dendritic F-actin structures did not localize to distal dendrites (Fig. 2 A and B). In contrast, microtubular structures in dendrites were not obviously altered by polyQ toxicity (Fig. 2 D and E). The absence of severe microtubular defects again distinguishes the dendrite phenotype of polyQ toxicity from dendritic changes attributed to caspase activation, which causes severing of microtubular structures in major branches (30). In addition, even though overall axonal morphology was unaffected by MJD-78Q expression, F-actin structures in the axon terminals of da neurons in the ventral nerve cord as well as neuromuscular junction (NMJ) were noticeably altered (Fig. 2F and Fig. S6), whereas stable microtubular structures were unaffected (Fig. 2F and Fig. S6). These findings show that polyQ toxicity from pathogenic SCA3 or SCA1 proteins preferentially disrupts F-actin structures in the distal processes of neurons such as dendritic and axonal terminals.
Fig. 2.
PolyQ toxicity from pathogenic SCA3 or SCA1 polyQ proteins disrupts dendritic F-actin but not microtubular structures. (A) MJD-78Q expression reduced F-actin structures (GMA) in dendrites of dorsal cluster da neurons (Left). Merged images of F-actin marker (green) with mRFP-labeled dendrites (red; Right). Yellow asterisks indicate an autofluorescent signal of the trachea. (Scale bar: 50 μm.) (B) Images of F-actin structures in dendrites of dorsal cluster da neurons expressing the denoted transgenes. (Scale bar: 50 μm.) (C) PolyQ-induced reduction in the normalized dendritic field area covered by F-actin structures in dorsal cluster da neurons. n = 4; **P < 0.01 (Student's unpaired t test) relative to MJD-27Q control. (D) Images of microtubular structures (tau-GFP) in dorsal cluster da neurons expressing the denoted transgenes. (Scale bar: 50 μm.) (E) Microtubule polarity as revealed by nod-GFP in dendrites of dorsal cluster da neurons. Asterisks indicate an autofluorescent signal of the trachea. (Scale bar: 50 μm.) (F) F-actin (GMA; Left) and microtubular (tau-GFP; Right) structures in axon terminals of class IV da neurons expressing the denoted transgenes. (Scale bars: 100 μm.)
Dendritic F-Actin Defects in PolyQ-Expressing Neurons May Result from Compromised Local Actin Regulatory Machinery.
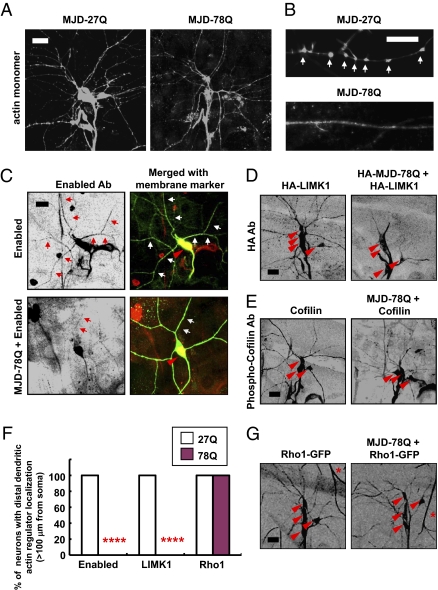
Next, we considered two possible models for polyQ-induced reduction in dendritic F-actin assembly. The observed F-actin defects may reflect a defect in actin regulatory machinery or, alternatively, polyQ proteins may trap or deplete actin monomers. To distinguish between these possibilities, we overexpressed actin monomers in da neurons expressing MJD-78Q. In these actin-overexpressing animals, we could still observe polyQ-induced dendrite defects (Fig. S7) despite the prominent actin distribution throughout the dendrites (Fig. 3A), suggesting it is unlikely that the polyQ-induced dendrite phenotype is associated with depletion or trapping of actin monomers. Despite the abundance of actin monomers in these neurons, they often failed to be assembled into local structures that are normally present within the main dendritic shaft at the base of filopodia in class III da neurons (marked with arrows in Fig. 3B). Thus, it seems more likely that the polyQ toxicity from a subset of polyQ proteins is associated with altered actin regulatory machinery in dendrites.
Fig. 3.
PolyQ toxicity reduces dendritic distribution of several overexpressed actin regulators, including Enabled, LIMK1, and phospho-Cofilin, but not overexpressed Rho1 GTPase. (A and B) Immunostaining of overexpressed actin monomers (actin-GFP) in dendrites of da neurons. Actin monomers were detected by anti-GFP immunostaining. Arrows indicate local actin structures in class III da neurons (B). (Scale bars: A, 50 μm; B, 10 μm.) (C–E and G) Dendritic distribution of overexpressed actin regulators was examined in class IV (Enabled) or dorsal cluster da neurons (HA-LIMK1, phospho-Cofilin, Rho1-GFP) expressing the denoted transgenes detected by either immunostaining (Enabled, LIMK1, and phospho-Cofilin) or live imaging of GFP-fused transgenic proteins (Rho1-GFP). Anti-HA staining also detected coexpressed HA-MJD-78Q proteins (D) that show a very strong tendency to be accumulated within the nucleus of da neurons. Arrows indicate dendritic distribution of overexpressed Enabled (Right, red) in class IV da neuron (C). Red arrowheads indicate cell bodies of class IV (C) and dorsal cluster (D, E, and G) da neurons. Asterisks indicate an autofluorescent signal of the trachea (G). (Scale bars: 20 μm.) (F) Quantitative analysis of polyQ-induced inhibition of dendritic localization of Enabled and LIMK1 but not Rho1 >100 μm from cell body. ****P < 0.0001 (Fisher's exact test) relative to MJD-27Q control; n = 10.
To explore this possibility, we examined dendritic distribution of several essential components of actin regulatory machinery, including Enabled/VASP, which is required for actin assembly; LIMK1, which regulates actin disassembly; and a phosphorylated form of Cofilin, a substrate of LIMK1 (31). Because the endogenous level of these proteins was too low for detection by antibody staining, we expressed tagged LIMK1, or untagged Enabled or Cofilin, in da neurons and examined their intracellular distribution. Notably, MJD-78Q expression significantly reduced dendritic distribution of the actin regulators involved in actin assembly and disassembly (Fig. 3 C–F), but had no detectable effect on the dendritic distribution of fluorescently tagged Rho1 GTPase, one of the upstream components modulating these actin regulators, in da neurons (Fig. 3 F and G). The altered distribution of multiple actin regulators (Fig. 3 C–E) correlates with the pattern of polyQ-induced F-actin defects detected preferentially in dendrites (Fig. 2 A and B), suggesting that polyQ-induced F-actin defects may involve compromised local actin regulatory machinery within dendrites.
PolyQ-Induced Dendrite Defects Can Be Partially Suppressed by Increasing Rac-PAK Signaling, Which Strongly Promotes F-Actin Formation.
The observed changes of actin regulators in polyQ-expressing neurons (Fig. 3) associated with dendrite defects could result from protein-level defects and/or mRNA-level defects of actin regulators. Given that pathogenic SCA3 and SCA1 proteins accumulate predominantly within the nucleus of Drosophila cells (14, 15), similar to what has been shown in human patients and transgenic mouse models (2, 3), it seems unlikely that polyQ aggregates directly trap actin regulators or actin monomers, because these actin regulators and actin monomers did not show obvious nuclear accumulation in polyQ-expressing da neurons (Fig. 3). We next explored the possibility that nuclear accumulation of toxic polyQ proteins may indirectly affect distribution of actin regulators by causing changes in the general nucleocytoplasmic protein shuttling or the nuclear retention of nuclear proteins, because many essential actin regulators localize to both the nucleus and the cytoplasm (32). Our analysis of neurons expressing fluorescently labeled ribosomal proteins (RpL11-GFP), which are shuttled between the nucleus and the cytoplasm (33), revealed no alteration of the subcellular distribution of ribosomal protein by polyQ toxicity (Fig. S8). In addition, the nuclear retention of nuclear-targeted dsRed proteins (Red Stinger) and the nuclear envelope structure marked by GFP-labeled Lamin (Lamin-GFP) were also not detectably altered by polyQ protein expression (Fig. S8). These findings raise the question of whether polyQ-induced dendrite defects may involve mRNA-level changes of actin regulators, such as defective supply and/or processing of actin-regulator mRNAs.
Notably, we have obtained evidence for the mRNA-level defects in polyQ-expressing da neurons that may affect dendrites; the dendritic distribution of FMR1-positive granules (FMR1-GFP) that normally contain mRNAs required for local F-actin formation (28, 34) was highly reduced in polyQ-expressing da neurons (Fig. S8). Moreover, we also found an interesting phenotypic similarity between the polyQ-induced dendritic changes involving altered dendritic F-actin structures and dendrite defects caused by genetic modulation of certain transcription factors such as Cut (27, 35) or RNA binding proteins such as Pumilio (36) and FMR1 (28), supporting the notion that nuclear polyQ proteins that interact with numerous transcription factors and RNA-binding proteins (4, 15, 37) can cause specific dendritic changes possibly through mRNA-level changes of actin regulators.
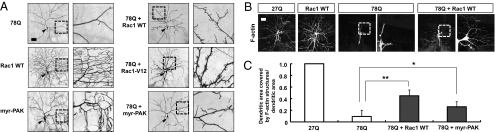
Next, we tested whether increasing the level of certain components of the actin regulatory machinery could mitigate polyQ-induced dendrite defects. Interestingly, overexpression of Rac1 or Rac2 GTPases, but not Profilin, Spir, Cappuccino, Enabled, LIMK1, or Cofilin, significantly promoted terminal dendrite formation in polyQ-expressing class IV da neurons (Fig. 4A and Tables S1 and S2). Overexpression of Rac1 could also partially restore dendritic F-actin structures in polyQ-expressing neurons (Fig. 4 B and C). Moreover, expression of a constitutively active form of Rac1, Rac1-V12, in polyQ-expressing neurons caused even greater enhancement of terminal dendrite formation (Fig. 4A and Table S1). Given that expressing Rac GTPases strongly promotes dendritic F-actin formation in control neurons as well as neurons expressing pathogenic polyQ proteins (Fig. 4B), robust activation of actin regulators due to increased Rac GTPases seems to be strong enough to partially overcome polyQ-induced inhibitory effects on local actin regulatory machinery. Furthermore, expression of a membrane-bound (active) form of PAK, a downstream effector of Rac GTPases (31), also partially suppressed terminal dendrite defects as well as dendritic F-actin defects in 40% of polyQ-expressing da neurons (Fig. 4 A and C and Table S1). Taken together, these results suggest that polyQ toxicity in dendrites is associated with defective F-actin structures resulting from compromised dendritic actin regulatory machinery that can be partially mitigated by increasing the upstream Rac-PAK signaling.
Fig. 4.
Amelioration of polyQ-induced dendrite defects by increasing Rac-PAK signaling. (A) Modulation of polyQ-induced dendrite defects by expressing Rac GTPases and myristoylated PAK (myr-PAK). Arrowheads indicate cell bodies. (Right) Magnified images for regions within the dashed boxes. (Scale bar: 50 μm.) (B) Images of dendritic F-actin structures (GMA) in dorsal cluster da neurons expressing denoted transgenes. (Right) Magnified images for regions within the dashed boxes. (Scale bar: 50 μm.) (C) Quantitative analysis of the normalized dendritic field area covered by F-actin structures in dorsal cluster da neurons expressing the denoted transgenes. *P < 0.05 and **P < 0.01 (Student's unpaired t test) relative to MJD-78Q, respectively; n = 3.
Discussion
For many polyQ diseases, patients often suffer motor symptoms and mental disorders for many years before neurodegeneration and cell loss become prominent during late stages of the disease (2), suggesting that polyQ-induced neuronal abnormalities other than cell death may contribute significantly to the disease pathogenesis. However, the cellular basis for various neuronal abnormalities preceding neuronal cell death, which underlies polyQ diseases, remains largely unexplored. In this study, we used the Drosophila da neurons as a model system to study neuronal defects other than apoptosis that result from polyQ toxicity.
We examined a subset of polyQ proteins (either truncated or full-length forms of mutated SCA3 and SCA1) and found that when overexpressed, they cause severe neuronal type-specific dendrite phenotypes through specific F-actin alterations in a similar way. Our Drosophila studies raise the question of whether dendrite alterations potentially caused by defects in regulation of the actin cytoskeleton occur before, or in the absence of, obvious neuronal degeneration in human patients carrying certain polyQ disease genes with expanded polyQ sequences such as SCA3 and SCA1. It will be interesting to further examine this possibility, for example, by using induced pluripotent stem cells (iPSC) derived from patients carrying these mutated polyQ disease genes.
Our observation revealed that polyQ-induced dendrite phenotypes vary among different types of da neurons (Fig. 1 and Fig. S1), but correlate well with their distinctive dendritic cytoskeletal architectures (Fig. S5). For instance, the F-actin–enriched terminal branches of class IV and the filopodia of class III da neuron dendrites were severely reduced, whereas dendrite branches with stable microtubular structures were not affected as much by polyQ toxicity. These observations are reminiscent of the range of dendritic abnormalities with variable severity observed in several different types of neurons from human patients and transgenic mouse models (6–10), including reduced arborization and inhibited formation of actin-rich spines.
In summary, our study reveals that a subset of polyQ proteins cause neuronal type-specific dendrite defects closely associated with specific alterations in the actin cytoskeleton, which play critical roles in neuronal morphogenesis (13). Given the importance of dendrite morphology in neuronal function (38) and the pathogenesis of other neurological diseases accompanied by dendritic changes (39, 40), the polyQ-induced dendrite defects discovered in our study may contribute to our understanding of the etiology and possible treatment of at least a subset of polyQ disorders associated with nuclear accumulation of polyQ proteins.
Materials and Methods
Fly Stocks.
The UAS-MJDFL-27Q and UAS-MJDFL-78Q fly lines were obtained from N. M. Bonini (University of Pennsylvania, Philadelphia). The UAS-GMA allele was obtained from D. P. Kiehart (Duke University, Durham, NC) and J. E. Brenman (University of North Carolina School of Medicine, Chapel Hill, NC). The UAS-chickadee (Profilin) fly lines were from L. Cooley (Yale University School of Medicine, New Haven, CT). The UAS-SCA1FL-30Q and UAS-SCA1FL-82Q-[M6] alleles were obtained from J. Botas (Baylor College of Medicine, Houston). The UAS-RpL11-GFP allele was obtained from P. J. DiMario (Louisiana State University, Baton Rouge, LA). The UAS-FMR1-GFP line was obtained from F. B. Gao (University of Massachusetts, Worcester, MA). Other fly lines were obtained from the Bloomington Stock Center. Flies were raised at 25 °C.
Immunocytochemistry.
Third instar larvae were fixed according to standard protocols (19). To detect overexpressed actin monomers and actin regulators, rabbit anti-GFP (1:1,000 dilution), mouse anti-Enabled (5G2, DSHB; 1:20 dilution), rat anti-HA (3F10, Roche; 1:100 dilution), and rabbit anti–phospho-Cofilin [from J. R. Bamburg (Colorado State University, Fort Collins, CO) and T. Uemura (Kyoto University, Kyoto, Japan); 1:100 dilution] antibodies were used.
Live Imaging, Genetic Interaction, and Statistical Analysis.
Details are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank N. M. Bonini, D. P. Kiehart, J. E. Brenman, J. Botas, T. Uemura, J. R. Bamburg, L. Cooley, P. J. DiMario, F. B. Gao, and the Bloomington Stock Center for providing fly stocks and reagents. We extend special thanks to Keun Hye Jeon for experimental design and criticism. This work was supported by National Institutes of Health Grants 2R37 NS040929 and R01 MH 084234 (to Y.-N.J.) and the National Science Foundation through Graduate Research Fellowship Grant DGE-0648991 (to J.A.B.). Y.-N.J. and L.Y.J. are investigators of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1113573108/-/DCSupplemental.
References
- 1.Paulson HL. Dominantly inherited ataxias: Lessons learned from Machado–Joseph disease/spinocerebellar ataxia type 3. Semin Neurol. 2007;27:133–142. doi: 10.1055/s-2007-971172. [DOI] [PubMed] [Google Scholar]
- 2.Zoghbi HY, Orr HT. Glutamine repeats and neurodegeneration. Annu Rev Neurosci. 2000;23:217–247. doi: 10.1146/annurev.neuro.23.1.217. [DOI] [PubMed] [Google Scholar]
- 3.Koeppen AH. The pathogenesis of spinocerebellar ataxia. Cerebellum. 2005;4:62–73. doi: 10.1080/14734220510007950. [DOI] [PubMed] [Google Scholar]
- 4.McCampbell A, Fischbeck KH. Polyglutamine and CBP: Fatal attraction? Nat Med. 2001;7:528–530. doi: 10.1038/87842. [DOI] [PubMed] [Google Scholar]
- 5.Kratter IH, Finkbeiner S. PolyQ disease: Too many Qs, too much function? Neuron. 2010;67:897–899. doi: 10.1016/j.neuron.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graveland GA, Williams RS, DiFiglia M. Evidence for degenerative and regenerative changes in neostriatal spiny neurons in Huntington's disease. Science. 1985;227:770–773. doi: 10.1126/science.3155875. [DOI] [PubMed] [Google Scholar]
- 7.Koeppen AH. The Purkinje cell and its afferents in human hereditary ataxia. J Neuropathol Exp Neurol. 1991;50:505–514. doi: 10.1097/00005072-199107000-00010. [DOI] [PubMed] [Google Scholar]
- 8.Clark HB, et al. Purkinje cell expression of a mutant allele of SCA1 in transgenic mice leads to disparate effects on motor behaviors, followed by a progressive cerebellar dysfunction and histological alterations. J Neurosci. 1997;17:7385–7395. doi: 10.1523/JNEUROSCI.17-19-07385.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guidetti P, et al. Early degenerative changes in transgenic mice expressing mutant Huntington involve dendritic abnormalities but no impairment of mitochondrial energy production. Exp Neurol. 2001;169:340–350. doi: 10.1006/exnr.2000.7626. [DOI] [PubMed] [Google Scholar]
- 10.Cemal CK, et al. YAC transgenic mice carrying pathological alleles of the MJD1 locus exhibit a mild and slowly progressive cerebellar deficit. Hum Mol Genet. 2002;11:1075–1094. doi: 10.1093/hmg/11.9.1075. [DOI] [PubMed] [Google Scholar]
- 11.Li XJ. The early cellular pathology of Huntington's disease. Mol Neurobiol. 1999;20:111–124. doi: 10.1007/BF02742437. [DOI] [PubMed] [Google Scholar]
- 12.Nishimura Y, et al. Selection of behaviors and segmental coordination during larval locomotion is disrupted by nuclear polyglutamine inclusions in a new Drosophila Huntington's disease-like model. J Neurogenet. 2010;24:194–206. doi: 10.3109/01677063.2010.514367. [DOI] [PubMed] [Google Scholar]
- 13.Jan YN, Jan LY. Branching out: Mechanisms of dendritic arborization. Nat Rev Neurosci. 2010;11:316–328. doi: 10.1038/nrn2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warrick JM, et al. Expanded polyglutamine protein forms nuclear inclusions and causes neural degeneration in Drosophila. Cell. 1998;93:939–949. doi: 10.1016/s0092-8674(00)81200-3. [DOI] [PubMed] [Google Scholar]
- 15.Fernandez-Funez P, et al. Identification of genes that modify ataxin-1–induced neurodegeneration. Nature. 2000;408:101–106. doi: 10.1038/35040584. [DOI] [PubMed] [Google Scholar]
- 16.Kazemi-Esfarjani P, Benzer S. Genetic suppression of polyglutamine toxicity in Drosophila. Science. 2000;287:1837–1840. doi: 10.1126/science.287.5459.1837. [DOI] [PubMed] [Google Scholar]
- 17.Warrick JM, et al. Suppression of polyglutamine-mediated neurodegeneration in Drosophila by the molecular chaperone HSP70. Nat Genet. 1999;23:425–428. doi: 10.1038/70532. [DOI] [PubMed] [Google Scholar]
- 18.Li LB, Yu Z, Teng X, Bonini NM. RNA toxicity is a component of ataxin-3 degeneration in Drosophila. Nature. 2008;453:1107–1111. doi: 10.1038/nature06909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grueber WB, Jan LY, Jan YN. Tiling of the Drosophila epidermis by multidendritic sensory neurons. Development. 2002;129:2867–2878. doi: 10.1242/dev.129.12.2867. [DOI] [PubMed] [Google Scholar]
- 20.Duvick L, et al. SCA1-like disease in mice expressing wild-type ataxin-1 with a serine to aspartic acid replacement at residue 776. Neuron. 2010;67:929–935. doi: 10.1016/j.neuron.2010.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nedelsky NB, et al. Native functions of the androgen receptor are essential to pathogenesis in a Drosophila model of spinobulbar muscular atrophy. Neuron. 2010;67:936–952. doi: 10.1016/j.neuron.2010.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warrick JM, et al. Ataxin-3 suppresses polyglutamine neurodegeneration in Drosophila by a ubiquitin-associated mechanism. Mol Cell. 2005;18:37–48. doi: 10.1016/j.molcel.2005.02.030. [DOI] [PubMed] [Google Scholar]
- 23.Williams DW, Truman JW. Cellular mechanisms of dendrite pruning in Drosophila: Insights from in vivo time-lapse of remodeling dendritic arborizing sensory neurons. Development. 2005;132:3631–3642. doi: 10.1242/dev.01928. [DOI] [PubMed] [Google Scholar]
- 24.Rumpf S, Lee SB, Jan LY, Jan YN. Neuronal remodeling and apoptosis require VCP-dependent degradation of the apoptosis inhibitor DIAP1. Development. 2011;138:1153–1160. doi: 10.1242/dev.062703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roman G, Endo K, Zong L, Davis RL. P[Switch], a system for spatial and temporal control of gene expression in Drosophila melanogaster. Proc Natl Acad Sci USA. 2001;98:12602–12607. doi: 10.1073/pnas.221303998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andersen R, Li Y, Resseguie M, Brenman JE. Calcium/calmodulin-dependent protein kinase II alters structural plasticity and cytoskeletal dynamics in Drosophila. J Neurosci. 2005;25:8878–8888. doi: 10.1523/JNEUROSCI.2005-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jinushi-Nakao S, et al. Knot/Collier and cut control different aspects of dendrite cytoskeleton and synergize to define final arbor shape. Neuron. 2007;56:963–978. doi: 10.1016/j.neuron.2007.10.031. [DOI] [PubMed] [Google Scholar]
- 28.Lee A, et al. Control of dendritic development by the Drosophila fragile X-related gene involves the small GTPase Rac1. Development. 2003;130:5543–5552. doi: 10.1242/dev.00792. [DOI] [PubMed] [Google Scholar]
- 29.Dutta D, Bloor JW, Ruiz-Gomez M, VijayRaghavan K, Kiehart DP. Real-time imaging of morphogenetic movements in Drosophila using Gal4-UAS-driven expression of GFP fused to the actin-binding domain of moesin. Genesis. 2002;34:146–151. doi: 10.1002/gene.10113. [DOI] [PubMed] [Google Scholar]
- 30.Lee HH, Jan LY, Jan YN. Drosophila IKK-related kinase Ik2 and Katanin p60-like 1 regulate dendrite pruning of sensory neuron during metamorphosis. Proc Natl Acad Sci USA. 2009;106:6363–6368. doi: 10.1073/pnas.0902051106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ng J, Luo L. Rho GTPases regulate axon growth through convergent and divergent signaling pathways. Neuron. 2004;44:779–793. doi: 10.1016/j.neuron.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 32.Jockusch BM, Schoenenberger CA, Stetefeld J, Aebi U. Tracking down the different forms of nuclear actin. Trends Cell Biol. 2006;16:391–396. doi: 10.1016/j.tcb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 33.Rosby R, et al. Knockdown of the Drosophila GTPase nucleostemin 1 impairs large ribosomal subunit biogenesis, cell growth, and midgut precursor cell maintenance. Mol Biol Cell. 2009;20:4424–4434. doi: 10.1091/mbc.E08-06-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reeve SP, et al. The Drosophila fragile X mental retardation protein controls actin dynamics by directly regulating profilin in the brain. Curr Biol. 2005;15:1156–1163. doi: 10.1016/j.cub.2005.05.050. [DOI] [PubMed] [Google Scholar]
- 35.Grueber WB, Jan LY, Jan YN. Different levels of the homeodomain protein cut regulate distinct dendrite branching patterns of Drosophila multidendritic neurons. Cell. 2003;112:805–818. doi: 10.1016/s0092-8674(03)00160-0. [DOI] [PubMed] [Google Scholar]
- 36.Ye B, et al. Nanos and Pumilio are essential for dendrite morphogenesis in Drosophila peripheral neurons. Curr Biol. 2004;14:314–321. doi: 10.1016/j.cub.2004.01.052. [DOI] [PubMed] [Google Scholar]
- 37.Bilen J, Bonini NM. Genome-wide screen for modifiers of ataxin-3 neurodegeneration in Drosophila. PLoS Genet. 2007;3:1950–1964. doi: 10.1371/journal.pgen.0030177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whitford KL, Dijkhuizen P, Polleux F, Ghosh A. Molecular control of cortical dendrite development. Annu Rev Neurosci. 2002;25:127–149. doi: 10.1146/annurev.neuro.25.112701.142932. [DOI] [PubMed] [Google Scholar]
- 39.Kaufmann WE, Moser HW. Dendritic anomalies in disorders associated with mental retardation. Cereb Cortex. 2000;10:981–991. doi: 10.1093/cercor/10.10.981. [DOI] [PubMed] [Google Scholar]
- 40.Knobloch M, Mansuy IM. Dendritic spine loss and synaptic alterations in Alzheimer's disease. Mol Neurobiol. 2008;37:73–82. doi: 10.1007/s12035-008-8018-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.